Abstract
Daphnia pulex is the first crustacean to have its genome sequenced. Availability of the genome sequence will have implications for research in aquatic ecology and evolution in particular, as addressed by a series of papers published recently in BMC Evolutionary Biology and BMC Genomics.
See research articles http://www.biomedcentral.com/1471-2148/9/78, http://www.biomedcentral.com/1471-2164/10/527, http://www.biomedcentral.com/1471-2148/9/79, http://www.biomedcentral.com/1471-2164/10/175, http://www.biomedcentral.com/1471-2164/10/172, http://www.biomedcentral.com/1471-2164/10/169, http://www.biomedcentral.com/1471-2164/10/170 and http://www.biomedcentral.com/1471-2148/9/243.
One of the major questions in evolutionary biology is to understand how species have adapted to different environments and how the underlying changes in morphology, physiology and behavior relate to modifications in the corresponding genes. The publication of the first crustacean genome sequence, that of Daphnia pulex [1,2], is part of an effort by the members of the Daphnia Genome Consortium to establish Daphnia as a model system for evolutionary environmental genomics. But can Daphnia rise to the challenge?
The vast number of publications on Daphnia in the literature prove that this animal is no newcomer to scientific research. Daphnia is most probably one of the best-studied subjects in ecology [3]. Populations can be found in freshwater environments ranging from huge lakes to small temporary pools and seasonally flooded depressions. The ecology of Daphnia has been studied from the point of view of its role as a primary consumer in aquatic food chains, its phenotypic plasticity, and its behavior, toxicology and the evolution of sexual and asexual reproduction. Extensive studies on the population genetics of Daphnia have addressed migration and gene flow, hybridization and inbreeding, among other topics. With the availability of the genome sequence, Daphnia research has now the potential to reach a new level. A number of papers on the D. pulex genome in relation to different aspects of Daphnia biology have been published in BMC Evolutionary Biology and BMC Genomics to accompany the genome release [4-11]. These constitute an initial exploration of the genome, and in this article I review some of the highlights and questions raised.
Daphnia ecology and life style
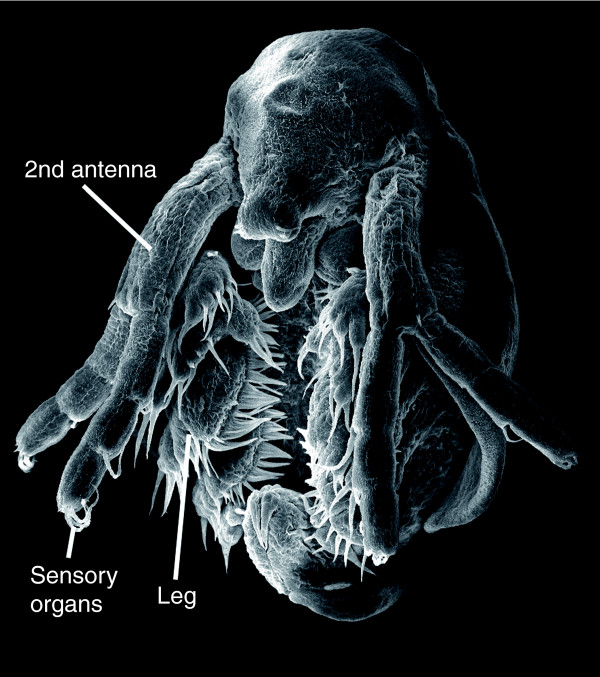
Daphnia are filter feeders that direct small suspended particles into their mouth by a water current produced by their leaf-like legs (Figure 1). Daphnia's common name of 'water flea' comes from its jump-like movement, which results from the beat of the large antennae used for swimming (Figure 1). In a normal growth season Daphnia generates diploid eggs by asexual reproduction (parthenogenesis). These eggs develop directly into larvae in the female brood chamber and are released into the water after about 3 days. In most species the larvae go through four to six larval stages before developing into sexually mature adults. However, the Daphnia life cycle is adapted to extreme environmental conditions such as cold winters or summer droughts. If triggered by external stimuli such as high population density and a scarcity of food, Daphnia can produce haploid resting eggs by meiosis; these require fertilization and a period of extended dormancy in order to develop [3]. Resting eggs are distributed by wind or animals and development is resumed in response to external stimuli (for example, rising temperature). Cyclic parthenogenesis, in which parthenogenesis and sexual reproduction alternate, is common in most Daphnia species, but lineages have been described that exclusively reproduce asexually (obligate parthenogenesis). Cyclic parthenogenetic Daphnia must contain the molecular tools for the production of both haploid gametes (by meiosis) and diploid eggs (by mitosis), the latter developing parthenogenetically into diploid zygotes. This makes Daphnia an ideal system to study the evolution of the molecular processes of parthenogenesis.
Figure 1.
Scanning electron micrograph of a Daphnia larva shortly before hatching. Photograph courtesy of Petra Ungerer.
In this regard, Eads and co-workers (Schurko et al. [4]) suggest that differences between sexual and asexual reproduction most probably relate to mechanisms that differ between meiosis and mitosis, such as kinetochore orientation, DNA recombination and sister-chromatid cohesion, and have screened the D. pulex genome for genes associated with meiosis. The authors report an inventory of 130 D. pulex genes that are homologous with known genes in other organisms and which represent more than 40 distinct protein-coding genes with diverse roles in meiosis. The majority of these genes are present in multiple copies, and Schurko et al. [4] speculate that the extra copies may be partly responsible for changes to these meiotic processes that enable parthenogenesis. Parthenogenetic species are present in all major animal phyla and future comparison of the genomes of cyclic and obligate parthenogenetic lineages will shed light on the evolution of the underlying molecular processes.
The offspring produced in parthenogenetic cycles are genetic clones of their mother [3]. This includes the males, as sex is environmentally determined in Daphnia. The existence of clonal reproduction is a powerful tool for quantitative genetic studies because it facilitates the analysis of genetic variation within and between populations. Genetic variation has been reported in Daphnia for a vast number of traits such as size, aging, behavior (for example, vertical migration, fish-escape behavior), morphology (for example, defensive spines, helmets), and the immune system (for example, resistance against parasites, immune responses), and the great number of duplicated genes in Daphnia seems to correlate with Daphnia's considerable phenotypic plasticity [3].
Predators and other enemies
Interestingly, several of the above-mentioned traits can be induced by environmental cues. Changes in both the behavior and the morphology of Daphnia can, for example, be affected by predator-borne chemical cues (kairomones). In the presence of fish kairomones, Daphnia magna gives rise to smaller offspring, whereas chemical cues from the phantom midge Chaoborus flavicans induce the generation of larger progeny. This has been shown to be an adaptive phenotypic plasticity that helps avoid predation as fish and midges prefer different sizes of prey. These observations raise the question of the nature of the molecular response to kairomones. Schwarzenberger et al. [5] have addressed this question by comparing the expression levels of genes involved in protein biosynthesis and catabolism in D. magna in the presence or absence of kairomones. Interestingly, they found that expression of the cyclophilin gene, which encodes an enzyme involved in protein folding, is upregulated in the presence of fish kairomones but downregulated by Chaoborus kairomones, which correlates with the opposite effects of these kairomones on progeny size. The authors used the D. magna cyclophilin sequence to search the D. pulex genome and identified 16 paralogs, which showed a very high variability. Future research will show whether the differences in cyclophilin expression levels can be linked to the observed phenotypic variations and if additional paralogs are involved in the process.
The first step in kairomone-mediated adaptive changes in behavior and morphology is obviously the reception of the chemical signal by specialized sensory structures of the prey. Our knowledge about chemoreception in aquatic organisms is fragmentary, however. In insects, a conserved chemoreceptor superfamily has been identified which can be subdivided into the gustatory (taste) receptor family and the odorant (smell) receptor family. It is obvious that the sensing of odorants will be different in water than in air as aquatic odorants are hydrophilic molecules dissolved in water whereas airborne odorants are mainly hydrophobic volatile molecules in gaseous form. Penalva-Arana et al. [6] have identified 58 orthologs of the insect gustatory receptor family in the D. pulex genome. Interestingly, they found no evidence of genes homologous with insect odorant receptor genes and suggest that the odorant receptor family evolved concomitantly with the transition from sea to land in the lineage leading to the insects.
Predators are not the only natural enemies of Daphnia. The study of parasites (viruses, bacteria and multicellular parasites) has also gained momentum as a result of their influence on Daphnia ecology and evolution [3]. Parasites can directly or indirectly affect host population dynamics, extinction, and maintenance of genetic diversity, among other features. It has been suggested that hosts continuously evolve to reduce parasite virulence, whereas parasites evolve to keep virulence as close as possible to an optimum level. Variation in resistance to infection has indeed been observed in natural fruit fly populations and has been associated with genetic polymorphisms [12].
All metazoans seem to have an innate immune system, and in insects, at least four different signaling pathways are involved in the immune response and mediate pathogen recognition, attack on the pathogen, and antiviral RNA interference, among other responses. McTaggart etal. [7] analyzed the D. pulex genome for genes related to the immune system and identified genes homologous with those in other arthropods. The authors found that the Toll signaling pathway, which is activated by the presence of pathogens, is conserved between insects and Daphnia. The activation of this pathway results in the production of antibacterial and antifungal proteins. These antimicrobial peptides could not be recovered from the D. pulex genome and thus seem to be less well conserved. In addition, McTaggart et al. [7] report considerable variation in gene family copy number in Daphnia and insects. These differences might reflect the evolutionary history of host-parasite interactions in the individual lineages. Further comparative studies are needed to uncover evolutionary changes in genes that mediate immune responses as well as taxon-specific expansions of gene families, which will contribute to our understanding of how host genes are evolving in response to parasites.
Although a vast number of ecto-and endoparasites have been described for Daphnia, the non-parasitic symbionts have not been analyzed in detail. Ebert and co-workers (Qi et al. [8]) have used metagenomics to address this question. Metagenomes genetic material recovered directly from environmental samples are sequenced and compared to the databases in order to characterize the biological community of a given habitat. One of the advantages of this approach is the recovery of DNA sequences from organisms that cannot be cultured. Ebert and co-workers [8] searched the shotgun sequences of three clones of three different Daphnia species (D. pulex, D. magna and D. pulicaria) for indications of bacterial and plastid symbionts and found sequences representing a large number of bacterial species in each dataset. The majority of the sequences were from the Proteobacteria but many other taxa were also detected. No clear evidence was found for the presence of symbiotic cyanobacteria or of plastids. Interestingly, the composition of the bacterial communities was similar at genus and higher taxonomic levels in all three Daphnia clones, but different bacterial species were present in individual clones. The D. pulex and D. pulicaria DNA used in this study was isolated from clones cultured in North America, whereas the D. magna cultures originated from a laboratory in Switzerland. Since contamination of the Daphnia cultures by cross-Atlantic exchange is unlikely, the authors suggest that the similarities between the symbiont communities in European and North American Daphnia samples indicate a long-term stability of symbiotic associations.
Environmental challenges
Daphnia species have been studied extensively because of their importance to aquatic ecosystems, and they show a striking ability to contend with environmental challenges. The availability of the D. pulex genome should now be able to provide insights into the adaptation to specific environmental conditions, from the ecological to the genetic level. On screening the D. pulex genome for genes involved in the biochemical response to toxicants, Baldwin et al. [9] identified 75 genes of the cytochrome P450 family, a protein family important in tolerance and resistance to environmental chemicals. The authors report that the same subgroups of cytochrome P450 genes are present in the Daphnia genome as in insects and nematodes, but they discovered distinct changes in the size and gene composition of each group. Dean and co-workers (Sturm et al. [10]) screened the Daphnia genome for the presence of members of the ABC transporter superfamily (ATP-binding cassette membrane transport proteins), which are also involved in biochemical defense against toxicants. They found that ABC family representation in Daphnia is as complex as in other metazoans, and that Daphnia most resembles the fruit fly in respect of its ABC transporter genes. Future studies on the expression and function of these genes will uncover their importance in the adaptation of Daphnia to environmental toxicants.
Daphnia and arthropod phylogeny
The D. pulex genome also has the potential to contribute to resolving long-standing debates on arthropod phylogeny. Current views of arthropod phylogenetic relationships are based mainly on two types of datasets - molecular genetic data and morphological characters - and this has led to partly contradictory concepts of arthropod phylogeny. There is now almost universal agreement that arthropods derive from a common ancestor, and that crustaceans and insects are sister groups [13]. However, some issues of arthropod relationships remain controversial, for example the question of whether insects, crustaceans and myriapods form a monophyletic group. Crustaceans show the greatest diversity of body organization and development among arthropods [14] and therefore the phylogenetic relationships within the crustaceans are far from being resolved. Several morphological and molecular studies have questioned the monophyly of crustaceans, and either Branchiopoda (such as Daphnia) or Malacostraca (lobster, shrimps) has been hypothesized to be the sister group to insects [15]. Some recent molecular analyses suggest a sister group relationship of myriapods (millipedes) and chelicerates (spiders) [16]. Interestingly, this suggestion is supported by recent morphological and molecular studies on the development of the nervous system that reveal a surprising degree of similarity between myriapods and chelicerates [17,18]. The morphological support for an insect-crustacean sister-group relationship is mainly based on the comparative analysis of neural characters in higher crustaceans (malacostracans) and insects. For example, in both insects and malacostracans, stem-cell-like neuroblasts have been detected that divide asymmetrically to generate the cells that contribute to the nervous system [14]. But are these neural characters representative of all crustacean groups? Are homologous genes required for the development and the function of the nervous system? With the availability of a branchiopod genome and the development of genetic tools for Daphnia these questions can now be addressed.
Furthermore, using genome sequences of a wide range of organisms, the origin and evolution of neural signaling pathways can be traced, which will broaden our understanding of the evolution of nervous systems. The neurotrophin signaling pathway plays a role in neural development, regeneration and neural plasticity in mammals. Analyzing the Daphnia genome, Wilson [11] shows for the first time that a neurotrophin and both a tyrosine receptor kinase (Trk) and a p75-type neurotrophin receptor (p75NTR) are present in a protostome, indicating that this pathway existed in the last common ancestor of protostomes and deuterostomes.
To conclude, the initial exploration of the D. pulex genome outlined above proves that with the availability of the genome sequence Daphnia research has entered a new era. New and long-standing questions in ecology and evolution can be addressed and it may finally be possible to link evolutionary and environmental adaptations to the underlying genetic processes.
Acknowledgements
I am grateful to Dieter Ebert for discussions and comments on the manuscript and I thank Petra Ungerer for the scanning electron micrograph of Daphnia. The work was supported by a grant to AS from the BBSRC.
References
- Daphnia Genome Consortium. Science. 2009. in press .
- wFleaBase Daphnia Genome Project. http://wfleabase.org/
- Ebert D. Ecology, Epidemiology, and Evolution of Parasitism in Daphnia. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information; 2005. http://www.ncbi.nlm.nih.gov/corehtml/pmc/homepages/bookshelf/daph.html [Google Scholar]
- Schurko AM, Logsdon JM Jr, Eads BD. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol Biol. 2009;9:78. doi: 10.1186/1471-2148-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenberger A, Courts C, von Elert E. Target gene approaches: gene expression in Daphnia magna exposed to predator-borne kairomones or to microcystin-producing and microcystin-free Microcystis aeruginosa. BMC Genomics. 2009;10:527. doi: 10.1186/1471-2164-10-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva-Arana DC, Lynch M, Robertson HM. The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol. 2009;9:79. doi: 10.1186/1471-2148-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart SJ, Conlon C, Colbourne JK, Blaxter ML, Little TJ. The components of the Daphnia pulex immune system as revealed by complete genome sequencing. BMC Genomics. 2009;10:175. doi: 10.1186/1471-2164-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Nong G, Preston JF, Ben-Ami F, Ebert D. Comparative metagenomics of Daphnia symbionts. BMC Genomics. 2009;10:172. doi: 10.1186/1471-2164-10-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, Marko PB, Nelson DR. The cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC Genomics. 2009;10:169. doi: 10.1186/1471-2164-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Cunningham P, Dean M. The ABC transporter gene family of Daphnia pulex. BMC Genomics. 2009;10:170. doi: 10.1186/1471-2164-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. The genome sequence of the protostome Daphnia pulex encodes respective orthologues of a neurotrophin, a Trk and a p75NTR: evolution of neurotrophin signalling components and related proteins in the bilateria. BMC Evol Biol. 2009;9:243. doi: 10.1186/1471-2148-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP. The complexity of genetic variation in a simple immune system. Trends Genet. 2005;21:197–199. doi: 10.1016/j.tig.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Richter S. The Tetraconata concept: hexapod-crustacean relationships and the phylogeny of Crustacea. Org Divers Evol. 2002;2:217–237. doi: 10.1078/1439-6092-00048. [DOI] [Google Scholar]
- Scholtz G. Baupläne versus Ground Patterns, Phyla versus Monophyla: Aspects of Patterns and Processes in Evolutionary Developmental Biology. Lisse: A. A. Balkema; 2004. [Google Scholar]
- Averof M, Akam M. Insect-crustacean relationships: insights from comparative developmental and molecular studies. Philos Trans R Soc London B Biol Sci. 1995;347:293–303. doi: 10.1098/rstb.1995.0028. [DOI] [Google Scholar]
- Hwang UW, Friedrich M, Tautz D, Park CJ, Kim W. Mitochondrial protein phylogeny joins myriapods with chelicerates. Nature. 2001;413:154–157. doi: 10.1038/35093090. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Weller M, Tautz D. Neurogenesis in the spider Cupiennius salei. Development. 2001;128:2673–2688. doi: 10.1242/dev.128.14.2673. [DOI] [PubMed] [Google Scholar]
- Dove H, Stollewerk A. Comparative analysis of neurogenesis in the myriapod Glomeris marginata (Diplopoda) suggests more similarities to chelicerates than to insects. Development. 2003;130:2161–2171. doi: 10.1242/dev.00442. [DOI] [PubMed] [Google Scholar]