Abstract
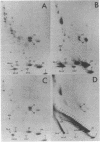
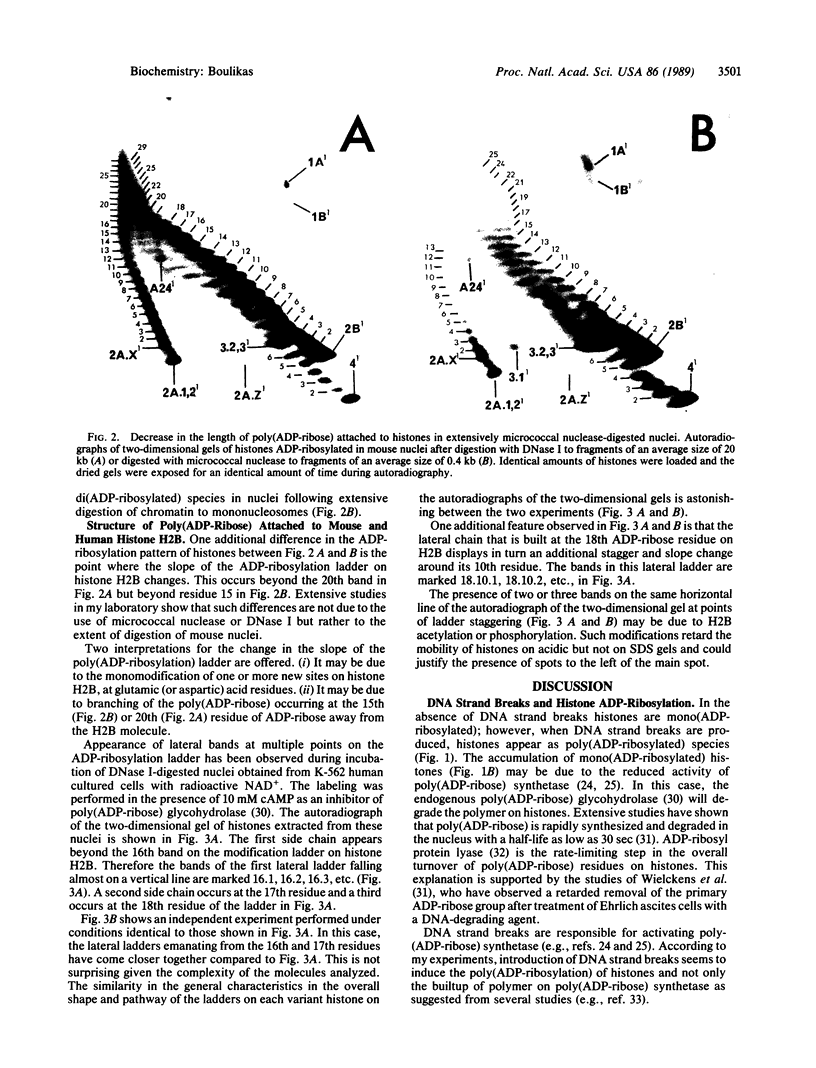
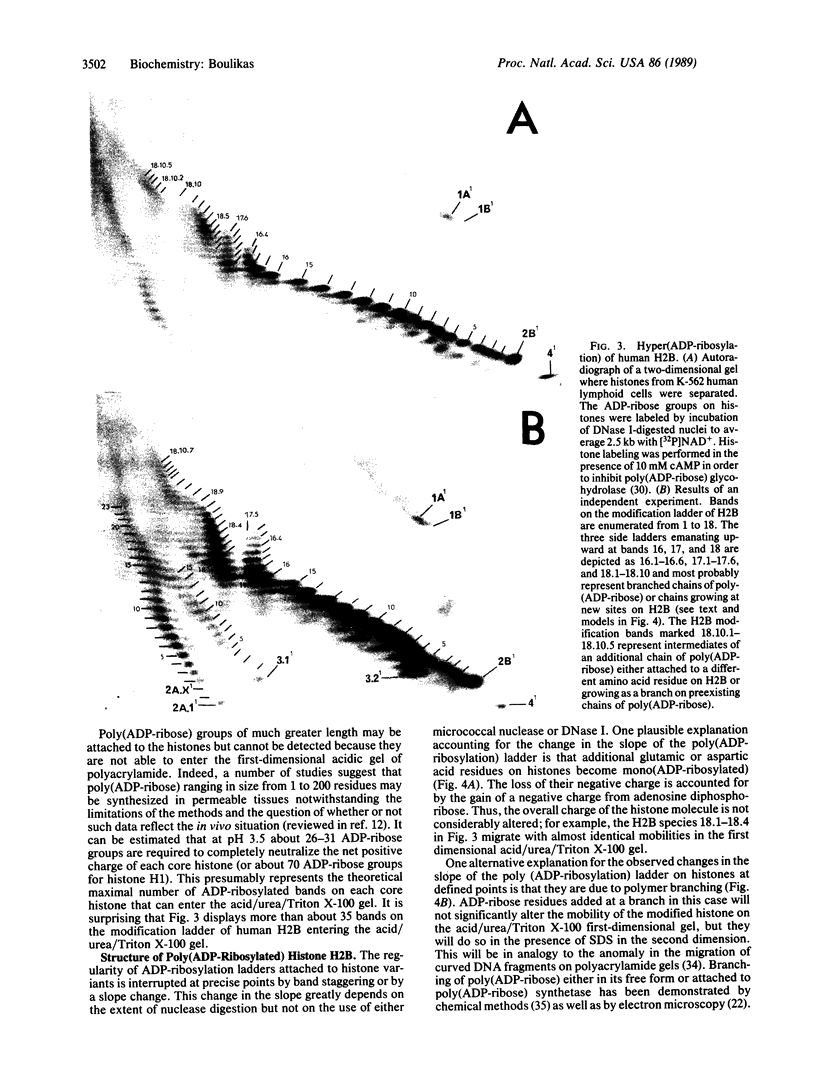
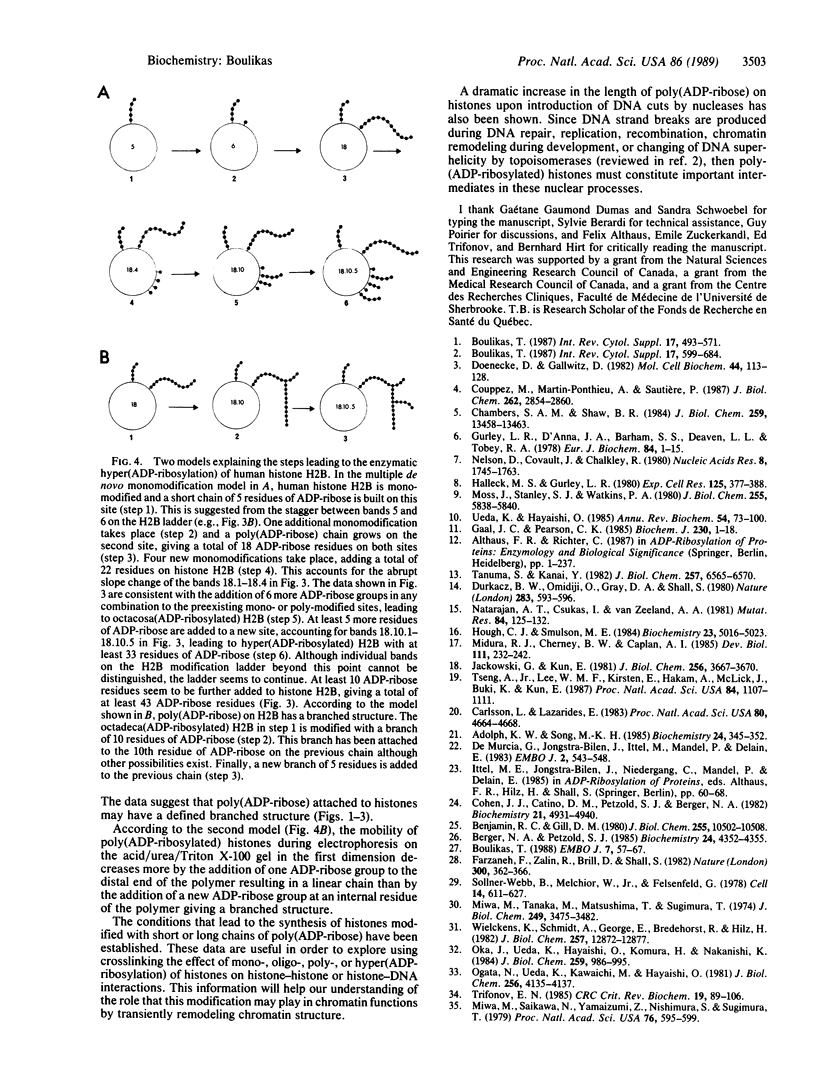
Histone ADP-ribosylation was studied using two-dimensional gel electrophoresis after cleavage of the nuclear DNA with nucleases. Modified histones carrying different numbers of ADP-ribose groups form a ladder of bands above each variant histone. Cellular lysates containing unfragmented DNA mainly synthesize mono(ADP-ribosylated) histones. Cleavage of the DNA with either DNase I or micrococcal nuclease to fragments of an average size of 10-20 kilobases (kb) dramatically induces the formation of poly(ADP-ribosylated) species of histones in nuclei. As the number of DNA strand breaks produced by either DNase I or micrococcal nuclease increases and a great number of DNA cuts is introduced (fragments of 0.4-0.2 kb), the size of the poly(ADP-ribose) chains on the histones decreases. Finally, in the presence of 10 mM cAMP as an inhibitor of poly(ADP-ribose) glycohydrolase, human lymphoid nuclei synthesize hyper(ADP-ribosylated) histone H2B with at least 40 ADP-ribose groups attached to it. Lateral ladders emanating at precise points of the linear ladder on hypermodified H2B can arise from branching of poly(ADP-ribose) or from multiple monomodifications of glutamic (or aspartic) acid residues. Branching or de novo monomodifications occur after a precise number of ADP-ribose groups have been added to a histone molecule. Poly(ADP-ribosylated) histones thus appear to be intermediates in nuclear processes involving DNA strand breaks.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolph K. W., Song M. K. Decrease in ADP-ribosylation of HeLa non-histone proteins from interphase to metaphase. Biochemistry. 1985 Jan 15;24(2):345–352. doi: 10.1021/bi00323a017. [DOI] [PubMed] [Google Scholar]
- Althaus F. R., Richter C. ADP-ribosylation of proteins. Enzymology and biological significance. Mol Biol Biochem Biophys. 1987;37:1–237. [PubMed] [Google Scholar]
- Benjamin R. C., Gill D. M. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem. 1980 Nov 10;255(21):10502–10508. [PubMed] [Google Scholar]
- Berger N. A., Petzold S. J. Identification of minimal size requirements of DNA for activation of poly(ADP-ribose) polymerase. Biochemistry. 1985 Jul 30;24(16):4352–4355. doi: 10.1021/bi00337a015. [DOI] [PubMed] [Google Scholar]
- Boulikas T. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 1988 Jan;7(1):57–67. doi: 10.1002/j.1460-2075.1988.tb02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L., Lazarides E. ADP-ribosylation of the Mr 83,000 stress-inducible and glucose-regulated protein in avian and mammalian cells: modulation by heat shock and glucose starvation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4664–4668. doi: 10.1073/pnas.80.15.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. A., Shaw B. R. Levels of histone H4 diacetylation decrease dramatically during sea urchin embryonic development and correlate with cell doubling rate. J Biol Chem. 1984 Nov 10;259(21):13458–13463. [PubMed] [Google Scholar]
- Cohen J. J., Catino D. M., Petzold S. J., Berger N. A. Activation of poly(adenosine diphosphate ribose) polymerase by SV 40 minichromosomes: effects of deoxyribonucleic acid damage and histone H1. Biochemistry. 1982 Sep 28;21(20):4931–4940. doi: 10.1021/bi00263a016. [DOI] [PubMed] [Google Scholar]
- Couppez M., Martin-Ponthieu A., Sautière P. Histone H4 from cuttlefish testis is sequentially acetylated. Comparison with acetylation of calf thymus histone H4. J Biol Chem. 1987 Feb 25;262(6):2854–2860. [PubMed] [Google Scholar]
- Doenecke D., Gallwitz D. Acetylation of histones in nucleosomes. Mol Cell Biochem. 1982 Apr 30;44(2):113–128. doi: 10.1007/BF00226895. [DOI] [PubMed] [Google Scholar]
- Durkacz B. W., Omidiji O., Gray D. A., Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980 Feb 7;283(5747):593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Farzaneh F., Zalin R., Brill D., Shall S. DNA strand breaks and ADP-ribosyl transferase activation during cell differentiation. Nature. 1982 Nov 25;300(5890):362–366. doi: 10.1038/300362a0. [DOI] [PubMed] [Google Scholar]
- Gaal J. C., Pearson C. K. Eukaryotic nuclear ADP-ribosylation reactions. Biochem J. 1985 Aug 15;230(1):1–18. doi: 10.1042/bj2300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978 Mar;84(1):1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- Halleck M. S., Gurley L. R. Histone H2A subfractions and their phosphorylation in cultured Peromyscus cells. Exp Cell Res. 1980 Feb;125(2):377–388. doi: 10.1016/0014-4827(80)90132-9. [DOI] [PubMed] [Google Scholar]
- Hough C. J., Smulson M. E. Association of poly(adenosine diphosphate ribosylated) nucleosomes with transcriptionally active and inactive regions of chromatin. Biochemistry. 1984 Oct 9;23(21):5016–5023. doi: 10.1021/bi00316a029. [DOI] [PubMed] [Google Scholar]
- Jackowski G., Kun E. Age-dependent variation of rates of polyadenosine-diphosphoribose synthesis by cardiocyte nuclei and the lack of correlation of enzymatic activity with macromolecular size distribution of DNA. J Biol Chem. 1981 Apr 25;256(8):3667–3670. [PubMed] [Google Scholar]
- Midura R. J., Cherney B. W., Caplan A. I. The relationship of nicotinamide adenine dinucleotide to the chondrogenic differentiation of limb mesenchymal cells. Dev Biol. 1985 Sep;111(1):232–242. doi: 10.1016/0012-1606(85)90448-8. [DOI] [PubMed] [Google Scholar]
- Miwa M., Saikawa N., Yamaizumi Z., Nishimura S., Sugimura T. Structure of poly(adenosine diphosphate ribose): identification of 2'-[1''-ribosyl-2''-(or 3''-)(1'''-ribosyl)]adenosine-5',5'',5'''-tris(phosphate) as a branch linkage. Proc Natl Acad Sci U S A. 1979 Feb;76(2):595–599. doi: 10.1073/pnas.76.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M., Tanaka M., Matsushima T., Sugimura T. Purification and properties of glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose). J Biol Chem. 1974 Jun 10;249(11):3475–3482. [PubMed] [Google Scholar]
- Moss J., Stanley S. J., Watkins P. A. Isolation and properties of an NAD- and guanidine-dependent ADP-ribosyltransferase from turkey erythrocytes. J Biol Chem. 1980 Jun 25;255(12):5838–5840. [PubMed] [Google Scholar]
- Natarajan A. T., Csukás I., van Zeeland A. A. Contribution of incorporated 5-bromodeoxyuridine in DNA to the frequencies of sister-chromatid exchanges induced by inhibitors of poly-(ADP-ribose)-polymerase. Mutat Res. 1981 Nov;84(1):125–132. doi: 10.1016/0027-5107(81)90056-7. [DOI] [PubMed] [Google Scholar]
- Nelson D., Covault J., Chalkley R. Segregation of rapidly acetylated histones into a chromatin fraction released from intact nuclei by the action of micrococcal nuclease. Nucleic Acids Res. 1980 Apr 25;8(8):1745–1763. doi: 10.1093/nar/8.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N., Ueda K., Kawaichi M., Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981 May 10;256(9):4135–4137. [PubMed] [Google Scholar]
- Oka J., Ueda K., Hayaishi O., Komura H., Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J Biol Chem. 1984 Jan 25;259(2):986–995. [PubMed] [Google Scholar]
- Sollner-Webb B., Melchior W., Jr, Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978 Jul;14(3):611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- Tanuma S., Kanai Y. Poly(ADP-ribosyl)ation of chromosomal proteins in the HeLa S3 cell cycle. J Biol Chem. 1982 Jun 10;257(11):6565–6570. [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Tseng A., Jr, Lee W. M., Jakobovits E. B., Kirsten E., Hakam A., McLick J., Buki K., Kun E. Prevention of tumorigenesis of oncogene-transformed rat fibroblasts with DNA site inhibitors of poly(ADP ribose) polymerase. Proc Natl Acad Sci U S A. 1987 Feb;84(4):1107–1111. doi: 10.1073/pnas.84.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- Wielckens K., Schmidt A., George E., Bredehorst R., Hilz H. DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins. J Biol Chem. 1982 Nov 10;257(21):12872–12877. [PubMed] [Google Scholar]
- de Murcia G., Jongstra-Bilen J., Ittel M. E., Mandel P., Delain E. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: electron microscopic visualization. EMBO J. 1983;2(4):543–548. doi: 10.1002/j.1460-2075.1983.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]