Abstract
Understanding the spatio-temporal subversion of host cell signaling by bacterial virulence factors is key to combating infectious diseases. Following a recent study by Buntru and co-workers published in BMC Biology, we review how fluorescence (Forster) resonance energy transfer (FRET) has been applied to studying host-pathogen interactions and consider the prospects for its future application.
See research article http://www.biomedcentral.com/1741-7007/7/81.
Pathogen-induced signaling in mammalian cells
The incidence of diseases caused by pathogenic bacteria is once again increasing with the resurgence of tuberculosis, the rise of nosocomial infections, and escalating resistance to antibiotics. Over the last two decades it has become increasingly apparent that many bacterial pathogens of substantial medical relevance engage in complex cross-talk with cells of their mammalian hosts. It is thus critical to decipher the molecular mechanisms of the underlying bacterial virulence weaponry, which includes not only the secreted exotoxins, like those that cause anthrax and cholera, but also the multiple 'effectors' injected directly into eukaryotic target cells via specialized nanomachines by pathogens like Escherichia coli O157:H7 and Yersinia. Such studies have already yielded new insights into the molecular basis of microbial pathogenesis, suggesting new avenues for the development of novel diagnostics, therapeutics and vaccines. These toxins and effectors can also be exploited as reagents to probe the pathways controlling key cellular processes such as signal transduction, cytoskeletal dynamics, intracellular trafficking and cytokinesis. An exciting new epoch of molecular, cellular and structural microbiology has therefore dawned.
In recent years, substantial research efforts have focused on the actions of such bacterial virulence proteins and the identity of their host targets, clearly an essential initial step. However, an appreciation of the spatio-temporal dynamics underlying the intricate molecular cross-talk that triggers complex events such as bacterial internalization into host cells remains a distant goal. Most biochemical and genetic approaches entail cell disruption or artificial protein localization and expression, and observations arise from population rather than single cell analyses. In addition, many signaling interactions are transient or of low-affinity and thus difficult to detect.
Fluorescence (Forster) resonance energy transfer (FRET) to detect direct interactions between labeled molecules
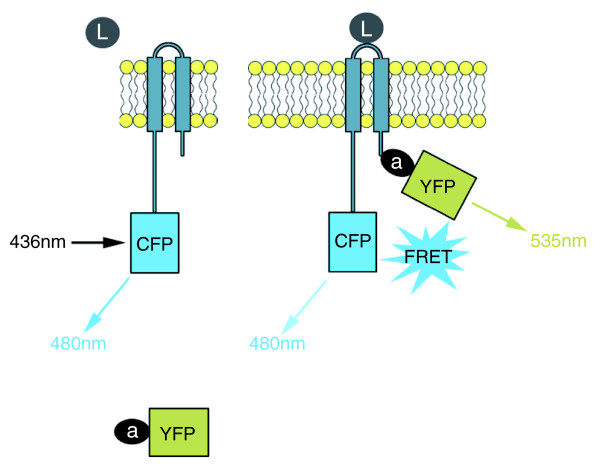
One powerful approach is fluorescence (Forster) resonance energy transfer (FRET) [1]. FRET utilizes two fluorophores, a donor and an acceptor, of which the donor emission spectrum overlaps with the acceptor absorption spectrum. If the two fluorophores are spatially segregated, excitation of the donor results in donor emission with high efficiency. In contrast, when in close proximity (usually 1-10 nm), excitation of the donor results in acceptor emission due to the overlap in spectra that allows resonance energy transfer between the donor and the acceptor probes (Figure 1). A modification of FRET termed acceptor photobleaching FRET involves selective photochemical destruction of the acceptor fluorophore, which, if the two fluorophores had previously been physically close enough for FRET, results in a release from donor quenching and an increase in donor emission. Technically, this is particularly advantageous as it reduces the requirements for compensation and calibration associated with standard FRET.
Figure 1.
Schematic representation of a hypothetical FRET experiment. In the resting state (left), a transmembrane receptor is fused to cyan fluorescent protein (CFP, donor). Adaptor protein (a) is fused to yellow fluorescent protein (YFP, acceptor). a-YFP is distal from receptor-CFP, so upon excitation at 436 nm, donor fluorescence at 480 nm is recorded. Upon binding to a ligand (L, right), a-YFP binds to receptor-CFP and the reduction in distance enables FRET. Upon equivalent excitation at 436 nm, donor fluorescence (480 nm) is reduced, but acceptor fluorescence at 535 nm is now recorded due to FRET. FRET can similarly be performed with two transmembrane or two soluble factors.
FRET is now widely applied in the context of cellular signaling to detect interactions such as the formation of protein complexes, and to detail molecular dynamics such as changes in protein conformation. When combined with fluorescence microscopy, it can simultaneously provide information about the location within a single cell in which an interaction is occurring in two and three dimensions. Given the power of this technique, its use in understanding host-pathogen interplay has, to date, been surprisingly limited. Here we review the principal studies that have utilized FRET to understand signaling events, in each case initiated at the plasma membrane by a bacterium, and we consider the prospects for future applications.
Probing the intermolecular interactions of bacterial exotoxins
Some pathogenic bacteria release exotoxins that poison mammalian target cells directly by disrupting their membrane integrity or indirectly using associated enzymatic activities that override cellular pathways. These virulence factors alone are often sufficient to cause disease, and their activities normally require toxin oligomerization and interaction with host targets.
Bastiaens and colleagues investigated one example of this [2]. Cholera toxin (CTX) has an oligomeric AB5 structure, comprising one enzymatically active CTX-A subunit embedded within an isopentamer of CTX-B moieties. The secreted holotoxin binds to the host cell surface via interactions between CTX-B and plasma membrane GM1 ganglioside. After internalization and processing, CTX-B and CTX-A dissociate, whereupon a liberated sub-fragment of CTX-A irreversibly ADP-ribosylates the α-subunits of heterotrimeric Gs proteins, leading to aberrant persistent activation of host adenylate cyclase. Compartmentalization and separation of the holotoxin were evaluated by measuring acceptor photobleaching FRET between CTX-B labeled with the sulfoindocyanine dye Cy3 and an antibody against CTX-A labeled with Cy5 in single cells, exploiting confocal laser scanning microscopy. The data illuminated that after holotoxin internalization, CTX-A is trafficked back towards the plasma membrane by retrograde transport, whereas the CTX-B multimer persists with the Golgi apparatus. This study was one of the first to apply FRET to a pathogen system and contributed to understanding the unexpected complexities of intracellular CTX transport.
A related study of Helicobacter pylori vacuolating cytotoxin (VacA), which binds and enters mammalian cells to induce cellular vacuolation, also addressed toxin multimerization. In this case FRET was monitored within cells co-expressing VacA and derivative proteins fused to cyan or yellow fluorescent protein, where association induces energy transfer between the VacA-CFP donor and VacA-YFP acceptor pairs [3]. These FRET data suggested that intermolecular interactions between discrete monomers are critical for intracellular activity of the toxin.
Probing pathogen-host cell interplay that triggers or prevents bacterial uptake
These early FRET-based studies of bacterial exotoxins focused on understanding the intermolecular interactions between toxin subunits rather than between toxins and their host targets. Another timely and ambitious application of FRET has been to examine the cellular signaling pathways underpinning the interaction of bacterial pathogens with both phagocytic and non-phagocytic host cells.
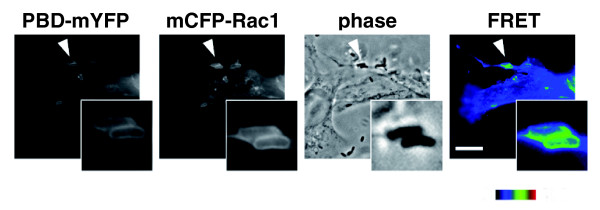
The Yersinia outer membrane protein invasin acts as a high affinity ligand for cellular β1-family integrins, transmembrane receptors involved in the formation of multi-protein structures termed focal adhesions that link the extracellular matrix to the intracellular cytoskeleton. Invasin-mediated clustering of integrins triggers host signaling on the cytoplasmic face of the plasma membrane. This requires the activation of the small GTPase Rac1, which subsequently binds downstream adaptors that promote cytoskeletal rearrangements and bacterial internalization. Ralph Isberg's laboratory demonstrated that Rac1 was activated at the site of internalization by normally non-phagocytic cells (Figure 2), detecting the presence of activated CFP-Rac1 by its ability to bind to a YFP-labeled domain of its downstream adaptor PAK1 [4]. Although Yersinia is capable of entry into non-phagocytic cells, the bacterium paradoxically also utilizes a specialized (type III) secretion system to translocate effector proteins that prevent its uptake by phagocytic immune cells. Isberg and co-workers used FRET to show that the concerted action of two effectors, YopE, which suppresses Rac1 activation, and YopT, which alters Rac1 membrane localization, generated two spatially distinct Rac1 populations, an active pool in the nucleus and an inactive pool in the cytoplasm, leading to cellular paralysis. Thus, FRET, being uniquely suited to investigating the location and activation state of host molecules, was integral to uncovering this multifaceted manipulation of GTPase-dependent signaling [5].
Figure 2.
Yersinia pseudotuberculosis binding to host cells leads to local activation of Rac1 GTPase. COS1 cells, expressing mCFP-Rac1 and mYFP fused to the p21 binding domain of Pak1 (PBD) were incubated with an effector-deficient Y. pseudotuberculosis for 20 minutes, then fixed. GTP-loaded Rac1 interaction with the mYFP-PBD brings fused mCFP and mYFP into close proximity, allowing energy transfer. This energy transfer is recorded microscopically as a corrected FRET image. CFP, YFP, and FRET images were captured using appropriate filter cube sets and the FRET image was corrected for bleed-through and cross-excitation. Scale bar (applicable to all images except insets) = 10 μm. Activation of Rac1 in response to bacterial binding is depicted here. Images courtesy of Sima Mohammadi and Ralph Isberg.
FRET can be combined with live cell imaging to reveal not only detailed interactions critical to manipulation of host cells by bacteria, as described above, but also the kinetics of those interactions, thereby establishing a specific sequence of events. Such a study resulted from a collaboration between the laboratories of Pascale Cossart and Joel Swanson, who combined FRET with live cell imaging to document the kinetics of signaling during Listeria cell entry [6]. Listeria monocytogenes employs outer membrane proteins termed internalins to hijack Rac1-dependent receptor-mediated endocytosis. Internalin B (InlB) binds the hepatocyte growth factor receptor (HGFR/c-Met) to stimulate actin reorganization. They used FRET to investigate the activation of two host factors critical for the entry process, Rac1 and phosphoinositide (PI)-3-kinase, which generates the bioactive signaling lipids phosphatidylinositol-3,4-bisphosphate [PI(3,4)P2] and phosphatidylinositol-3,4,5-triphosphate [PI(3,4,5)P3]. Rac1 activation was monitored similarly to that described above. To monitor activation of PI-3-kinase, the investigators co-expressed YFP and CFP derivatives of the pleckstrin-homology (PH) domain of the serine/threonine kinase Akt, which specifically interacts with these phosphoinositides. Bacterial attachment resulted in a localized FRET signal, as PI(3,4)P2 and PI(3,4,5)P3 were sufficiently concentrated at the plasma membrane to permit a FRET signal between the bound Akt PH domains. Their kinetic analysis of living cells showed that activation of PI3-kinase and concomitant generation of 3'-phosphoinositides at bacterial entry sites occurs upstream of Rac1 activation, which in turn is critical for F-actin assembly. Thus, FRET enabled both the spatial and temporal mapping of lipid- and protein-based signaling at the plasma membrane.
Most recently, in their study in BMC Biology [7], Hauck and co-workers combined live fluorescence microscopy and FRET techniques to study events triggered by the association of Neisseria gonorrhoeae surface (Opa) proteins with the mammalian transmembrane receptor CEACAM3. Uptake of CEACAM3-bound bacteria depends on an immunoreceptor tyrosine-based activation motif (ITAM)-like sequence within the cytoplasmic domain of the receptor, which is rapidly phosphorylated upon ligand binding. This is engaged by multiple host signaling proteins that contain a Src-homology 2 (SH2) domain, including the Src-family kinase (SFK) Hck. Using acceptor photobleaching FRET, as well as other approaches, Hauck and colleagues demonstrated that Hck and CEACAM3 transiently but directly interact specifically at sites of bacterial attachment [7]. Although the interaction of these two mammalian signaling proteins was predicted from earlier biochemical studies, this investigation confirmed the prediction and also revealed the dynamic nature of the association in living cells.
FLIM-FRET: it's easy to FRET
Measuring FRET can be simplified considerably by determining the fluorescence lifetime rather than fluorescence intensities. Energy transfer from the donor to the acceptor accelerates the decay of donor fluorescence, allowing energy transfer efficiency to be determined directly from measurements of fluorescence lifetime. Only a single measurement is required and, because many sources of artifact and noise that modulate fluorescence intensity such as sample absorption and variation in laser intensity have no effect on fluorescence lifetime, the requirements for calibration are minimal. For these reasons, fluorescence lifetime imaging (FLIM) is rapidly becoming the preferred method for making FRET measurements. This technique has been utilized to investigate plant cell invasion by the ascomycete powdery mildew fungus [8], but has yet to be applied to any human pathogen.
In addition, FLIM-FRET facilitates the use of FRET as a 'spectroscopic ruler' to measure the physical distance between donor and acceptor fluorophores, a technique utilized by Latz and colleagues [9] to decipher signaling associated with immune recognition of CpG DNA, a pathogen-associated molecular pattern (PAMP), by the Toll-like receptor TLR9. To determine how engagement of the ectodomain of TLR9 might transmit an intracellular signal, they expressed both CFP-TLR9 and YFP-TLR9 in cultured cells and measured the distance between adjacent TLR9 cytoplasmic domains using FLIM-FRET. They calculated that receptor engagement resulted in a decrease in the intermolecular distance from 7.0 nm to less than 5.4 nm, a change apparently sufficient to trigger immune signaling by recruiting the downstream adaptor molecule MyD88.
Future perspectives
The investigation of pathogen-induced signal transduction in mammalian cells is a rich vein to tap for understanding not only the pathogenesis of infectious agents, but also fundamental features of mammalian cell signaling. As illustrated here, FRET-based techniques that enable the description of the activation state of key signaling molecules, their cellular location, proximity to each other, and the timing of their interactions, have already proved to be powerful tools to decipher the spatiotemporal features of critical signaling events. Given the fundamental importance of microbial-host cell communication and the growing capacity for its application, FRET should be more widely employed by investigators of infectious agents. Making FLIM-capable instruments more widely available and accessible for use with samples containing live infectious agents will be needed to realize this welcome development.
Acknowledgements
We thank Ralph Isberg for helpful discussion and Sima Mohammadi for Figure 2. This was supported by NIH R01 AI46454 to JML. RDH is a Royal Society University Research Fellow.
References
- Förster T. Intermolecular energy migration and fluorescence. Ann Phys. 1948;2:55–75. doi: 10.1002/andp.19484370105. [DOI] [Google Scholar]
- Bastiaens PHI, Majoul IV, Verveer PJ, Söling H-D, Jovin TM. Imaging the intracellular trafficking and state of the toxin AB5 quaternary structure of cholera toxin. EMBO J. 1996;15:4246–4253. [PMC free article] [PubMed] [Google Scholar]
- Willhite DC, Ye D, Blanke SR. Fluorescence resonance energy transfer microscopy of the Helicobacter pylori vacuolating cytotoxin within mammalian cells. Infect Immun. 2002;70:3824–3832. doi: 10.1128/IAI.70.7.3824-3832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KW, Isberg RR. Yersinia pseudotuberculosis spatially controls activation and misregulation of host cell Rac1. PLoS Pathog. 2005;1:e16. doi: 10.1371/journal.ppat.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KW, Mohammadi S, Isberg RR. The polybasic region of Rac1 modulates bacterial uptake independently of self-association and membrane targeting. J Biol Chem. 2008;283:35954–35965. doi: 10.1074/jbc.M804717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seveau S, Tham TN, Payrastre B, Hoppe AD, Swanson JA, Cossart P. A FRET analysis to unravel the role of cholesterol in Rac1 and PI 3-kinase activation in the InlB/Met signalling pathway. Cell Microbiol. 2007;9:790–803. doi: 10.1111/j.1462-5822.2006.00832.x. [DOI] [PubMed] [Google Scholar]
- Buntru A, Zimmermann T, Hauck CR. Fluorescence resonance energy transfer (FRET) based subcellular visualization of pathogen-induced host receptor signalling. BMC Biol. 2009;7:81. doi: 10.1186/1741-7007-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA. 2005;102:3135–3140. doi: 10.1073/pnas.0500012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]