Abstract
A light-activated reaction analog has been developed to mimic the catalytic reaction cycle of Δ5-3-ketosteroid isomerase in order to probe the functionally relevant protein solvation response to the catalytic charge transfer. Δ5-3-ketosteroid isomerase from Pseudomonas putida catalyses a C-H bond cleavage and formation through an enolate intermediate. Conversion of the ketone substrate to the enolate intermediate is simulated by a photoacid bound to the active site oxyanion hole. In the ground state, the photoacid electrostatically resembles the enolate intermediate while the low pKa excited state resembles the ketone starting material. Time-resolved fluorescence experiments with photoacids coumarin 183 and equilenin show the active site of Δ5-3-ketosteroid isomerase to be largely unperturbed by the light-activated reaction. The small solvation response for the photoacid at the active site as compared with a simple solvent suggests the active site does not significantly change its electrostatic environment during the catalytic cycle. Instead, the reaction takes place in an electrostatically preorganized environment.
Introduction
The role of electrostatic interactions at the active site of enzymes has been widely discussed. 1–5 Both time-averaged and dynamic aspects of electrostatic interactions are potentially important; in this paper we specifically consider whether there is a solvation response at the active site of the enzyme Δ5-3-ketosteroid isomerase (KSI) following a sudden electrostatic perturbation that simulates the catalytic reaction coordinate.
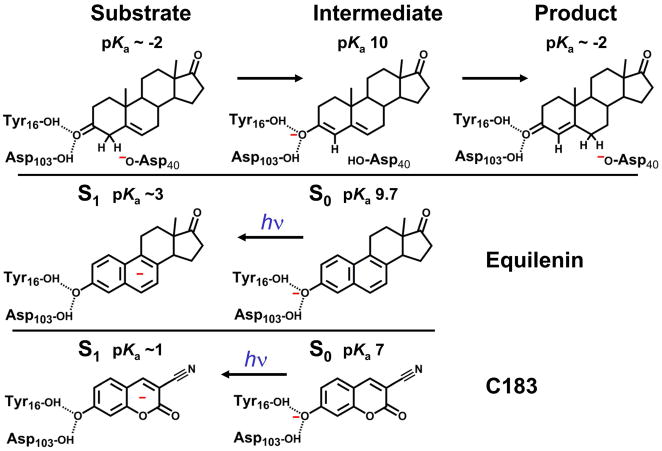
KSI from Pseudomonas putida catalyses a C-H bond cleavage and formation through an enolate intermediate, as shown in Figure 1.6 The general base Asp40 abstracts a proton from the 4 position of the steroid ring to form an enolate stabilized by the hydrogen bond donating residues Tyr16 and Asp103, which are positioned deep within the active site and form a so-called “oxyanion hole.” Protonated Asp40 then transfers its proton to the 6 position of the steroid ring to complete the reaction. While often described as a hydrophobic active site, the large number of polar and potentially charged amino acids in the oxyanion hole suggests that a more precise description is warranted. These amino acids (and others) collectively create a time-averaged electric field that is heterogeneous.7 In this paper we explore whether these polar amino acids can respond dynamically to the charge perturbation that occurs during the course of the reaction or whether the active site is electrostatically preorganized.
Figure 1.
The enzymatic reaction for ketosteroid isomerase (top) is compared to the equilenin (middle) and C183 (bottom) photocycle.
Time-resolved fluorescence experiments are often used to probe the changing solvent environment around chromophore excited-states. Solvation dynamics of excited-state fluorophores has been extensively studied in simple solvents8–10 and to a more limited extent in proteins11–17. In a simple, polar solvent time-resolved fluorescence spectra show a red shift in time as the excited-state dipole becomes better solvated, often called a dynamic Stokes shift. The magnitude of the shift reflects the solvation capacity of the medium, while the duration at earliest times (sub-ps) involves inertial motions of the solvent, and on a somewhat longer timescale (ps) is best described by the dielectric relaxation time.8–10 By contrast, solvation dynamics experiments conducted in proteins exhibit a wide range of solvation capacities and relaxation timescales from the femtosecond to nanosecond regime, where the details depend on the location of probe.13 This heterogeneity in different protein environments suggests that the dielectric relaxation properties of an enzymatic active site may be subject to evolutionary pressure to influence reactivity. However, with the exception of one measurement on the chromophore in mPlum, a GFP variant,14 such experiments have been limited to generic measures of protein solvation, with no specific connection to motions that may be relevant to function.
The strategy utilized to determine solvation along the reaction coordinate in KSI is described schematically in Figure 1. Both equilenin and coumarin 183 (C183) bind tightly in the oxyanion hole and chemically resemble the intermediate in the catalytic cyclic. Both are photoacids, that is, upon photoexcitation, the pKa of the hydroxyl group changes substantially. This light-activated change in electron density around the photoacid hydroxyl group simulates the change in substrate pKa during the catalytic cycle. Such a large change in dipole moment associated with photoexcitation produces a substantial solvation response in simple solvents that can be probed quantitatively by measuring the dynamic Stokes shift in the emission spectrum.8–10 The experiments described in the following measure the magnitude of the solvation response to this charge displacement when these molecules are bound at the active site of KSI.
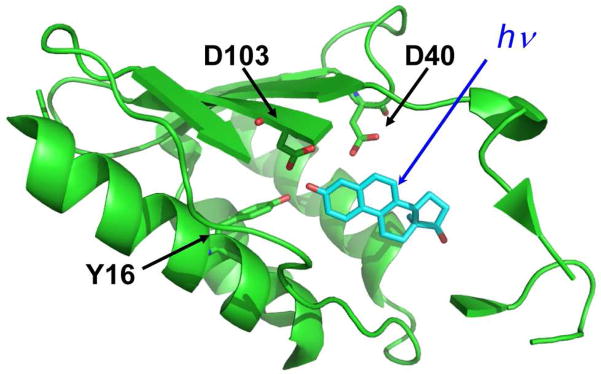
Much work by other labs has determined how substituted naphthols, especially equilenin, bind to KSI. Crystal structures of equilenin bound to KSI show the probe hydroxyl group hydrogen bonds to the oxyanion hole residues as depicted in Figure 2.18 The oxyanion hole is a critically important area of the active site. Mutation of either Tyr16 or Asp103 to non-polar residues reduces the catalytic efficiency by 103.4 and 102.1, respectively.19 Two other tyrosine residues (Tyr57 and Tyr32) are close to Tyr 16 and Asp 103, making the oxyanion hole itself potentially quite polar; the unusual properties of this site are described in detail elsewhere 20. Furthermore, the oxyanion hole is the most likely area of the active site to be affected by electrostatic perturbation (or to induce an electrostatic perturbation on the substrate) because the remainder of the active site is composed of hydrophobic residues. The charge transfer associated with excitation of the photoacid results in a large change in electron density around the probe hydroxyl group as evident by the change in pKa. This movement of charge is an ideal tool for studying the activity of the protein oxyanion hole during catalysis because a similar charge transfer occurs upon the conversion of substrate ketone to enolate during catalysis.6
Figure 2.
Crystal structure 1oh0 of equilenin (blue) bound to KSI from Pseudomonas putida (green).16 Two β-stands that form one side of the active site pocket have been removed for viewing purposes.
We can also frame these experiments in a broader context to test aspects of competing theories of enzymatic catalysis in KSI. Some researchers have proposed an electrostatically preorganized active site that should be a relatively fixed environment over the reaction coordinate.2,21–24 Others have suggested protein motions as a general feature of catalytic rate enhancement.25–27 If the active site is characterized by electrostatic preorganization, then the electric field produced by the protein environment should be invariant over the reaction coordinate, and the solvation response to a charge transfer related to catalysis should be minimal. Alternatively, if protein motions are responsible for activating catalysis, then the same motions should induce changes in the electric field around the substrate during catalysis. If changes in the electric field induce catalysis, then the light driven reaction should induce protein motion, that is, a solvation response. Therefore, direct observation of the solvation response within the active site from a probe whose transition resembles the reaction coordinate can distinguish between a preorganized active site and one that relies on protein motion to induce catalysis.
Methods
Protein expression
All experiments were conducted with KSI from Pseudomonas putida with the common mutation, D40N, to enhance binding. Protein was over expressed by standard microbiology techniques and purified by affinity chromatography as previously reported.3 Electrospray mass spectrometry confirmed that the protein was expressed correctly.
Steady-state fluorescence
Steady-state fluorescence experiments were conducted with a Fluorolog-3 fluorimeter from Horiba Jobin Yvon. Room temperature and 77 K protein spectra were obtained with 0.5mM equilenin, 1 mM KSI D40N in pH 7, 40 mM potassium phosphate buffer and 50% glycerol. Room temperature and 77 K aqueous samples were conducted with 0.5 mM equilenin in 0.5 M NaOH and 50% glycerol (see below and ref (28) for discussion of the relevant protonation state of equilenin in the protein). Low temperature experiments were conducted in an immersion cryostat with a 50 μm path length sample, while room temperature experiments were conducted in a 1 mm quartz cuvette. Parallel steady-state fluorescence experiments were not possible for C183 because its binding affinity to KSI was found to be greatly reduced in the presence of cryoprotectant (DMSO, glycerol, or sucrose).
Time-resolved fluorescence spectroscopy
Descriptions of our set ups for fluorescence upconversion and time-correlated single photon counting (TCSPC) have been reported elsewhere.29–31 In the current work, the fluorescence upconversion instrument response was measured to be ~140 fs, while the TCSPC instrument response was measured to be ~22 ps. Fluorescence upconversion was performed exclusively with C183 as insufficient laser intensity is available on our setup at the wavelength of the equilenin absorption band. C183 was excited with 400 nm, 120 pJ pulses of light. All experiments were preformed with 46 μM C183 in pH 7, 40 mM potassium phosphate buffer. Protein-bound C183 experiments were obtained in the presence of 0.65 mM KSI D40N.
For equilenin, time-resolved fluorescence measurements were only possible using TCSCP because the excitation light at 365 nm (~60 pJ/pulse) available from our laser system provides insufficient power in the gate beam to observe an upconversion signal. All experiments were performed with 0.5 mM equilenin. Measurements in aqueous buffers were conducted in pH 12 CAPS buffer with 50% DMSO to ensure solubility (see below and ref (28) for discussion of the relevant protonation state of equilenin in the protein). Protein-bound equilenin experiments were conducted in pH 7 potassium phosphate buffer with 1.0 mM KSI D40N. In all TCSPC experiments involving equilenin, the buffer was purged with argon for one hour before sample preparation. After addition of protein and ligand, the sample was purged with argon for an additional 30 minutes.
As described previously, time-resolved fluorescence spectra were reconstructed from individual decay curves.8–10 Individual fluorescence decays were measured in 5 nm increments across the fluorescence emission band. The intensity of each fluorescence decay was normalized to the steady-state intensity at the corresponding emission wavelength. Steady-state spectra used to normalize the individual decays were taken from samples composed identically to those used in the time-resolved experiments and excited at the same wavelength. The time-resolved spectra were fit to a log-normal line shape at each time point to determine the peak position as function of time.
Results/Discussion
Light-activated reaction analog
Excitation of a photoacid does resemble the charge transfer that occurs during KSI catalysis, but it is not a perfect analog. Here we describe the strengths and weaknesses of our model system and discuss the choice of photoacids.
Probably the largest distinction between the actual reaction and the reaction analog is the exact direction and magnitude of the difference dipole between electronic states in the model and the reactant (or product) and intermediate in the catalytic cycle. During the actual reaction, a formal charge is transferred between the oxyanion hole and residue D40, as shown in Figure 1. In the reaction analog, charge is transferred between the oxyanion hole and the conjugated ring system of the dye. Thus, the direction of charge transfer from the oxyanion hole is rotated by ~ 45° between the two systems. Since we are dealing with a specific solvent environment and a specific transition, it may be that degrees of freedom that might be accessed in response to charge migration in the course of catalysis are somewhat different from those accessed by the light-driven analog.
The magnitude of the change in electron density around the oxyanion hole is smaller in the reaction analog than in the actual reaction. As shown in Figure 1, the actual reaction converts a ketone to enolate, and this change represents a ΔpKa of ~12 units. As discussed below, the ΔpKa produced by the reaction analog is ~ 6–7 pKa units. While the change is less than in the actual reaction, the change in electron density during the reaction analog represents a significant perturbation to the protein, especially considering the ΔpKa between the substrate and transition state, which, while not known with great confidence, has been suggested to be ~ 9 units.32
Finally, the reaction analog does not model all aspects of the catalytic reaction. Specifically, the proton transfer to Asp40 is not reproduced. The reaction analog reproduces the change in electron density on the substrate around the oxyanion hole, and the solvation dynamics we probe are related to changes in and around the oxyanion hole.
Note that the analog ground state resembles the reaction intermediate while the analog excited state resembles the reaction starting material. Therefore, the light-activated reaction actually simulates the reaction running backwards from the intermediate to the substrate. However, microscopic reversibility applies here, and therefore, whatever process the protein undergoes is observable in either direction. Additionally, the excited state resembles the product ketone as well as the starting material ketone, and therefore, the light driven reaction equally well simulates the reaction between the intermediate and product.
Photoacids
The choice of photoacid is critical to ensure that the experiment is meaningful and technically possible. The two photoacids chosen for these experiments, equilenin and C183, are shown in Figure 1. Equilenin is a particularly convenient choice because it has been studied in KSI for many years as a reaction intermediate analog. Previous work has shown that it binds to KSI with 1 nM affinity,3 and crystal structures have been solved to determine its orientation when bound to KSI as shown in Figure 2.18 Critically, the molecule is not susceptible to solvation from bulk solvent outside the active site of KSI. The active site of KSI is a deep pocket, but a ligand bound in the pocket is still solvent exposed at the end opposite the hydroxyl group. In the case of equilenin, a non-polar region that is not conjugated to the π system is exposed to the solvent. We have shown in separate work that equilenin binds at the active site in approximately a 50/50 mixture of the protonated and deprotonated form;28 deconvolution of the spectra of these forms shows that only the deprotonated form is excited at 365 nm, and the deprotonated form at high pH is therefore used for comparison. Unfortunately, equilenin is not suitable for ultra-fast fluorescence upconversion experiments since the molecule does not absorb well in regions accessible with the doubled output of the Ti:sapphire laser and because it is relatively susceptibility to photodestruction. With our setup, the power output is severely reduced at 730 nm (necessary for excitation at 365 nm) from the optimal 800 nm. As a result, the maximum excitation power and gate beam power are reduced. Because fluorescence upconversion is a non-linear process, a diminished gate beam dramatically reduces the potential for using upconversion. As a result, experiments with equilenin were limited to TCSPC.
C183 was found to be a suitable probe for the faster fluorescence upconversion experiments because its absorption is further to the red than equilenin and it is relatively photochemically stable. C183 does bind to KSI as demonstrated by a change in fluorescence lifetime upon binding. Unbound C183 has a fluorescence lifetime of greater than 1 ns, but the lifetime is reduced to tens of picoseconds when bound at the active site of KSI. The reduction in lifetime is not unique to the protein environment as a similar quenching of the C183 anion is observed in hydrophobic mixtures of 2:1:0.01 (v/v/v) toluene:phenol:NaOH saturated in water. Quenching is only observed in low dielectric, hydrogen-bonding solvents; the reason for quenching is unknown, and we do not speculate on the mechanism. While the reduced fluorescence lifetime limits the observational time window, it also provides a clear observable to distinguish bound from unbound ligand. The addition of equilenin (Kd ~ 1 nM) to solutions of C183 bound to KSI D40N restores the solution phase C183 fluorescence lifetime. Because equilenin is known to bind to the KSI active site, displacement of C183 by equilenin strongly suggests C183 also binds to the active site. Conversely, when equilenin is pre-bound to KSI, C183 does not bind.
Much work has focused on the binding of naphthols and phenols to the KSI active site, and those experiments show the binding affinity of the hydroxyl group to the oxyanion hole is at least an order of magnitude greater than that of a carbonyl group.3,33 Also, the infrared absorption spectrum of C183 bound to KSI shows the nitrile stretch at a frequency unperturbed from the buffer value. This strongly suggests that the nitrile is exposed to buffer when bound to KSI, and this is consistent with the hydroxyl group on C183 binding to the oxyanion hole. Given the affinity of hydroxyl binding and the unperturbed nitrile stretch, we can say with high certainty that C183 does bind with its hydroxyl group bound to the oxyanion hole. The solution pKa of C183 and measured proton affinity of the active site suggest that it binds as its anionic form.28 Consistent with this, electronic and infrared (looking at the characteristic nitrile stretch) absorption spectrum of C183 when bound to the protein is similar to the absorption spectrum of C183 in basic solution. As a result, solvation experiments conducted on C183 in the absence of protein focused on the solvation response of the C183 anion.
The electron accepting cyano and carbonyl groups on C183 help produce the fluorescence properties that make the upconversion experiment possible. However, because these groups are solvent exposed to some extent when bound to KSI they also expose the conjugated system to the solvent, and solvation of these groups by buffer contributes to the observed solvation response. As a result, the solvation response observed in C183 systems can only be considered an upper bound of what occurs during catalysis.
ΔpKa is obtained for photoacids by the Förster cycle,34,35 Equation 1:
| (1) |
where Na is Avogadro’s number, h is Plank’s constant, c is the speed of light, R is the gas constant, T is the temperature of the system, and (ν̄ArO− − ν̄ArOH) is the difference in transition energy for the deprotonated and protonated states in cm−1. Absorption and fluorescence emission spectra are generally used to determine (ν̄ArO− − ν̄ArOH) for both the S0-S1 transition and S1-S0 transition, and the average value is used in Equation (1). Ideally, the transition energies are the 0-0 transition energies; in practice, the 0-0 energies are difficult to determine, and the peak energies are used instead. Using the Förster cycle, the ΔpKa was found to be ~ 5.2 for C183 with ground state pKa ~ 7. ΔpKa for equilenin was found to be ~ 7 with ground state pKa = 9.7. The measured ΔpKa for equilenin is similar to the previously measured ΔpKa of 7 for 2-naphthol,36 which is structurally similar to equilenin.
In summary, equilenin and C183 are very good, but not perfect probes for the charge-transfer reaction that occurs during catalysis. This is a rare case in which a process very closely related to the reaction coordinate can be triggered with light and the response of the protein observed and compared with a simple solvent. Each dye poses experimental challenges and has limitations due either to its photophysical or photochemical properties, and in the case of C183, partial exposure to bulk solvent.
Steady-state fluorescence
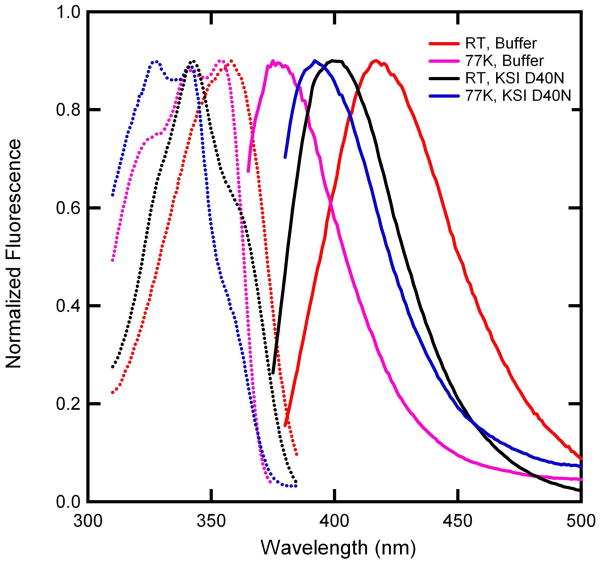
The temperature dependence of the fluorescence excitation and emission spectra of equilenin in buffer and bound to KSI are shown in Figure 3. In buffer, a blue shift of ~ 40 nm (2500 cm−1) is observed between room temperature and 77 K. Temperature dependent Stokes shifts have long been attributed to a reduced solvation capacity at lower temperatures;37 at 77 K the glycerol/water buffer is a frozen glass and the solvation capacity is greatly reduced. Fluorescence spectra from equilenin bound to KSI are also temperature dependent; however, the degree of the blue shift is a relatively modest ~ 8 nm (400 cm−1). The temperature dependent spectra are consistent with a solvation response occurring in both solvent environments with the buffer environment displaying a significantly greater solvation capacity.
Figure 3.
Normalized steady-state fluorescence excitation and emission spectra of equilenin. Emission from KSI bound equilenin at room temperature (solid, black) and 77 K (solid, blue) was observed from 350 nm and 365 nm excitation, respectively. Corresponding excitation spectra (broken line with matching color) were observed from 400 nm emission. Emission from equilenin in buffer at room temperature (solid, red) and 77 K (solid, purple) was observed from excitation at 360 nm and 350 nm, respectively. Excitation spectra (broken lines) of the room temperature sample (red) were observed from emission at 420 nm while the 77 K sample (purple) was observed from 390 nm emission. The protein-bound spectra were taken in pH 7, 40 mM potassium phosphate buffer while the unbound spectra were determined in 0.5 M NaOH. All spectra were obtained in 50% glycerol.
Time-resolved fluorescence
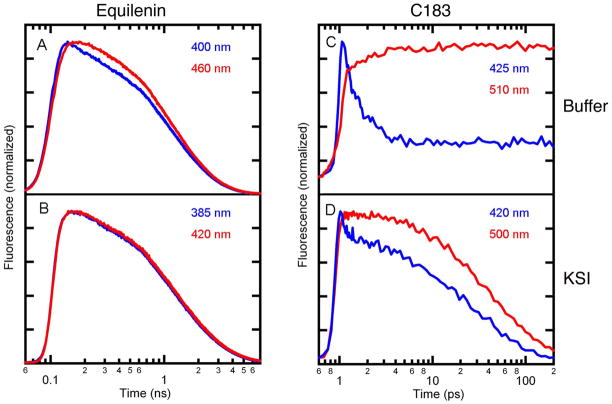
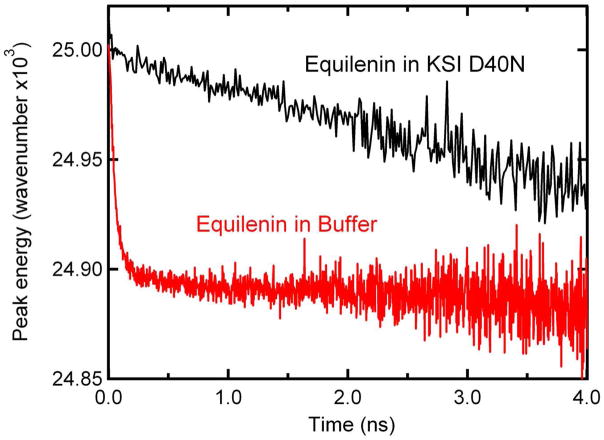
As discussed in the Methods section, TCSPC was used to measure the time-resolved emission spectra of equilenin in buffer and bound to KSI, both in anionic form.28 As shown in Figure 4A, the fluorescence decays on the red and blue sides of the equilenin emission spectrum in buffer exhibit different kinetics. The fast decay on the blue side of the band with accompanying rise on the red side is consistent with a solvation response. When equilenin is bound to KSI, the fluorescence decays on the red and blue sides are nearly identical (Fig. 4B). However, close examination shows the bluer wavelength does decay faster than the red. Individual decays taken at 5 nm increments were used to construct time-resolved emission spectra, and the fluorescence emission maximum was determined as a function of time, as shown in Figure 5. The observation window of 4 ns is the time until the fluorescence decays were reduced to 10% of their initial intensity. Equilenin in buffer shows the expected rapid solvation response typical in simple solvents. The magnitude of the solvation response was measured to be ~ 100 cm−1. The solvation response was best reproduced by fitting to multiple exponentials; however, fitting the response to a single exponential allows for an estimation of the solvent relaxation timescale. When fit to a single exponential, the response was found to occur with a lifetime of ~ 50 ps. Note that the instrument response time is ~ 22 ps, and the bulk of the solvation response probably occurs within the instrument response, as found with many similar dyes in simple solvents 9,10. Comparison of the steady-state fluorescence spectra at 77K and room temperature spectra suggests a solvation response as large as 2500 cm−1 is possible. Figure 5 shows almost no solvation at times greater than 1 ns, so the bulk of solvation likely occurs within the instrument response.
Figure 4.
Fluorescence decay curves of equilenin (A and B) and C183 (C and D) in buffer and bound to KSI measured on the blue and red side of the emission bands. The time-resolved fluorescence intensity of equilenin in buffer (A) and KSI D40N (B) was measured with TCSPC, which provides a ~ 22 ps instrument response. For the purposes of graphing, time zero for the TCSPC was set to 100 ps. Time-resolved fluorescence intensity of C183 in buffer (C) and KSI D40N (D) was measured by fluorescence upconversion, which provides a ~150 fs instrument response. Time zero for upconversion experiment was set to 1 ps.
Figure 5.
Time-resolved peak emission energy for equilenin in KSI (black) and buffer (red). For purposes of graphing, the data from equilenin in buffer (red) has been shifted up in energy by 1150 cm-1. Time-resolved fluorescence was observed with TCSPC, which has an instrument response function of ~22 ps.
In contrast to the rapid spectral shift observed for equilenin in buffer, the protein-bound sample undergoes a red shift of ~ 70 cm−1 over a broad time scale during the 4 ns observation window. Comparison of the fluorescence spectra of equilenin bound to KSI at room temperature and 77 K suggests a solvation response as large as 400 cm−1 is possible. The 4 ns observation window limits a quantitative fitting of the red shift; however, the red shift is happening on the tens-to-hundreds of nanosecond time scale.
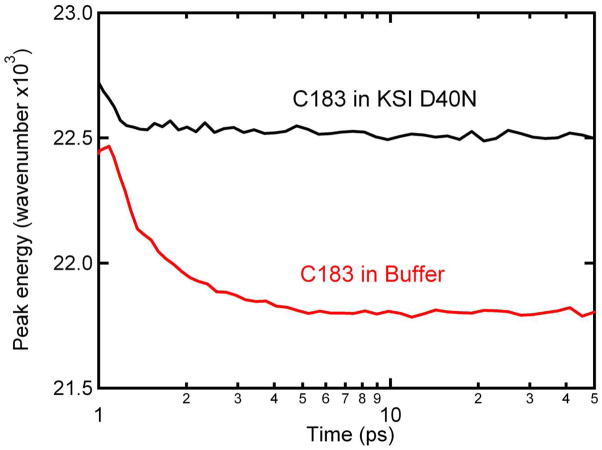
Fluorescence upconversion could be performed with C183 bound to the KSI active site. The fluorescence lifetime of C183 was dramatically reduced upon binding to KSI. While this limited the observation window to 100 ps, it also was used to demonstrate ~100% binding of C183 to KSI active site. As shown in Figure 4C, fluorescence from C183 in buffer undergoes a rapid decrease in fluorescence intensity on the blue side of the emission spectrum concurrent with a rise in fluorescence intensity on the red side, and this shift in fluorescence intensity is consistent with a solvation response. C183 bound to the KSI active site also shows a rapid shift of the fluorescence intensity (Figure 4D). However, the magnitude of the shift is much reduced from the buffer value. As shown in Figure 6, the solvation of excited C183 in buffer resulted in an ~ 750 cm−1 red shift occurring on the timescale of a few ps. This observation is consistent with small molecule solvation in a simple, low-viscosity, polar solvent. Solvation within KSI resulted in ~ 200 cm−1 shift occurring with similar kinetics. As noted above, the cyano and carbonyl groups of C183 are solvent exposed even when C183 is bound to the active site. The fast but small solvation response observed for C183 bound to KSI is consistent with water solvation of the solvent exposed electron accepting groups, and the solvation response observed from C183 can only be considered an upper bound for solvation possible within KSI.
Figure 6.
Time-resolved peak emission energy for C183 in KSI (black) and buffer (red). Time-resolved fluorescence was observed with fluorescence upconversion, which has an instrument response function of ~150 fs. For the purposes of graphing, time-zero was set to 1 ps. Note that the horizontal axis is log time and covers a very different range than in Figure 5.
Rigidity and preorganization
The solvation response for equilenin and C183 is significantly diminished when bound to KSI, and the diminished solvation response signifies an electrostatically rigid active site. The active site is often considered a hydrophobic environment due to the large number of hydrophobic residues lining much of the active site and the propensity for inhibitors with increasing hydrophobicity to bind with greater affinity.3,28 The purported hydrophobic environment of the active site may seem to necessarily limit the solvation response observed in KSI. However, the hydrogen bonding oxyanion hole could provide a significant solvation effect if these hydrogen bonding groups are able to reorganize after the substrate is excited. Asp103 and Tyr16 form hydrogen bonds to equilenin, and if these hydrogen bonds were able to rearrange in response to the charge shift upon excitation, then they would produce a dynamic Stokes shift. The conventional solvation response is described as the inertial response and reorientation of solvent dipoles around the excited-state dipole. However, if the fluorophore is capable of hydrogen bonding to the solvent, then motion between and around the hydrogen bonding members can dominate the solvation response. Indeed, the solvation response of 2-naphthol in simple, protic solvents is largely attributed to the rearrangement of solvent hydrogen bonds to the hydroxyl moiety.36 In our earlier work on mPlum (derived from mRFP 38), the solvation response was shown to result entirely from a single hydrogen bond interaction between the chromophore and the protein environment,14 and the observed solvation response was larger and some two orders of magnitude faster than that observed with equilenin in KSI.
The presence of the oxyanion hydrogen bonds but lack of a bulk-solvent-like solvation response suggests an enzymatic role for the electrostatically rigid nature of the active site. Two orthogonal theories of catalytic rate enhancement revolve around the mechanical and electrostatic stability of the protein active site. Some researchers have proposed special networks of coupled protein motion as a general mechanism of catalytic rate enhancement.25–27 A recent computational study specifically examining the conformational changes in KSI has found conformational changes of the oxyanion hole residues and Asp40 during the catalytic cycle.39 Our experiment is not well optimized to examine the reorganization of Asp40; however, the lack of large solvation response suggests the oxyanion hole residues do not reorganize during the catalytic cycle. Warshel and other researchers have proposed a preorganized active site in which the electrostatic environment is largely invariant over the reaction coordinate.2, 21–24 The small dynamic Stokes shift observed in the KSI active site indicates only minimal dielectric reorganization occurs during the light-driven reaction analog. The lack of a significant dielectric reorganization during the reaction analog suggests that the active site does not significantly reorganize during the catalytic cycle. And, the lack of electrostatic reorganizing during the catalytic cycle suggests an electrostatically preorganized active site.
The dynamic Stokes shift can only be observed for approximately 4 ns with equilenin and 100 ps with C183; however, these are relevant timescales to observe catalytically relevant protein motion. The only motion allowed to the protein is thermal motion and the only thermal motions that are unique to organized solvents are low frequency vibrational modes (breathing modes) that encompass most or all of the protein. Protein breathing modes have been observed in molecular dynamics simulations to occur on timescales ranging from 0.1 to 10 ps in proteins of similar size as KSI including trypsin inhibitor, crambin, ribonuclease, and lysozyme.40 The timescales observed for these vibration modes suggest a solvation response in the picosecond regime if a protein breathing mode or combination of modes is responsible for catalytic rate enhancement. However, as discussed above, the solvation response observed with the equilenin light-driven reaction analog is in the 10-to-100s of nanosecond regime. Therefore, protein breathing modes are unlikely to be coupled to the reaction coordinate.
The lack of a significant solvation response in KSI suggests a preorganized active site. When the light-driven reaction analog occurs in the KSI active site, the electrostatic environment changes little. If the protein is optimized to induce an electrostatic environment to affect the substrate, then inducing the change in the substrate should induce a change in the protein. Alternatively, a preorganized active site would always be positioned to induce transition state formation. An electrostatically preorganized active site would resist changing its electrostatic environment as the substrate is bound and reacts, and this resistance to electronic perturbation is observed during the light-driven reaction analog in the KSI active site.
Acknowledgments
This work was supported in part by a grant from the National Institute of Health GM27738. Plasmids containing the KSI gene and affinity column resin were graciously provided by the Daniel Herschlag lab at Stanford University. We are grateful for extended discussions with members of the Herschlag lab as well as Aaron Fafarman in our lab.
References
- 1.Perutz MF. Science. 1978;201:1187. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 2.Warshel A. J Bio Chem. 1998;273:27035. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- 3.Kraut DA, Sigala PA, Pybus B, Liu CW, Ringe D, Petsko GA, Herschlag D. PLoS Biol. 2006;4:e99. doi: 10.1371/journal.pbio.0040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warshel A, Sharma PK, Chu ZT, Aqvist J. Biochemistry. 2007;46:1446. doi: 10.1021/bi061752u. [DOI] [PubMed] [Google Scholar]
- 5.Kamerlin SCL, Sharma PK, Chu ZT, Warshel A. Proc Natl Acad Sci USA. 2010 early Edition. [Google Scholar]
- 6.Pollack RM. Bioorg Chem. 2004;32:341. doi: 10.1016/j.bioorg.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Sigala PA, Fafarman AT, Bogard PE, Boxer SG, Herschlag D. J Am Chem Soc. 2007;129:12104. doi: 10.1021/ja075605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Zwan G, Hynes JT. J Phys Chem. 1985;89:4181. [Google Scholar]
- 9.Horng ML, Gardecki JA, Maroncelli M. J Phys Chem. 1995;99:17311. [Google Scholar]
- 10.Stratt RM, Maroncelli M. J Phys Chem. 1996;100:12981. [Google Scholar]
- 11.Homoelle BJ, Edington MD, Diffey WM, Beck WF. J Phys Chem B. 1998;102:3044. [Google Scholar]
- 12.Pierce D, Boxer SG. J Phys Chem. 1992;96:5560. [Google Scholar]
- 13.Abbyad P, Shi X, Childs W, McAnaney TB, Cohen BE, Boxer SG. J Phys Chem B. 2007;111:8269. doi: 10.1021/jp0709104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbyad P, Childs W, Shi X, Boxer SG. Proc Natl Acad Sci USA. 2007;104:20189. doi: 10.1073/pnas.0706185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong D. Adv Chem Phys. 2009;143:83. [Google Scholar]
- 16.Kao YT, Saxena C, He TF, Guo L, Sancar A, Zhong D. J Am Chem Soc. 2008;130:13132. doi: 10.1021/ja8045469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warshel A, Chu ZT, Parson WW. Science. 1989;246:112. doi: 10.1126/science.2675313. [DOI] [PubMed] [Google Scholar]
- 18.Kim SW, Cha SS, Cho HS, Kim JS, Ha NC, Cho MJ, Joo S, Kim KK, Choi KY, Oh BH. Biochemistry. 1997;36:14030. doi: 10.1021/bi971546+. [DOI] [PubMed] [Google Scholar]
- 19.Choi G, Ha NC, Kim SW, Kim DH, Park S, Oh BH, Choi KY. Biochemistry. 2000;39:903. doi: 10.1021/bi991579k. [DOI] [PubMed] [Google Scholar]
- 20.Fafarman AT, Sigala PA, Herschlag D, Boxer SG. doi: 10.1073/pnas.1111566109. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warshel A. Proc Natl, Acad Sci USA. 1978;75:5250. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannon WR, Benkovic SJ. J Biol Chem. 1998;273:26257. doi: 10.1074/jbc.273.41.26257. [DOI] [PubMed] [Google Scholar]
- 23.Feierberg I, Åqvist J. Biochemistry. 2002;41:15728. doi: 10.1021/bi026873i. [DOI] [PubMed] [Google Scholar]
- 24.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson M. Chem Rev. 2006;106:3210. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 25.Hammes G. Nature. 1964;204:342. doi: 10.1038/204342a0. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal PK, Billeter SR, Rajagopalan PTR, Benkovic SJ, Hammes-Schiffer S. Proc Natl Acad Sci USA. 2002;99:2794. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammes-Schiffer S, Benkovic S. Annual Reviews. 2006;75:519. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 28.Childs W, Boxer SG. Biochemistry. 2010 doi: 10.1021/bi100074s. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chattoraj M, King BA, Bublitz GU, Boxer SG. Proc Natl Acad Sci USA. 1996;93:8362. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley RJ, Boxer SG. J Phys Chem. 1995;99:859–863. [Google Scholar]
- 31.McAnaney TB, Zeng W, Doe CFE, Bhanji N, Wakelin S, Pearson DS, Abbyad P, Shi X, Boxer SG, Bagshaw CR. Biochemistry. 2005;44:5510. doi: 10.1021/bi047581f. [DOI] [PubMed] [Google Scholar]
- 32.Zeng B, Pollack RM. J Am Chem Soc. 1991;113:3838. [Google Scholar]
- 33.Kuliopulos A, Mildvan AS, Shortle D, Talalay P. Biochemsitry. 1989;28:149–159. doi: 10.1021/bi00427a022. [DOI] [PubMed] [Google Scholar]
- 34.Förster T. Z Elektrochem. 1950;54:531. [Google Scholar]
- 35.Weller A. Prog React Kinet. 1961;1:187. [Google Scholar]
- 36.Solntsev KM, Huppert D, Agmon N. J Phys Chem A. 1998;102:9599. [Google Scholar]
- 37.Ronne C, Thrane L, Astrand PO, Wallquist A, Mikkelsen KV, Keiding SR. J Chem Phys. 1997;107:5319. [Google Scholar]
- 38.Wang L, Jackson WC, Steinbach PA, Tsien RY. Proc Natl Acad Sci USA. 2004;101:16745. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravorty DK, Soudackov AV, Hammes-Schiffer S. Biochemistry. 2009;48:10608. doi: 10.1021/bi901353v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levitt M, Sander C, Stern PS. J Mol Biol. 1985;181:423. doi: 10.1016/0022-2836(85)90230-x. [DOI] [PubMed] [Google Scholar]