Abstract
Hepatocellular carcinoma (HCC) is one of the most frequent and malignant diseases worldwide. Epidemiological studies have clearly demonstrated that chronic hepatitis B virus (HBV) infection is a major etiological factor in the development of HCC. The pathogenesis of HBV-associated HCC has been studied extensively, and the molecular changes associated with malignant transformation have been identified. The predominant carcinogenic mechanisms of HBV-associated HCC are chronic inflammation and the effects of cytokines in the development of fibrosis and liver cell proliferation. An important role is also played by the integration of HBV DNA into host cellular DNA, which disrupts or promotes the expression of cellular genes that are important in cell growth and differentiation. Especially, HBx protein is a transactivating protein that promotes cell growth, survival, and the development of HCC. Continued investigation of the mechanisms underlying hepatocarcinogenesis will refine our current understanding of the molecular and cellular basis for neoplastic transformation in the liver. Prevention of HBV infections and effective treatments for chronic hepatitis B are still needed for the global control of HBV-associated HCC. This review summarizes the current knowledge on the mechanisms involved in HBV-associated hepatocarcinogenesis.
Keywords: Hepatocellular carcinoma, Hepatitis B virus, Hepatitis B x protein (HBx), Transactivator
INTRODUCTION
Hepatocellular carcinoma (HCC) is currently the fifth most common cancer worldwide and the fourth leading cause of cancer-related deaths. The number of new cases is estimated to be more than 500,000 per year, accounting for 4% of all newly diagnosed cancers.1 More than 80% of HCC cases occur in developing countries, especially in Southeast Asia and sub-Sahara Africa, but the incidence is increasing in economically developed regions, including Japan, Western Europe, and the United States.1,2 In South Korea, HCC remains the second most common malignancy in men and the fourth in women. The annual number of new cases is estimated to be 11,000, comprising 8,300 men and 2,700 women.3 The overall prognosis of HCC is poor, which is due to many patients at presentation already being in an advanced and unresectable state, for which the median survival is less than 6 months.4 The high mortality may be partially attributable to the noncapsular part of the liver being lacking in sensory fibers, with symptoms therefore only presenting in advanced HCC.4 A small proportion of patients are eligible for liver resection, which results in a 5-year survival of about 40%.4 However, even with surgical resection the recurrence rates can be as high as 50% at 2 years.5 Fortunately these outcomes are rapidly changing, since 30-40% of patients are now diagnosed at the initial stages when curative treatments can be optimally applied.5 Therefore, estimates of outcome need to take into account the stage at diagnosis. The major causes of HCC include hepatitis B, hepatitis C, alcoholic liver disease, nonalcoholic steatohepatitis, and other rarer forms of cirrhosis. At least 90% of HCC cases occur in patients with chronic liver disease, and most have cirrhosis.1 It has been recently estimated that about 53% of HCC cases worldwide are related to hepatitis B virus (HBV).1
Of the approximately 2 billion people who have been infected worldwide, more than 350 million are chronic carriers of HBV.1,2 The prevalence of carriers exhibits marked geographic variations, ranging from 10 to 20% in Southeast Asia and sub-Sahara Africa to less than 1% in northern Europe and the United States.1,2 Approximately 75% of chronic carriers live in Asia and the western Pacific.1,2 Carriers of the hepatitis B surface antigen (HBsAg) constitute 5.1% of males and 4.1% of females in South Korea.6 Where HBsAg is endemic, the most common route of transmission is perinatal or the infection is acquired during the preschool years.1,2 Where HBsAg is not endemic, most HBV infections are acquired by horizontal transmission in adolescents and young adults in relatively well-defined high-risk groups, including injecting drug users, homosexual males, health-care workers, and patients receiving regular blood transfusions or hemodialysis.1,2 The frequency of viral persistence following acute HBV infection is related to age, gender, and immune deficiency: 90% of infants under 1 year of age, 30% of children aged 1-5 years, and 10% of adults; twofold higher in men than in women; and immune-deficient individuals, such as those with HIV infection and renal insufficiency requiring hemodialysis.7 Typically 15-40% of infected patients will develop cirrhosis, liver failure, or HCC.2 In addition, exposure to other hepatotoxins (e.g., alcohol and aflatoxin B1 [AFB1]) in the individual with HBV infection hastens both the development of cirrhosis and HCC.8 AFB1 is produced by the fungus Aspergillus flavus and is metabolized to an epoxide, which is mutagenic.8 AFB1 exposure is highest in southern China and southern Africa, with patients with HCC from these regions tending to have a specific p53 mutation in codon 249 involving a G-to-T transversion (arginine to serine) in the third nucleotide.8 In contrast, codon 249 mutations are rare in Korea, where AFB1 exposure is low.9 Also, coinfection with hepatitis C virus and HBV increases the risk of developing HCC by at least twofold.8
The pathogenesis of HBV-associated HCC has been studied extensively, and the molecular changes that occur during malignant transformation have been identified. However, the mechanisms underlying how HBV causes carcinogenic changes in the liver are still unclear. Studying HBV is difficult due to the lack of cell culture systems for virus propagation in vitro. Much of what we know about the replication and expression of HBV is derived from investigations of closely related hepadnaviruses such as the woodchuck hepatitis virus and the Beechey ground squirrel hepatitis virus.10 As with most viral diseases, HBV-infected host cells are exposed to multiple opposing signals mediated by growth hormones, immune cytokines, and the effects of adjacent cells such as lymphocytes or Kupffer cells. Hepatocytes must convert these signals into a defined response - such as proliferation, differentiation, or death - via signal transduction. Moreover, the infected hepatocytes must modulate signal transduction pathways leading to growth, inflammation, or cell death in order to maximize the symbiotic survival of both the virus and the cell in a process that often progresses to cirrhosis and HCC.7 Furthermore, early HCC progresses to advanced HCC, which is a larger tumor with malignant potential, and moderately or poorly differentiated.7 Both the development and progression of HCC are believed to be caused by the accumulation of genetic changes as well as genes involved in different regulatory pathways, such as cell-cycle control, apoptosis, adhesion, and angiogenesis.4 This review summarizes the current knowledge on the mechanisms involved in HBV-associated hepatocarcinogenesis.
EPIDEMIOLOGIC EVIDENCE OF A LINK BETWEEN HBV INFECTION AND HCC
Epidemiological studies have clearly demonstrated that chronic HBV infection is a major etiological factor in the development of HCC. The incidence of HCC and the prevalence of HBV serological markers follow the same general geographic distribution pattern.2 HCC is more common than other types of cancer only in regions where HBV is endemic.2,8 In addition, the duration of HBV infection and the severity of the underlying chronic hepatitis correlate with an increased risk of HCC. The yearly incidence of HCC was reportedly 0% in inactive carriers, approximately 0.3% in patients with chronic HBV infection, and 1.5-6.6% in patients with compensated cirrhosis.2,8,11,12 Moreover, HCC can occur even in the presence of convalescent HBV infection.2 The risk of HCC is several-fold higher in patients positive for hepatitis B core or surface antibody than in HBV-naive controls, although the risk is still much lower than that in those with active infection.13,14 Importantly, a study from Taiwan demonstrated that a universal hepatitis B vaccination reduced the incidence of HCC in children.11 A study in South Korea suggested that immunization with the hepatitis B vaccine, even in adulthood, can reduce the risk of HCC.15 HBsAg carriers constitute 2.1% of males and 2.7% of females among Korean patients younger than 20 years.6 Several studies have provided strong evidence of a relation between viral replication and the risk of HCC.2,12 The relative risk of HCC is much higher in patients positive for hepatitis B e antigen (HBeAg) than in inactive HBsAg carriers.12,16 Furthermore, HBV DNA has been identified as the most important predictor of the development of HCC in HBsAg-positive patients with diverse clinical conditions.16 Taken together, these data suggest that earlier seroconversion of HBeAg and sustained suppression of viral load confer a lower risk of HCC development in HBV carriers. Large numbers of persons are infected with HBV or will continue to become infected, irrespective of vaccination, and hence therapy for the underlying liver disease may be the best approach for preventing HCC.
HBV can be classified into eight genotypes (A-H). HBV genotypes B and C are prevalent in the Far East and Southeast Asia, and the clinical relevance of HBV genotypes has become increasingly recognized.17 In Korea, genotype C prevails predominantly among chronic HBV carriers, irrespective of the clinical stages of their liver disease.18 In general, the disease activity is more aggressive and the risk of HCC and its recurrence after curative resection are higher in patients with HBV genotype C than in those with genotype B.17
STRUCTURE OF HBV GENOME
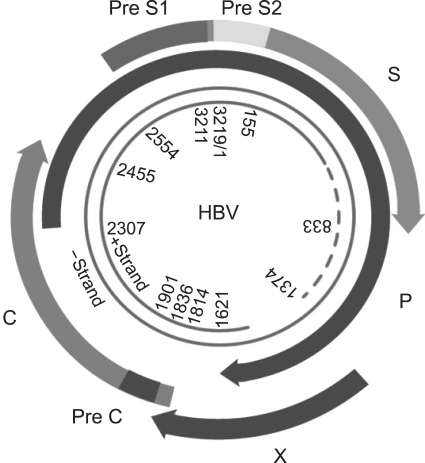
HBV belongs to the family of hepadnaviruses. The infectious virion (also known as Dane particle) is a 42-nm-diameter spherical particle containing an envelope and a nucleocapsid. The HBV genome comprises relaxed circular, partially double-stranded DNA of approximately 3,200 base pairs (Fig. 1). There are four partially overlapping open reading frames encoding the envelope (pre-S/S), core (precore/core), polymerase (P), and X proteins.19 Transcription is initiated by four promoters: preC/pregenomic, S1, S2, and X. In addition, two enhancers (enhancers I and II) play important roles in the regulation of viral gene transcription. Transcription, which is unidirectional, results in the formation of preC/pregenomic, preS1, preS2/S, and X mRNA. All HBV primary transcripts are modified by 3' polyadenylation and the addition of a 5' cap. The single polyadenylation signal terminates transcription at a common 3' end for all the transcripts, while the 5' ends are variable and are determined by the location of each of the HBV promoters initiating transcription. The 3.5-kb pregenomic mRNA serves as the template for the core and polymerase proteins in addition to being the template for reverse transcription. The preC transcript is slightly longer than the pregenomic RNA and is the template for translation of the precore/core protein. This protein is processed in the endoplasmic reticulum and secreted as HBeAg. The 2.4- and 2.1-kb mRNAs encode the large and the middle and major surface proteins, respectively, and the small 0.9-kb transcript is the template for translation of HBV X protein (HBx). The polymerase protein functions as a reverse transcriptase as well as a DNA polymerase.7,19
Fig. 1.
Schematic of the organization of the hepatitis B virus (HBV) genome. The HBV genome comprises relaxed circular, partially double-stranded DNA of approximately 3,200 base pairs. There are four partially overlapping open reading frames encoding the envelope (pre-S/S), core (precore/core), polymerase (P), and X proteins.
The mechanism underlying liver injury is still unclear, but it is believed that host immune responses accompanying HBV infection induce tissue damages, since HBV is not directly cytotoxic.7 The chronic HBV infection is related to an inefficient cytotoxic-T-lymphocyte (CTL) response to viral components critical to protective immunity.7 In other words, if the T-cell response is strong enough, HBV can be eliminated from the liver; if not, a procarcinogenic effect can be induced by permanently triggering necroinflammatory disease without resulting in a final eradication of HBV from the liver.
CURRENT CONCEPTS OF HBV-ASSOCIATED HEPATOCARCINOGENESIS
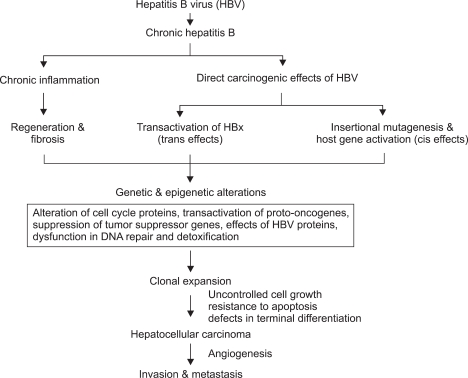
Despite there being clear evidence for an etiological association between HBV infection and HCC, the complexity of the mechanism means that the distinct underlying molecular pathway or molecules are not yet known. HCC exhibits a high degree of genetic heterogeneity, suggesting that multiple molecular pathways are involved in the genesis of subsets of HCC. There are two general concepts associated with the mechanism of HBV-associated hepatocarcinogenesis (Fig. 2).4,13 At present, HBV-associated carcinogenesis can be viewed as a multifactorial process that includes both direct and indirect mechanisms that might act synergistically. One involves chronic necroinflammation of hepatocytes, cellular injury, mitosis, and hepatocyte regeneration.7 Chronic HBV infection causes inflammation with the release of free radicals (reactive nitrogen or oxygen species), chemokines, and cytokines resulting in DNA damage, cell proliferation, fibrosis, and angiogenesis.7,10 Also, continuous necrosis of hepatocytes during chronic HBV infection accompanied by rapid regeneration may lead to the accumulation of mutations and selection of cells with a malignant phenotype.10 Regenerative hepatocytes in chronic liver diseases give rise to hyperplastic hepatocyte nodules, and these progress to dysplastic nodules, which are thought to be the direct precursor of HCC.7 Several observations support this theory, such as that the period of inflammation in HBV infection is proportional to the risk of developing HCC. HBV-related HCC develops 10-30 years after infection with the virus, suggesting that HBV is not an acute oncogenic agent.7 That is, the long latency of HCC development after the initial HBV infection suggests that the virus acts indirectly. Patients with cirrhosis are more prone to develop HCC than those that have active chronic hepatitis or HBV infection without cirrhosis.13,14 Chronic HBV leading to cirrhosis remains the most important precancerous etiologic factor, with 70-90% of HBV-associated HCCs developing on a background of cirrhosis.2,8,13 In addition, integration of the virus DNA does not occur in 20% of patients with HCC associated with HBV.13 However, 10-20% of cases of HCC develop in persons without cirrhosis.2,8 That is, the phenomenon of HBV-induced HCC occurring in the context of a noncirrhotic liver in chronically infected children and young adults strongly supports a specific role for HBV integration in liver carcinogenesis.
Fig. 2.
Mechanism of HBV-associated hepatocarcinogenesis. The predominant carcinogenic mechanism of HBV-associated hepatocellular carcinoma (HCC) is through the process of cirrhosis, but direct oncogenic effects of HBV may also contribute. The neoplastic transformation of hepatocytes results from the accumulation of genetic damage during the cellular proliferation that occurs in the injured liver in response to stimulation by growth factors and cytokines. HCCs exhibit numerous genetic abnormalities (e.g., chromosomal deletions, rearrangements, aneuploidy, gene amplifications, and mutations) and epigenetic alterations (e.g., modulation of DNA methylation). These genetic and epigenetic alterations combine to activate positive mediators of cellular proliferation (e.g., cellular proto-oncogenes and their mitogenic signaling pathways) and inactivate negative mediators of cellular proliferation (e.g., tumor suppressor genes), resulting in cells with autonomous growth potential.
The other pathway evokes the direct oncogenic potential of HBV via chromosomal integration (cis-activation) or transactivation of cellular genes.13 The integration of HBV DNA into the host genome occurs at the early stages of clonal tumor expansion.13 There are several lines of evidence that HBV leads directly to HCC. Persistent HBV replication is associated with a high frequency of integration of HBV sequences into the human host genome.13 HBV DNA reportedly transactivates many cellular genes associated with cell proliferation, and induced HCC in a transgenic mouse.10 Almost all HBV-associated HCCs harbor chromosomally integrated HBV DNA. In many cases, these integrated viral genomes are characterized by rearrangements and/or partial deletions,4,13 but the insertion location is random and is not usually near other important genes.20 Since integration of the virus DNA does not occur in 20% of patients with HBV-associated HCC,13,21 this concept cannot also explain the clinical observation that asymptomatic HBV carriers with extensive viral replication rarely develop HCC.2,21 Furthermore, some patients develop HCC during occult HBV infection that is not even detectable in the serum.22 Therefore, the cause of HBV-associated HCC could be a combination of generalized processes that lead to chronic hepatic disease and those specific ones related to HBV integration.
INTEGRATION OF HBV DNA
HBV shares a replication strategy that includes the reverse transcription of an RNA intermediate.19,23 HBV DNA integration into cellular DNA is not necessary for viral replication but allows for persistence of the viral genome.23 HBV DNA sequences have been shown to be integrated into cellular DNA in HCC tissue and can also be identified in nontumorous tissue obtained from patients with chronic hepatitis.13 Moreover, integration of the viral DNA can occur during the acute or early stages of infection and can persist in patients irrespective of whether there is detectable HBsAg in their serum.24 HBV integrations produce a wide range of secondary genetic alterations within the host genome, including deletions, translocations, the production of fusion transcripts, and generalized genomic instability.4,13 That is, alterations in the number of chromosomes are found in HCC and are designated as either aneuploidy (the gain or loss of whole chromosomes) or loss of heterozygosity (LOH[the gain or loss of chromosome sections or single genes]).13 Several studies investigating the chromosomal integrity of HCC have identified deletions in portions of chromosomes. Losses in chromosomes 1p, 4q, 5q, 6q, 8p, 9p, 13q, 16p, 16q, and 17p are evident in 25-45% of patients, while gains occur in chromosomes 1p, 6p, 8q, and 17q in 30-55% of patients.4,25,26 Many of these chromosomal segments contain known tumor suppressor genes such as p53, retinoblastoma protein (Rb), cyclin D1, and p16.4
Several HBV genes have been found in infected tissues, including hepatitis B X gene, truncated pre-S2/S, and a novel spliced transcript of HBV referred to as hepatitis B spliced protein (HBSP).27 Research over the past decade has suggested that HBx plays a pivotal role in hepatocarcinogenesis related to HBV. HBx is a multifunctional regulator that modulates transcription, signal transduction, cell-cycle progression, protein degradation pathways, apoptosis, and genetic stability by interacting with host factors either directly or indirectly.28-30 Transgenic mice have provided insight into the mechanisms of hepatocarcinogenesis, but the results have been inconsistent, with HBx promoting liver tumor formation in some transgenic mice31,32 but not in others.10,33,34 However, HBx acts as a cofactor in both carcinogen- and c-Myc-induced hepatocarcinogenesis.10 More than 95% of patients with HBV-associated cirrhosis and dysplasia are positive for HBx, and 70% of patients with HBV-associated HCC produce HBx.8,35 Therefore, it appears likely that HBx contributes to the initiation of tumor formation in the liver during chronic active hepatitis and cirrhosis. Integrated HBV sequences frequently have a carboxy-terminal deleted X gene, resulting in translation of a truncated HBx.20 These deletion mutants appear to lose their capacity for oncogene-induced apoptosis and cell-cycle arrest, leading to an increased tendency for unregulated cell proliferation that enhances the transforming capacity of the protein.36,37
Another HBV gene product that has been reported to possess transactivational properties is a truncated form of the pre-S2/S gene, referred to as MHBst.13 Truncated pre-S2/S sequences are frequently found in HBV DNA integration sites in HCC.13,38 Specific activation of mitogen-ctivated protein kinase (MAPK) and extracellular-signal-egulated kinase (ERK) signaling by the truncated pre-S2/S protein activates transcription factors such as AP-1 and nuclear factor-(B (NF-kB).38 Furthermore, the pre-S2/S activators increase the proliferation rate of hepatocytes by activating this signaling cascade.39 The pre-S mutants also induce oxidative DNA damage via endoplasmic reticulum (ER) stress signaling pathways. The oxidative DNA damage caused by pre-S mutants result in genomic instability and mutation of liver cells, and ultimately lead to HCC.38,40 Cytopathologically, overproduction of HBV envelope proteins (pre-S2/S) - particularly L and possibly M - results in their intracellular accumulation and may predispose the cell to stress, which in turn may lead to the development of HCC.40 Pre-S2/S mutants that overaccumulate envelope polypeptides within the cell have also been observed in association with advancing liver disease and may be partially responsible for ground-glass hepatocytes and perhaps even HCC lesions.41 In addition, the overexpression of LHBs protein in transgenic mice has been shown to be cytopathic, possibly leading to liver injury, regenerative hyperplasia, chronic inflammation, oxidative DNA damage, hepatocyte aneuploidy, and eventually progression to HCC.42
HBSP is encoded by one of the spliced RNAs of HBV,43 and its expression induces apoptosis without cell-cycle block. HBSP expression has been correlated with viral replication and the onset of hepatic fibrosis.43
ROLES OF HBx IN HEPATOCARCINOGENESIS
The 154-amino-acid viral gene product 'X' is the viral function that is probably most frequently implicated in oncogenesis. It is named 'X' because of uncertainty about its function, but it does appear to be important for HBV replication.27 The accumulated data suggest that HBx is a multifunctional regulatory protein that communicates either directly or indirectly with a variety of host targets and mediates many opposing cellular functions, including cell-cycle regulation, transcriptional regulation, signaling, encoding of cytoskeleton and cell adhesion molecules, as well as oncogenes and tumor suppressor genes.28-30
1. Transcriptional transactivation
Intracellular localization studies have shown that HBx is present predominantly in the cytoplasm, with lesser amounts in the nucleus.28 The function of HBx differs with the localization: cytoplasmic HBx modulates intracellular signal transduction cascades,28,30 and nuclear HBx may interfere directly with transcription factors or exert a transcription-factor-like function. Thus, HBx stimulates signal transduction pathways such as the MAPK/ERK pathway in the cytoplasm, and also behaves as a transcriptional transactivator that up-regulates the expression of proto-oncogenes such as c-Myc and c-Jun, transcriptional factors such as NF-kB, AP-1, AP-2, the RPB5 subunit of RNA polymerase II, TATA-binding protein, and ATF/cAMP-response element-binding protein (CREB), as well as other viral genes such as HBV enhancers in the nucleus.26-28,44,45
More target proteins that directly interact with HBx are being identified, including Smad4 and nuclear-factor-activated T cells in tumor necrosis factor (TNF)-α signaling, COX-2,46 and retinoid X receptor in phosphoenolpyruvate carboxykinase expression.28 Moreover, several host genes are also reportedly indirectly affected by HBx, such as the up-regulation of interleukin (IL)-6 and IL-8, the induction of nitric oxide synthetase, and Fas ligand.30,45 NF-kB and AP-1 are reportedly directly or indirectly affected by HBx. NF-kB activation by HBx includes direct interaction with inhibitory proteins, such as the p105 precursor and I(B kinase, and the induction of oxidative stress by mitochondria-associated HBx.28,47 In addition, HBx activates components of the MAPK/ERK, stress-activated protein kinase (SAPK)/c-Jun N-terminal kinase (JNK), and protein kinase C (PKC) signaling pathways to regulate NF-kB and AP-1-dependent transcription (Fig. 3).28 Irrespective of the mechanism, HBx-induced NF-kB and AP-1 activity ultimately accelerates cell-cycle progression, enhances proliferation, and may repress apoptosis.
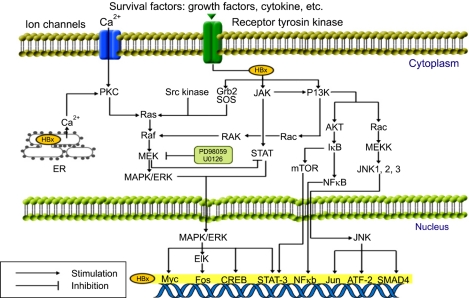
Fig. 3.
Signaling pathways in HBV-associated hepatocarcinogenesis. HBx activates components of the Ras/Raf/MAPK (mitogen-activated protein kinase), SAPK (stress-activated protein kinase)/JNK (c-Jun N-terminal kinase), PI3K (phosphatidylinositol 3-kinase)/AKT (protein kinase B), PKC (protein kinase C), and JAK (Janus kinase)/STAT kinase pathways in the cytoplasm.
2. Regulation of signaling pathways
The complex signaling networks are largely mediated by growth factors, cytokines, and hormones. Such factors can enhance or inhibit cell proliferation, as well as induce a series of differentiated responses in appropriate target cells. The interaction of a growth factor with its receptor by specific binding in turn activates a cascade of intracellular biochemical events that is ultimately responsible for the biological responses observed. The eventual transmission of biochemical signals to the nucleus affects the expression of genes involved in mitogenic responses.48 This involves various complex intracellular signaling pathways, with a key aspect being the significant cross-talk between these signaling pathways, such as up- or downregulation of one of them possibly triggering coordinated responses in another. Thus, inhibition of one component of a signal-transduction pathway may be compensated for in the cell by up-regulation of another pathway.48 These pathways are frequently inappropriately activated in cancer cells by either inappropriate expression of an oncogene coding for a growth factor, a growth-factor receptor, or components of intracellular signaling pathways. Thus, it is clear that disruption of signal transduction pathways is a common event in human cancer and provides a target for therapeutic intervention. Similarly, signal transduction in hepatocytes converts these signals into the defined responses such as proliferation, differentiation, or death. Moreover, the infected hepatocytes modulate signal transduction pathways leading to growth, inflammation, or cell death in order to maximize the symbiotic survival of both the virus and the cell, in a process that often progresses to cirrhosis and HCC.26,27,44,45 The focus of this review is on the roles that signal transduction pathways play in liver cells undergoing pathological changes during an HBV infection.
Several studies have investigated the effects of HBx on signaling pathways such as those involving MAPK/ERK, phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT), PKC, SAPK/JNK, Janus kinase (JAK) signal transducer, and activator of transcription factor (STAT) (Fig. 3). These pathways can overlap or interact with one another to generate unique constitutive or prolonged activations that increase cell proliferation and survival.26,27,44,45
1) Ras/Raf/MAPK/ERK signaling pathway
In general, activation of the ERK members of the MAPK family (ERK or p42/p44 MAPK) promotes cell survival, while the SAPK, JNK, and p38 MAPK members of this family promote cell death.27,44 MAPK pathways can be described by the successive activation of three kinase families: (1) MAPKKK (MAPK kinase kinase) phosphorylates and activates MAPK kinase (MEK), (2) which then activates by phosphorylating MAPK, with (3) MAPK phosphorylating numerous proteins driving the biological effects of the pathway. Activation of the MAPK pathway is initiated by the activation of Ras. One of the main targets of Ras is the serine/threonine kinase Raf, which is a MAPKKK. Activated Ras recruits Raf to the plasma membrane where it is phosphorylated and activated, with activated Raf in turn phosphorylating MEK1 and MEK2, which subsequently phosphorylate MAPKs ERK1 and ERK2 in the MAPK pathway.27,44 Activated ERK1/2 translocates to the nucleus to activate a variety of transcription factors including ELK-1, c-Fos, c-Jun, c-Myc, and STAT-3, which are involved in regulating cell proliferation and differentiation.30,44,48
HBx has been shown to up-regulate the MAPK signaling cascade. HBx activation of the Ras/Raf/MAPK pathway also accelerates the entry of cells into the S phase, and is causally associated with transformation.27,44 Also, HBx activates both the p38 MAPK and JNK pathways in the HBx-mediated apoptosis that occurs in response to weak apoptotic signals.48 HBx increases the levels of epidermal growth factor receptor by transactivating its promoter, and subsequently activates the Ras/Raf/MAPK signaling cascade.44,48 Furthermore, HBx activates AP-1 and NF-kB transcription factors via endogenous PKC.44
2) JAK/STAT signaling pathway
The JAK/STAT pathway is activated by cytokines and growth factors, and is involved in multiple cell functions including differentiation, proliferation, and apoptosis.44,48 In this pathway, the cytokines induce phosphorylation of JAKs (JAK1, JAK2, JAK3, and Tyk2), followed by activation of STATs.44,48 Nuclear localization of STATs also results in the activation of STAT target genes. HBx can also activate the JAK/STAT pathway, leading to activation of STAT-regulated genes.44,48 Because of its association with mitochondria and oxidative stress, HBx constitutively induces the transcription factors STAT-3 and NF-kB.44,48 These data suggest that activation of the JAK/STAT pathways is essential for HCC development.
3) PI3K/AKT signaling pathway
Growth factors activate the PI3K/AKT signaling pathway via the corresponding receptor tyrosine kinases. After receptor dimerization, PI3K is recruited to the plasma membrane where its catalytic subunit generates lipid second messengers (phosphoinositide phosphates PIP2 and PIP3) at the inner surface of the plasma membrane. Phosphoinositide-dependent protein kinase-1 then acts in concert with PIP2 and PIP3 to phosphorylate and activate AKT.48,49 AKT regulates the activity of several transcription factors, including CREB, members of the Fork Head family, and Ets-2. AKT promotes cell-cycle progression, cell survival, and tumor cell invasion.48,49 HBx down-regulates transforming growth factor (TGF)-β-induced apoptosis in hepatocytes by stimulating PI3K activity.49 This activated PI3K signaling pathway is attributed to an antiapoptotic mechanism.49-52 Taken together, these data indicate that the HBV inhibits apoptotic death via an HBx/PI3K/AKT pathway.
4) WNT/β-catenin signaling pathway
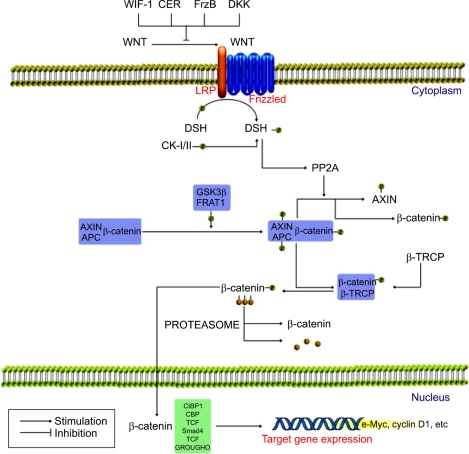
WNT binds to its receptor, Frizzled, which ultimately leads to inactivation of glycogen synthase kinase 3β (GSK3β) via activation of the Disheveled (DSH)(Fig. 4). This leads to hypophosphorylation of β-catenin and its release from a complex with adenomatosis polyposis coli (APC) and AXIN, with ensuing nuclear translocation of β-catenin and the control of transcription by binding to various target genes.53-56 Recent studies showed that HBx could activate WNT/β-catenin signaling by stabilizing cytoplasmic β-catenin in HCC.53,54 An alternative mechanism for WNT activation in HCC is that HBx represses E-cadherin expression at the transcriptional level via hypermethylation of the E-cadherin promoter by activating DNA methyltransferase 1.57 Mutations of AXIN1, another factor in the WNT/β-catenin signaling pathway, have been found in a substantial proportion of HCCs, with β-catenin accumulation in the absence of mutation of the β-catenin gene.58 Furthermore, up-regulation of Frizzled-7 receptors in association with activation of the WNT/β-catenin pathway is common in HCC.56 After all, HBx is associated with decreased expression of E-cadherin, accumulation of β-catenin in the cytoplasm and nucleus, and increased cell migration, which may make important contributions to hepatocarcinogenesis.
Fig. 4.
WNT/β-catenin signaling pathway. WNT signaling leads to a stabilization of β-catenin, the chief downstream effector of the WNT pathway. In the activated state WNT binds to its receptor, Frizzled, which ultimately leads to inactivation of GSK3β (glycogen synthase kinase 3β) via activation of the DSH (Disheveled). This leads to hypophosphorylation of β-catenin and its release from a complex with APC (adenomatosis polyposis coli) and AXIN, with ensuing nuclear translocation of β-catenin where it binds to the various target genes.
3. Regulation of apoptosis
Apoptosis (programmed cell death) is a cell-suicide mechanism that enables organisms to eliminate unneeded or aging cells. Defects in apoptotic cell death contribute to neoplastic diseases by preventing or delaying normal cell turnover, thereby promoting cell accumulation. Defects in apoptosis also facilitate tumor progression by rendering cancer cells resistant to death mechanisms relevant to metastasis, hypoxia, growth-factor deprivation, chemotherapy, and irradiation.
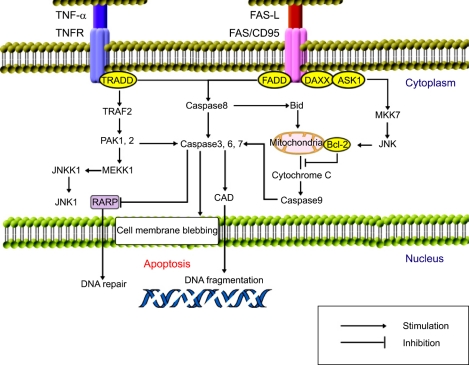
HBx is involved in apoptosis, including with p53, Rb, Fas-associated death domain protein (FADD), TRADD (TNF-receptor-associated death domain protein), and NF-kB in HBV-associated HCC.29,59 Hepatocyte apoptosis can be induced via the death-receptor-dependent pathway (extrinsic pathway) or the mitochondria-dependent pathway (intrinsic pathway) (Fig. 5). In brief, with respect to the extrinsic pathway, it is thought that ligation of death receptors such as Fas and TNF receptor, and the TRAIL (TNF-related apoptosis-inducing ligand) leads to activation of apoptosis via the recruitment of factors such as FADD and procaspase-8 (an initiator caspase) via the formation of a death-inducing signaling complex (DISC) and activation of caspase-3. Several counter-regulatory proteins, such as FLIP (FADD-like interleukin-1-converting enzyme-inhibiting protein) and inhibitor of apoptosis protein (IAP), are also present at the level of the DISC and caspase-8 activation.27,30 In contrast, activation of the intrinsic pathway is regulated by the interaction of various antiapoptotic compounds (e.g., B-cell CLL/lymphoma 2 [Bcl-2], and Mcl-1 [myeloid cell leukemia sequence 1]) and/or proapoptotic compounds (e.g., Bcl-2/Bcl-XL-associated death promoter [Bad], Bcl-2-associated X protein [Bax], Bim [Bcl-2 interacting mediator of cell death], and BH3-interacting-domain-death agonist [Bid]) of the Bcl-2 family of proteins, which control the permeability/flux and function of mitochondrial ions. Dysfunction if this process culminates in the release of various proteins that contribute to apoptosome formation (cytochrome C, Smac/Diablo, apaf-1, and caspase-9) and the eventual activation of caspase-3/7 (an executioner caspase).59
Fig. 5.
Regulation of apoptosis. Apoptosis can be induced via the death-receptor-dependent pathway (extrinsic pathway) or the mitochondria-dependent pathway (intrinsic pathway). The extrinsic pathway is initiated in the liver by death ligands such as TNF, Fas ligand (FasL, CD95L), and TRAIL (TNF-related apoptosis-inducing ligand), following their binding to their relevant death receptors. In contrast, the intrinsic pathway is triggered by a variety of intracellular stressors such as DNA damage, growth-actor deprivation, and metabolic disturbances from the extracellular matrix and/or surrounding cells.
The survival or apoptosis of cells depends upon the balance of various extracellular and/or intracellular stimuli. HBx is located in mitochondria and causes loss of the mitochondrial membrane potential and subsequently induces mitochondria-dependent cell death.60 Hepatitis viruses also modulate other antiapoptotic signals such as STAT-3 and NF-kB via ER stress and the generation of reactive oxygen species,61 where the latter ultimately leads to the activation of STAT-3 and NF-kB.62 HBx may modulate calcium homeostasis by inhibiting cytosolic calcium-dependent Pyk2 (proline-rich tyrosine kinase-2), an Src kinase activator, or calcium signaling mediated by mitochondrial calcium channels.63 HBx also inhibits TGF-β- and FasL-induced apoptosis by activating the PI3K pathway.49,51 In addition, the activation of PI3K by HBx, the phosphorylation of the downstream factors AKT and Bad, and down-regulation of caspase-3 have been observed.50,52
Inhibition of apoptosis may result from the failure of p53 - in the presence of HBx - to up-regulate genes (e.g., p21WAF1, Bax, or Fas) that are involved in apoptotic pathways.64 A p53 mutation is present in 30-60% of patients with HCC.65 HBx can bind to the C-terminus of p53 forming a protein-protein complex, thereby inactivating several critical p53-dependent activities, including p53-mediated apoptosis.66 Additional studies have indicated that HBx sequesters p53 in the cytoplasm and prevents it from entering the nucleus.67,68 Moreover, HBV protects against apoptotic death via an HBx/PI3K/AKT/Bad pathway and by inactivating caspase-3 activity that is at least partially p53 independent in liver cells.50,69 The proapoptotic activity of HBx overcomes or bypasses the inhibitory effect of Bcl-2 against Fas cytotoxicity.70 HBx also up-regulates the expression of survivin, which is a member of the IAP family.71 The level of the proapoptotic protein Bid might be reduced by a mechanism associated with HBx.72 However, HBx promotes the apoptosis of hepatocytes by regulating the expressions of Fas/FasL, Bax/Bcl-2, Bcl-xL, and c-Myc genes in a dose-dependent manner.28,73-75 Accordingly, HBx can modulate both preapoptotic and antiapoptotic pathways, depending on the diverse clinical conditions. The imbalance of increased antiapoptosis and decreased proapoptosis seen in HCC is critical to the uncontrolled growth of tumor cells.
The expressions of HSP27, HSP70, and HSP90 are also commonly up-regulated in HBV-associated HCC.76 These heat-shock proteins are important in the development of cancer, including regulation of apoptosis and modulation of the immune response.77,78
4. Regulation of the cell cycle
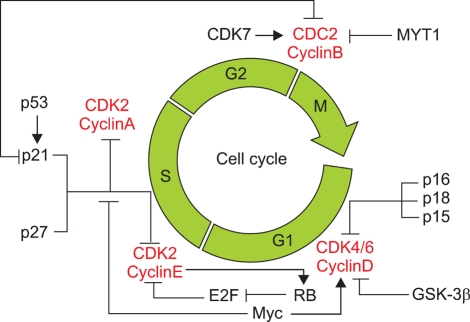
The cell-division cycle can be divided into two functional phases, S and M, and two preparatory phases, G1 and G2. The molecular mechanisms associated with these cell-cycle checkpoints involve the transient inactivation of a series of specific cyclin-dependent kinases (CDKs) and their respective regulatory cyclin subunits. Other regulators are the diverse members of the family of proteins known as CDK inhibitors (CDKIs), which can block the activation of CDKs. Two distinct classes of CDKIs have been described: (1) those that inhibit multiple CDKs, which includes p21CIP1, p27KIP1, and p57KIP2; and (2) those that specifically inhibit cyclin D, CDK4, or CDK6, which includes p16INK4, p15INK4B, p18INK4C, and p19INK4D.48 The cell cycle is regulated by the activities of cyclins, CDKs, and CDKIs, with one or more of these checkpoint controls being altered in most (if not all) human cancers at some stage in their progression to invasive cancer.
Several studies have shown that various types of alterations to cell-cycle regulators are found in HCC. CDKs, cyclins, and CDKIs generally function within several defined pathways, including the p21WAF1/p27KIP1/cyclin E/CDK2, p16INK4A/cyclin D1/CDK4/CDK6/Rb/E2F, and p14ARF/MDM2/p53 pathways (Fig. 6).48,79 HBx increases the rate and level of activation of CDK2, where the rate is associated with cyclins E and A, and the level with cyclin B.80 HBx also causes cyclin D1 overexpression,81 the Rb gene is a key player in the G1/S checkpoint system, and HBxAg transactivates the Rb promoter.48 Increased levels of phosphorylated Rb result in the release of E2F from Rb and cell-cycle progression.48,64 As tumors develop and progress, the Rb gene is inactivated by point mutations and LOH,82,83 resulting in elevated E2F activity. Furthermore, HBx represses the transcription of p21WAF1.84 Interestingly, the reduction or loss by methylation of p16INK4A, p14ARF, p15INK4B, and p27KIP1 promoters have been detected in HCC.85-89 Ultimately, HBx has been shown to stimulate cell-cycle progression by accelerating the transit through the G1/S and G2/M checkpoints.
Fig. 6.
Regulation of the cell cycle. The cell-division cycle can be divided into two functional phases, S and M, and two preparatory phases, G1 and G2. The molecular mechanisms associated with these cell-cycle checkpoints involve the transient inactivation of a series of specific cyclin-dependent kinases (CDKs) and their respective regulatory cyclin subunits. Other regulators are the diverse members of the family of proteins known as CDK inhibitors (CDKIs), which can block activation of CDKs.
The mitotic checkpoint genes monitor the proper assembly of the mitotic spindle, and block the onset of anaphase unless all of the chromosomes are stably attached to microtubules. Components of mitotic checkpoint genes are members of the BUB family (BUB1, BUBR1, and BUB3) and the MAD family (MAD1 and MAD2). Aneuploidy partly results from mutations in mitotic checkpoint genes, with decreased mitotic checkpoint protein leading to a mitotic checkpoint defect in HCC.90
5. Regulation of angiogenesis
Angiogenesis, the growth of new capillary blood vessels, is central to the growth of cancers. bFGF (basic fibroblast growth factor), TGFα, and vascular endothelial growth factor (VEGF), which are secreted by many tumors, all have angiogenic properties.64,91 Angiogenesis might be particularly important in hepatocarcinogenesis since HCCs are often highly vascular tumors.91 HBx is strongly implicated in the angiogenesis and metastasis that occurs during hepatocarcinogenesis. HBx induces angiogenesis via up-regulation of VEGF transcription, which is a potent angiogenic mechanism.92,93 More recent studies have provided evidence that HBx can induce the expression of VEGF via stabilization of hypoxia inducible factor-1α (HIF-1α) by enhancing its association with CBP (CREB-binding protein), thereby leading to angiogenesis during hepatocarcinogenesis.92,93 It has also been suggested that metastatic tumor antigen 1 (MTA1) plays a role in angiogenic processes as a stabilizer of HIF-1α.94,95 MTA1 induces the deacetylation of HIF-1α by increasing the expression level of histone deacetylase 1.96 MTA1 overexpression increases the transcriptional activity and stability of HIF-1α protein. Accordingly, MTA1 overexpression in HCC contributes to the extrahepatic metastasis as well as vascular invasion. A large recent study by our group has shown that MTA1 expression is closely associated with larger tumors, worse histologic differentiation, microvascular invasion, frequent postoperative recurrence, and poor patient survival.97 Therefore, the expression levels of MTA1 in HCC might be an important prognostic marker after curative surgery. Other factors appear to also be involved, such as angiopoietin-2.98
6. Regulation of telomere function
One difference between replicating and senescent cells is in the length of specialized tails at the end of the chromosomes, called telomeres. The telomere length is maintained by an enzyme complex called telomerase, which is a ribonucleoprotein complex that contains several proteins and RNA. The catalytic component of this complex is a reverse transcriptase (hTERT) that uses the RNA in the complex as a template for reverse transcription to replicate the DNA sequences in the telomere.48,71 Defects in the maintenance of telomeres and the impairment of telomerase activity are responsible for the development of many human cancers. Telomeres are shorter in tumors than in normal tissue, suggesting that telomeres are involved in HCC development.26 Activation of telomerase is considered to be a major mechanism underlying the development of HCC. There is recent evidence that telomere dysfunction leading to telomere-based chromosomal instability is present during the early stages of hepatocarcinogenesis, while telomerase activation occurs during HCC progression.48,99 hTERT is the rate-limiting determinant for regulating telomerase activity, and integrating the HBV genome into the hTERT promoter region and HBV enhancer can cis-activate transcription of the hTERT gene.100,101 In addition, telomeric repeat-binding factor 1 (TRF1), TRF2, and TRF1-interacting nuclear protein 2 (TIN2) are involved in telomere maintenance,100 and the expressions of TRF1, TRF2, TIN2 mRNA, and TRF1 protein gradually increase as hepatocarcinogenesis progresses.102
7. Inhibition of DNA-mismatch repair
DNA repair mechanisms are important for maintaining DNA integrity and preventing oncogenesis. In humans, more than 70 genes are involved in the five major DNA repair pathways: nucleotide excision repair (NER), base excision repair, mismatch repair, homologous recombinational repair, and nonhomologous end joining. Defective DNA-mismatch repair can lead to the accumulation of mutations and microsatellite instability in the cellular genome and thus increase the chance of malignant transformation. 65 Defects in DNA-mismatch repair genes also appear to play a role in HCC.103 DNA alterations resulting from exogenous genotoxic factors or normal replication processes are corrected by several repair pathways. HBx inhibits the repair of damaged hepatocyte DNA,28 which may be mediated by interaction with p53 or by binding to the damaged DNA-binding protein, which is a highly conserved protein implicated in DNA repair and cell-cycle regulation that plays an accessory role in NER.103,104 HBx also represses the transcription-repair factor TFIIH, thereby impairing TFIIH-related DNA repair mechanisms.105 Therefore, HBx acts as a cofactor in hepatocarcinogenesis by preventing the cell from efficiently repairing damaged DNA, thus leading to an accumulation of DNA mutations and, eventually, cancer.
8. Modulation of adhesion-deadhesion balance
Cells in tissues are attached to both one another and to the extracellular matrix (ECM). Disruption of these adhesions increases cell motility and the potential invasiveness of cells through the ECM.106 Transformed malignant cells produce a variety of lytic enzymes that degrade the ECM and allow cancer cells to invade tissues, lymphatic channels, and the vasculature. These proteases include plasminogen activator, cathepsins, adamalysin-related membrane proteases, and several matrix metalloproteases (MMPs).107 HBx may play a role in tumor spreading by modulating the adhesion-deadhesion balance of the cells in the primary tumor site and favoring integrin-mediated cell migration.107 HBx disrupts adherens junctions and decreases cellular adhesion to the ECM, resulting in modulation of cell adhesion and cell motility.108 HBx also enhances the migratory phenotype of hepatoma cells by up-regulating MMP-1 and MMP-9.107,109 Moreover, HBx represses several cell adhesion molecules and cytoskeleton proteins, including E-cadherin,57 integrin,108 fibronectin,110 CD47,111 and the hyaluronan receptor (CD44).106 Genes regulating the composition of the ECM and the cytoskeleton such as tubulin-α1, MMP-14, osteonectin/SPARC, and RhoA are also up-regulated in HCC.112 These genes play important roles in cell motility and invasion, which suggests that HBxAg promotes hepatocellular remodeling prior to tumor formation and in metastasis during tumor progression. HBx also regulates proteasomes, and thus controls the degradation of cellular and viral proteins.113
GENETIC AND EPIGENETIC ALTERATIONS
Numerous genetic and epigenetic alterations have been identified as being responsible for the activation of carcinogenic pathways in HCC.5 Most studies into the mechanisms of tumorigenesis have focused on the genetic changes, with various epigenetic changes being increasingly found in HCC recently.5 The term "epigenetics" covers all phenomena that control the functional state of DNA without changing the DNA sequence (i.e., without inducing genetic mutation).5 Epigenetic changes include methylation and acetylation of histone proteins and methylation of DNA.5,64 Several genes other than the above-mentioned p53, p21, Rb, AXIN1, and cyclin D1 are also involved in hepatocarcinogenesis,5,28 including the activation of insulin growth factor (IGF) via IGF-2 overexpression and M6P/IGF-2R-inactivated mutations.114 Other genetic and epigenetic changes include mutations of Smad2/4,28 HCCS1 (HCC suppressor 1),28 PTEN (phosphatase and tensin homolog deleted on chromosome 10),28,115 WWOX (WW domain containing oxidoreductase),116 and T cell factor 1 (TCF-1),28 and gene silencing by hypermethylation of GSTP128,117 and SOCS-1 (suppressor of cytokine signaling-1).28 Moreover, the expression of IGF-binding protein 3 (IGFBP-3) is down-regulated in HCC. IGFBP-3 mediates a wide variety of growth suppression signals in the IGF-receptor-dependent pathway, including TGF-β, retinoic acid, TNF-α, and p53.118,119 Transcriptional silencing by hypermethylation of the delete-in-liver-cancer-1 (DLC-1) gene, a putative tumor suppressor gene mapped to 8p21.3-22, is frequent in HCC, suggesting that inactivation of DLC-1 plays a role in hepatocarcinogenesis.28 DLC-2, one of the most frequently deleted chromosome arms at 13q, is significantly underexpressed in HCC.120 BDNF (brain-derived neutrophilic factor) is also involved in hepatocarcinogenesis,121 and the methylation of the genome is maintained by DNA methyltransferases (DNMTs).122 HBx expression increases the total DNMT activities by up-regulating DNMT1, DNMT3A1, and DNMT3A2, and selectively promoting the regional hypermethylation of specific tumor suppressor genes including RASSF1A, GSTP1, and CDKN2B.122
FIBROGENESIS AND CHRONIC HBV INFECTION
Liver injury is associated with the conversion of hepatic stellate cells (HSCs) to a myofibroblast-like phenotype. Activated HSCs are the major source of fibrillar collagens in cirrhotic injury. HSCs also have the capacity to remodel this matrix as they express MMPs and their specific inhibitors, TIMPs (tissue inhibitors of metalloproteinases). Chronic HSC activation induced by HBV replication could contribute to fibrogenesis and the increased proliferation of hepatocytes. This increased production of the ECM and hepatocyte turnover coupled with activation of the MAPK pathway may ultimately lead to HCC. The WNT/β-catenin pathways and MMT might be directly associated with hepatocarcinogenesis.123 Other factors that could contribute to disease and are known to be elevated during cirrhosis are TGF-β, gelatinases, fibroblast-activating proteins, and members of the interferon response pathway.123 Thus, several factors can lead to HCC - directly or indirectly and alone or in combination.
SINGLE-NUCLEOTIDE POLYMORPHISMS AND HEPATOCARCINOGENESIS
The DNA sequences of any two humans exhibit approximately 99.9% homology, yet substantial and often medically relevant phenotypic differences exist between such individuals. A significant proportion of these phenotypic differences are caused by this relatively small amount of genetic variation interacting with environmental factors. A clinically important element of phenotypic variation relates to susceptibility to disease and response to therapy.124-127 Variations in inherited DNA sequences between individuals can be due to the deletion or addition of bases, or to variable lengths of repeated sequences within or between genes. However, the most common type of DNA sequence variant is the single-nucleotide polymorphism (SNP), in which a single base in a sequence is replaced by a different nucleotide. SNPs on average appear approximately every 200 to 300 base pairs in the human genome, many of which cause functional changes by affecting transcription-factor binding sites, influencing splicing or stability of messenger RNA, or altering the amino acid sequence of the protein.128,129 The outcomes of HBV infection do not appear to be determined by viral strains. Instead, allelic variants in human genome are likely to affect the viral hepatitis progression after infection.129,130 Thus, it is conceivable that genetic differences significantly influence the progression of HBV infection. It has been reported that several genetic polymorphisms of TNF-α, IL-10, IL-6, TGF-β1, IGF-2, CTL antigen (CTLA)-4, and NFKB1A can influence the outcomes of chronic HBV infections.131,132 Screening of these polymorphisms might be clinically useful for identifying the risk of HCC, and hence in designing effective HCC surveillance programs for patients with chronic HBV infection.131 Furthermore, future studies of SNPs will not only provide insight into the pathogenesis of HCC, but may also provide a novel rationale for new methods of diagnosis and therapeutic strategies.
CONCLUSION
Hepatocarcinogenesis is a multistep process involving different genetic alterations that ultimately lead to malignant transformation of hepatocytes. Chronic HBV infection is a major risk factor for HCC, with the etiologic role of the HBV in hepatocarcinogenesis being well established from basic, epidemiological, and clinical research studies. The pathogenesis of HBV-associated HCC has been studied extensively, and the molecular changes that occur during malignant transformation have been identified. Several mechanisms are involved in HBV-related carcinogenesis, with chronic inflammation of the liver and increased hepatocyte proliferation being important contributing factors to the development of HCC. Multiple factors including damage caused by inflammatory cytokines, mutations incurred during liver regeneration, defects in DNA repair, integration of viral DNA into the host cell genome, host genomic instability, activation of cellular oncogenes, and induction of signaling pathways have been implicated as causes of HCC. New genomic technologies and approaches are resulting in the rapid accumulation of useful genetics data. New technologies such as gene expression profiling and proteomics may elucidate molecular tumor markers and identify molecules involved in hepatocarcinogenesis. However, our understanding of the molecular mechanisms of hepatocarcinogenesis is still only rudimentary. Considerable efforts are currently focused on unraveling the molecular pathogenesis of HCC in order to design better treatments, or even to prevent the disease altogether.
ACKNOWLEDGEMENTS
We thank M.S. Eui-Kyu Noh for helpful discussions on this manuscript. This work was supported by the Ulsan University Hospital Biomedical Research Center Promotion Fund (grant no. UUH-2006-13).
References
- 1.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Lok AS. Prevention of hepatitis B virus-related hepatocellular carcinoma. Gastroenterology. 2004;127:S303–S309. doi: 10.1053/j.gastro.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 3.Annual Report on the Case of Death Statistics (Based on Vital Registration) Korean National Statistical office Web site. Available at: http://www.nso.go.kr.
- 4.Cha C, Dematteo RP. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol. 2005;19:25–37. doi: 10.1016/j.bpg.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. J Clin Oncol. 2005;23:2892–2899. doi: 10.1200/JCO.2005.03.196. [DOI] [PubMed] [Google Scholar]
- 6.Lee DH, Kim JH, Nam JJ, Kim HR, Shin HR. Epidemiological findings of hepatitis B infection based on 1998 National Health and Nutrition Survey in Korea. J Korean Med Sci. 2002;17:457–462. doi: 10.3346/jkms.2002.17.4.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganem D, Prince AM. Hepatitis B virus infection-natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 8.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Park NH, Chung Y-H, Youn KH, et al. Close correlation of p53 mutation to microvascular invasion in hepatocellular carcinoma. J Clin Gastroenterol. 2001;33:397–401. doi: 10.1097/00004836-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Singh M, Kumar V. Transgenic mouse models of hepatitis B virus-associated hepatocellular carcinoma. Rev Med Virol. 2003;13:243–253. doi: 10.1002/rmv.392. [DOI] [PubMed] [Google Scholar]
- 11.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 12.Yang HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:168–174. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]
- 13.Brechot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56–S61. doi: 10.1053/j.gastro.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H, Iwasaki Y, Nouso K, et al. Possible contribution of prior hepatitis B virus infection to the development of hepatocellular carcinoma. J Gastroenterol Hepatol. 2005;20:850–856. doi: 10.1111/j.1440-1746.2005.03823.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee MS, Kim DH, Kim H, et al. Hepatitis B vaccination and reduced risk of primary liver cancer among male adults: a cohort study in Korea. Int J Epidemiol. 1998;27:316–319. doi: 10.1093/ije/27.2.316. [DOI] [PubMed] [Google Scholar]
- 16.Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678–686. doi: 10.1053/j.gastro.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Chan HL, Hui AY, Wong ML, et al. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494–1498. doi: 10.1136/gut.2003.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae SH, Yoon SK, Jang JW, et al. Hepatitis B virus genotype C prevails among chronic carriers of the virus in Korea. J Korean Med Sci. 2005;20:816–820. doi: 10.3346/jkms.2005.20.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeger C, Mason WS. Hepatitis B Virus Biology. Microbiology and Molecular Biology Reviews. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Lau SH, Sham JS, Wu MC, Wang T, Guan XY. Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene. 2004;23:142–148. doi: 10.1038/sj.onc.1206889. [DOI] [PubMed] [Google Scholar]
- 21.Brechot C, Gozuacik D, Murakami Y, Paterlini-Brechot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) Semin Cancer Biol. 2000;10:211–231. doi: 10.1006/scbi.2000.0321. [DOI] [PubMed] [Google Scholar]
- 22.Pollicino T, Squadrito G, Cerenzia G, et al. Hepatitis B virus maintains its pro-oncogenic properties in the case of occult HBV infection. Gastroenterology. 2004;126:102–110. doi: 10.1053/j.gastro.2003.10.048. [DOI] [PubMed] [Google Scholar]
- 23.Robinson WS. Molecular events in the pathogenesis of hepadnavirus-associated hepatocellular carcinoma. Annu Rev Med. 1994;45:297–323. doi: 10.1146/annurev.med.45.1.297. [DOI] [PubMed] [Google Scholar]
- 24.Murakami Y, Minami M, Daimon Y, Okanoue T. Hepatitis B virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis B virus surface antigen. J Med Virol. 2004;72:203–214. doi: 10.1002/jmv.10547. [DOI] [PubMed] [Google Scholar]
- 25.Zhang SH, Cong WM, Xian ZH, Wu MC. Clinicopathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol. 2005;11:3034–3039. doi: 10.3748/wjg.v11.i20.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong CH, Chan SK, Chan HL, Tsui SK, Feitelson M. The molecular diagnosis of hepatitis B virus-associated hepatocellular carcinoma. Crit Rev Clin Lab Sci. 2006;43:69–101. doi: 10.1080/10408360500410407. [DOI] [PubMed] [Google Scholar]
- 27.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 28.Pang R, Tse E, Poon RT. Molecular pathways in hepatocellular carcinoma. Cancer Lett. 2006;240:157–169. doi: 10.1016/j.canlet.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 29.Peng Z, Zhang Y, Gu W, et al. Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol. 2005;26:467–473. [PubMed] [Google Scholar]
- 30.Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim C-M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 32.Yu DY, Moon HB, Son JK, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 33.Lee TH, Finegold MJ, Shen RF, DeMayo JL, Woo SL, Butel JS. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64:5939–5947. doi: 10.1128/jvi.64.12.5939-5947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perfumo S, Amicone L, Colloca S, Giorgio M, Pozzi L, Tripodi M. Recognition efficiency of the hepatitis B virus polyadenylation signals is tissue specific in transgenic mice. J Virol. 1992;66:6819–6823. doi: 10.1128/jvi.66.11.6819-6823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami Y, Saigo K, Takashima H, et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162–1168. doi: 10.1136/gut.2004.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu H, Bonura C, Giannini C, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803–7810. [PubMed] [Google Scholar]
- 37.Terradillos O, Billet O, Renard CA, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 38.Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683–688. doi: 10.1111/j.1349-7006.2006.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagano JS, Blaser M, Buendia MA, et al. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol. 2004;14:453–471. doi: 10.1016/j.semcancer.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387–7392. doi: 10.1128/jvi.71.10.7387-7392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tai PC, Suk FM, Gerlich WH, Neurath AR, Shih C. Hypermodification and immune escape of an internally deleted middle-envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology. 2002;292:44–58. doi: 10.1006/viro.2001.1239. [DOI] [PubMed] [Google Scholar]
- 42.Chisari FV, Klopchin K, Moriyama T, et al. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- 43.Soussan P, Tuveri R, Nalpas B, et al. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J Hepatol. 2003;38:343–348. doi: 10.1016/s0168-8278(02)00422-1. [DOI] [PubMed] [Google Scholar]
- 44.Panteva M, Korkaya H, Jameel S. Hepatitis viruses and the MAPK pathway: is this a survival strategy? Virus Res. 2003;92:131–140. doi: 10.1016/s0168-1702(02)00356-8. [DOI] [PubMed] [Google Scholar]
- 45.Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol. 2007;13:74–81. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng AS, Chan HL, Leung WK, et al. Expression of HBx and COX-2 in chronic hepatitis B, cirrhosis and hepatocellular carcinoma: implication of HBx in upregulation of COX-2. Mod Pathol. 2004;17:1169–1179. doi: 10.1038/modpathol.3800196. [DOI] [PubMed] [Google Scholar]
- 47.Chan CF, Yau TO, Jin DY, Wong CM, Fan ST, Ng IOL. Evaluation of nuclear factor-kappa B, urokinase-type plasminogen activator, and HBx and their clinicopathological significance in hepatocellular carcinoma. Clin Cancer Res. 2004;10:4140–4149. doi: 10.1158/1078-0432.CCR-03-0574. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Zhang H, Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J Lab Clin Med. 2006;147:58–66. doi: 10.1016/j.lab.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein inhibits transforming growth factor-beta-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem. 2000;275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 50.Lee YI, Kang-Park S, Do S-I, Lee YI. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling sascade. J Biol Chem. 2001;276:16969–16977. doi: 10.1074/jbc.M011263200. [DOI] [PubMed] [Google Scholar]
- 51.Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Brechot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848–4859. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 52.Shin EC, Shin JS, Park JH, Kim H, Kim SJ. Expression of fas ligand in human hepatoma cell lines: role of hepatitis-B virus X (HBX) in induction of Fas ligand. Int J Cancer. 1999;82:587–591. doi: 10.1002/(sici)1097-0215(19990812)82:4<587::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Cha MY, Kim CM, Park YM, Ryu WS. Hepatitis B virus X protein is essential for the activation of Wnt/beta-catenin signaling in hepatoma cells. Hepatology. 2004;39:1683–1693. doi: 10.1002/hep.20245. [DOI] [PubMed] [Google Scholar]
- 54.Ding Q, Xia W, Liu JC, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 56.Merle P, Kim M, Herrmann M, et al. Oncogenic role of the frizzled-7/beta-catenin pathway in hepatocellular carcinoma. J Hepatol. 2005;43:854–862. doi: 10.1016/j.jhep.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 57.Lee JO, Kwun HJ, Jung JK, Choi KH, Min DS, Jang KL. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene. 2005;24:6617–6625. doi: 10.1038/sj.onc.1208827. [DOI] [PubMed] [Google Scholar]
- 58.Satoh S, Daigo Y, Furukawa Y, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 59.Yin XM, Ding WX. Death receptor activation-induced hepatocyte apoptosis and liver injury. Curr Mol Med. 2003;3:491–508. doi: 10.2174/1566524033479555. [DOI] [PubMed] [Google Scholar]
- 60.Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem. 2003;278:22071–22078. doi: 10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- 61.Lee YI, Hwang JM, Im JH, et al. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem. 2004;279:15460–15471. doi: 10.1074/jbc.M309280200. [DOI] [PubMed] [Google Scholar]
- 62.Waris G, Huh KW, Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol Cell Biol. 2001;21:7721–7730. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 64.Feitelson MA. Parallel epigenetic and genetic changes in the pathogenesis of hepatitis virus-associated hepatocellular carcinoma. Cancer Lett. 2006;239:10–20. doi: 10.1016/j.canlet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 65.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 66.Lin Y, Nomura T, Yamashita T, Dorjsuren D, Tang H, Murakami S. The transactivation and p53-interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res. 1997;57:5137–5142. [PubMed] [Google Scholar]
- 67.Wang XW, Gibson MK, Vermeulen W, et al. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 68.Shintani Y, Yotsuyanagi H, Moriya K, et al. Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J Gen Virol. 1999;80:3257–3265. doi: 10.1099/0022-1317-80-12-3257. [DOI] [PubMed] [Google Scholar]
- 69.Kim YC, Song KS, Yoon G, Nam MJ, Ryu WS. Activated ras oncogene collaborates with HBx gene of hepatitis B virus to transform cells by suppressing HBx-mediated apoptosis. Oncogene. 2001;20:16–23. doi: 10.1038/sj.onc.1203840. [DOI] [PubMed] [Google Scholar]
- 70.Terradillos O, de La Coste A, Pollicino T, et al. The hepatitis B virus X protein abrogates Bcl-2-mediated protection against Fas apoptosis in the liver. Oncogene. 2002;21:377–386. doi: 10.1038/sj.onc.1205110. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Dong N, Yin L, et al. Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. J Med Virol. 2005;77:374–381. doi: 10.1002/jmv.20466. [DOI] [PubMed] [Google Scholar]
- 72.Chen GG, Lai PBS, Chan PKS, et al. Decreased expression of Bid in human hepatocellular carcinoma is related to hepatitis B virus X protein. Eur J Cancer. 2001;37:1695–1702. doi: 10.1016/s0959-8049(01)00182-4. [DOI] [PubMed] [Google Scholar]
- 73.Zhang SJ, Chen HY, Chen ZX, Wang XZ. Possible mechanism for hepatitis B virus X gene to induce apoptosis of hepatocytes. World J Gastroenterol. 2005;11:4351–4356. doi: 10.3748/wjg.v11.i28.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim WH, Hong F, Jaruga B, et al. Hepatitis B virus X protein sensitizes primary mouse hepatocytes to ethanol- and TNF-alpha-induced apoptosis by a caspase-3-dependent mechanism. Cell Mol Immunol. 2005;2:40–48. [PubMed] [Google Scholar]
- 75.Miao J, Chen GG, Chun S-Y, Lai PPS. Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl-xL expression. Cancer Lett. 2006;236:115–124. doi: 10.1016/j.canlet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 76.Lim SO, Park SG, Yoo JH, et al. Expression of heat shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in hepatitis B virus-related hepatocellular carcinomas and dysplastic nodules. World J Gastroenterol. 2005;11:2072–2079. doi: 10.3748/wjg.v11.i14.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mese H, Sasaki A, Nakayama S, et al. Prognostic significance of heat shock protein 27 (HSP27) in patients with oral squamous cell carcinoma. Oncol Rep. 2002;9:341–344. [PubMed] [Google Scholar]
- 78.Concannon CG, Orrenius S, Samali A. Hsp27 inhibits cytochrome c-mediated caspase activation by sequestering both pro-caspase-3 and cytochrome C. Gene Expr. 2001;9:195–201. doi: 10.3727/000000001783992605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Semczuk A, Jakowicki JA. Alterations of pRb1-cyclin D1-cdk4/6-p16 (INK4A) pathway in endometrial carcinogenesis. Cancer Lett. 2004;203:1–12. doi: 10.1016/j.canlet.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 80.Bouchard M, Giannakopoulos S, Wang EH, Tanese N, Schneider RJ. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol. 2001;75:4247–4257. doi: 10.1128/JVI.75.9.4247-4257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein A, Guhl E, Tzeng YJ, et al. HBX causes cyclin D1 overexpression and development of breast cancer in transgenic animals that are heterozygous for p53. Oncogene. 2003;22:2910–2919. doi: 10.1038/sj.onc.1206539. [DOI] [PubMed] [Google Scholar]
- 82.Niketeghad F, Decker HJ, Caselmann WH, et al. Frequent genomic imbalances suggest commonly altered tumour genes in human hepatocarcinogenesis. Br J Cancer. 2001;85:697–704. doi: 10.1054/bjoc.2001.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edamoto Y, Hara A, Biernat W, et al. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer. 2003;106:334–341. doi: 10.1002/ijc.11254. [DOI] [PubMed] [Google Scholar]
- 84.Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Research. 2004;32:2202–2213. doi: 10.1093/nar/gkh553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X, Hui AM, Sun L, et al. p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin Cancer Res. 2004;10:7484–7489. doi: 10.1158/1078-0432.CCR-04-1715. [DOI] [PubMed] [Google Scholar]
- 86.Pascale RM, Simile MM, Calvisi DF, et al. Role of HSP90, CDC37, and CRM1 as modulators of P16 (INK4A) activity in rat liver carcinogenesis and human liver cancer. Hepatology. 2005;42:1310–1319. doi: 10.1002/hep.20962. [DOI] [PubMed] [Google Scholar]
- 87.Lei PP, Zhang ZJ, Shen LJ, Li JY, Zou Q, Zhang HX. Expression and hypermethylation of p27 kip1 in hepatocarcinogenesis. World J Gastroenterol. 2005;11:4587–4591. doi: 10.3748/wjg.v11.i29.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukai K, Yokosuka O, Imazeki F, et al. Methylation status of p14ARF, p15INK4b, and p16INK4a genes in human hepatocellular carcinoma. Liver Int. 2005;25:1209–1216. doi: 10.1111/j.1478-3231.2005.01162.x. [DOI] [PubMed] [Google Scholar]
- 89.Qin Y, Liu JY, Li B, Sun ZL, Sun ZF. Association of low p16INK4a and p15INK4b mRNAs expression with their CpG islands methylation with human hepatocellular carcinogenesis. World J Gastroenterol. 2004;10:1276–1280. doi: 10.3748/wjg.v10.i9.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 91.Lee S-W, Lee YM, Bae S-K, Murakami S, Yun Y, Kim K-W. Human hepatitis B virus X protein is a possible mediator of hypoxia-induced angiogenesis in hepatocarcinogenesis. Biochem Biophys Res Commun. 2000;268:456–461. doi: 10.1006/bbrc.2000.2093. [DOI] [PubMed] [Google Scholar]
- 92.Yoo Y-G, Cho S, Park S, Lee M-O. The carboxy-terminus of the hepatitis B virus X protein is necessary and sufficient for the activation of hypoxia-inducible factor-1[alpha] FEBS Letters. 2004;577:121–126. doi: 10.1016/j.febslet.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 93.Moon EJ, Jeong CH, Jeong JW, et al. Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia-inducible factor-1&alpha. FASEB J. 2004;18:382–384. doi: 10.1096/fj.03-0153fje. [DOI] [PubMed] [Google Scholar]
- 94.Moon HE, Cheon H, Chun KH, et al. Metastasis-associated protein 1 enhances angiogenesis by stabilization of HIF-1alpha. Oncol Rep. 2006;16:929–935. [PubMed] [Google Scholar]
- 95.Kim SH, Jeong JW, Park JA, et al. Regulation of the HIF-1alpha stability by histone deacetylases. Oncol Rep. 2007;17:647–651. [PubMed] [Google Scholar]
- 96.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J. 2006;25:1231–1241. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rhu SH, Chung YH, Lee HS, et al. Metastatic tumor antigen 1 is closely associated with frequent post-operative recurrence and poor survival in patients with hepatocellular carcinoma. Hepatology. 2007 doi: 10.1002/hep.22124. in press. [DOI] [PubMed] [Google Scholar]
- 98.Mitsuhashi N, Shimizu H, Ohtsuka M, et al. Angiopoietins and Tie-2 expression in angiogenesis and proliferation of human hepatocellular carcinoma. Hepatology. 2003;37:1105–1113. doi: 10.1053/jhep.2003.50204. [DOI] [PubMed] [Google Scholar]
- 99.Farazi PA, Glickman J, Jiang S, Yu A, Rudolph KL, DePinho RA. Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res. 2003;63:5021–5027. [PubMed] [Google Scholar]
- 100.Zhang X, Dong N, Zhang H, You J, Wang H, Ye L. Effects of hepatitis B virus X protein on human telomerase reverse transcriptase expression and activity in hepatoma cells. J Lab Clin Med. 2005;145:98–104. doi: 10.1016/j.lab.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 101.Ferber MJ, Montoya DP, Yu C, et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene. 2003;22:3813–3820. doi: 10.1038/sj.onc.1206528. [DOI] [PubMed] [Google Scholar]
- 102.Oh BK, Kim YJ, Park C, Park YN. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am J Pathol. 2005;166:73–80. doi: 10.1016/S0002-9440(10)62233-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mathonnet G, Lachance S, Alaoui-Jamali M, Drobetsky EA. Expression of hepatitis B virus X oncoprotein inhibits transcription-coupled nucleotide excision repair in human cells. Mutat Res. 2004;554:305–318. doi: 10.1016/j.mrfmmm.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 104.Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jaitovich-Groisman I, Benlimame N, Slagle BL, et al. Transcriptional regulation of the TFIIH transcription repair components XPB and XPD by the hepatitis B virus x protein in liver cells and transgenic liver tissue. J Biol Chem. 2001;276:14124–14132. doi: 10.1074/jbc.M010852200. [DOI] [PubMed] [Google Scholar]
- 106.Lara-Pezzi E, Serrador JM, Montoya MC, et al. The hepatitis B virus X protein (HBx) induces a migratory phenotype in a CD44-dependent manner: possible role of HBx in invasion and metastasis. Hepatology. 2001;33:1270–1281. doi: 10.1053/jhep.2001.1270. [DOI] [PubMed] [Google Scholar]
- 107.Lara-Pezzi E, Gomez-Gaviro MV, Galvez BG, et al. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J Clin Invest. 2002;110:1831–1838. doi: 10.1172/JCI200215887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lara-Pezzi E, Majano PL, Yanez-Mo M, et al. Effect of the hepatitis B virus HBx protein on integrin-mediated adhesion to and migration on extracellular matrix. J Hepatol. 2001;34:409–415. doi: 10.1016/s0168-8278(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 109.Chung TW, Lee YC, Kim CH. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERKs and PI-3K/AKT pathways: Involvement of invasive potential. FASEB J. 2004;18:1123–1125. doi: 10.1096/fj.03-1429fje. [DOI] [PubMed] [Google Scholar]
- 110.Matsuo M, Sakurai H, Ueno Y, Ohtani O, Saiki I. Activation of MEK/ERK and PI3K/Akt pathways by fibronectin requires integrin alphav-mediated ADAM activity in hepatocellular carcinoma: a novel functional target for gefitinib. Cancer Sci. 2006;97:155–162. doi: 10.1111/j.1349-7006.2006.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshida H, Tomiyama Y, Oritani K, et al. Interaction between Src homology 2 domain bearing protein tyrosine phosphatase substrate-1 and CD47 mediates the adhesion of human B lymphocytes to nonactivated endothelial cells. J Immunol. 2002;168:3213–3220. doi: 10.4049/jimmunol.168.7.3213. [DOI] [PubMed] [Google Scholar]
- 112.Delpuech O, Trabut JB, Carnot F, Feuillard J, Brechot C, Kremsdorf D. Identification, using cDNA macroarray analysis, of distinct gene expression profiles associated with pathological and virological features of hepatocellular carcinoma. Oncogene. 2002;21:2926–2937. doi: 10.1038/sj.onc.1205392. [DOI] [PubMed] [Google Scholar]
- 113.Cui F, Wang Y, Wang J, et al. The up-regulation of proteasome subunits and lysosomal proteases in hepatocellular carcinomas of the HBx gene knockin transgenic mice. Proteomics. 2006;6:498–504. doi: 10.1002/pmic.200500218. [DOI] [PubMed] [Google Scholar]
- 114.Fan ZR, Yang DH, Cui J, Qin HR, Huang CC. Expression of insulin like growth factor II and its receptor in hepatocellular carcinogenesis. World J Gastroenterol. 2001;7:285–288. doi: 10.3748/wjg.v7.i2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma DZ, Xu Z, Liang YL, et al. Down-regulation of PTEN expression due to loss of promoter activity in human hepatocellular carcinoma cell lines. World J Gastroenterol. 2005;11:4472–4477. doi: 10.3748/wjg.v11.i29.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Park SW, Ludes-Meyers J, Zimonjic DB, Durkin ME, Popescu NC, Aldaz CM. Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br J Cancer. 2004;91:753–759. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niu ZS, Wang M. Expression of c-erbB-2 and glutathione S-transferase-pi in hepatocellular carcinoma and its adjacent tissue. World J Gastroenterol. 2005;11:4404–4408. doi: 10.3748/wjg.v11.i28.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hanafusa T, Shinji T, Shiraha H, et al. Functional promoter upstream p53 regulatory sequence of IGFBP3 that is silenced by tumor specific methylation. BMC Cancer. 2005;5:9. doi: 10.1186/1471-2407-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yumoto E, Nakatsukasa H, Hanafusa T, et al. IGFBP-3 expression in hepatocellular carcinoma involves abnormalities in TGF-beta and/or Rb signaling pathways. Int J Oncol. 2005;27:1223–1230. [PubMed] [Google Scholar]
- 120.Ching YP, Wong CM, Chan SF, et al. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem. 2003;278:10824–10830. doi: 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- 121.Yang ZF, Ho DW, Lam CT, et al. Identification of brain-derived neurotrophic factor as a novel functional protein in hepatocellular carcinoma. Cancer Res. 2005;65:219–225. [PubMed] [Google Scholar]
- 122.Park IY, Sohn BH, Yu E, et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 123.Wu CG, Salvay DM, Forgues M, et al. Distinctive gene expression profiles associated with Hepatitis B virus x protein. Oncogene. 2001;20:3674–3682. doi: 10.1038/sj.onc.1204481. [DOI] [PubMed] [Google Scholar]
- 124.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 125.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 126.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33:S238–S244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 127.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 128.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 129.Juran BD, Lazaridis KN. Genomics and complex liver disease: Challenges and opportunities. Hepatology. 2006;44:1380–1390. doi: 10.1002/hep.21453. [DOI] [PubMed] [Google Scholar]
- 130.Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 131.Kim YJ, Lee HS. Single nucleotide polymorphisms associated with hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Intervirology. 2005;48:10–15. doi: 10.1159/000082089. [DOI] [PubMed] [Google Scholar]
- 132.Kim YJ, Yoon JH, Kim CY, et al. IGF2 polymorphisms are associated with hepatitis B virus clearance and hepatocellular carcinoma. Biochem Biophys Res Commun. 2006;346:38–44. doi: 10.1016/j.bbrc.2006.05.080. [DOI] [PubMed] [Google Scholar]