Abstract
Early stages in the development of chronic lymphocytic leukemia (CLL) have not been explored mainly due to the inability to study normal B-cells in route to transformation. In order to determine such early events of leukemogenesis, we have used a well established mouse model for CLL. Over-expression of human TCL1, a known CLL oncogene, in murine B-cells leads to the development of mature CD19+/CD5+/IgM+ clonal leukemia with a similar disease phenotype seen in human CLL. Herein, we review our recent study using this TCL1 murine model for CLL and corresponding human CLL samples in a cross-species epigenomics approach to address the timing and relevance of epigenetic events occurring during leukemogenesis. We were able to demonstrate that the mouse model recapitulates epigenetic events very similar to what has been reported for human CLL and thus provides an exciting new tool to study early epigenetic events. Epigenetic alterations are seen at a time of three month after birth, much earlier than the phenotypically visible disease which occurs around 11 month of age. An early event in gene silencing is the inactivation of transcription factor Foxd3 expression through an NF-κB mediated process in animals with one month of age.
Keywords: CLL, genetics, methylation, epigenetics, TCL1
Introduction
CLL is characterized by several genetic abnormalities and laboratory features predictive of rapid disease progression and shortened survival 1, 2. CLL progression from early stage CLL to refractory disease is commonly associated with clonal evolution, as defined by multiple genetic abnormalities 3, 4. While long-term longitudinal follow-up of CLL patients is part of many studies, no study has effectively identified early initiating features in CLL. Identification of one or more initiating events that occur prior to development of multiple genetic abnormalities could provide insight into the etiology of CLL and also establish the rationale for pharmacologic targeting to prevent the development or progression of the disease.
Epigenetic alterations human CLL
Epigenetic alterations in cancer genomes have been recognized as major contributors to the malignant phenotype in several types of cancer. Epigenetic alterations do not change the DNA sequences and are transmitted to daughter cells. Two main epigenetic alterations, DNA methylation and modifications of chromatin proteins, have been described. These epigenetic alterations are interrelated and it is thought that they cooperate in the silencing of genes through the change in chromatin conformation [for a review see Ref. 5]. The role of aberrant DNA methylation in CLL is not clear 6. In a global screen for CpG island methylation hypermethylation was found in CLL patients with a mean number of 4.8% CpG islands affected 7. These genes include novel tumor suppressor genes, such as DAPK1, SFRP1, ID4 and genes involved in apoptosis 8–11. Additionally, cell cycle regulators CDKN2A, CDKN2B12–14 as well as prognostic markers ZAP70, TWIST2 have been found methylated in CLL patients15, 16.
Early epigenetic events in the progression to CLL in a mouse model
The question emerging now is: what is the relevance of these gene silencing events for the leukemic process? Are these all required events in order for the transformation into a malignant clone or are many of the events secondary events accumulating during leukemogenesis? We have chosen to determine early events in this process under the assumption that those should represent required epigenetic changes. Due to the lack of defined early pre-leukemic stages of human CLL and given the similarity of the Eµ-TCL1 transgenic model of CLL to human CLL 17, 18, we decided to perform a comprehensive epigenetic study with TCL1 mice at the period of polyclonal/oligoclonal expansion of premalignant CLL transformation to seek the potential targets for early therapy. In these mice, initial expansion of non-clonal B lymphocytes occurs at approximately three months and is followed by progression to a mature B-cell leukemia at 9–11 months, with immuno-phenotypic and clinical characteristics of human CLL.
We have now performed a genome-wide scan for aberrant CpG island methylation in B-cells collected at multiple time points towards the progression to CLL and found aberrant DNA methylation in cells harvested at three month after birth at a time where no disease phenotype was visible. DNA methylation levels increased from 0.4%, 0.6%, 1.2%, and 1.9% (3, 5, 7 and 9 months respectively) to 3.9% in Eµ-TCL1 mice with advanced CLL 19. Most interesting epigenetic target genes were comparable to those in human CLL. We tested ten of the early targets identified in the mouse model and found nine of them methylated and silenced in human CLL. The similarity of this murine CLL model and human CLL methylation patterns therefore provides further justification for using this system.
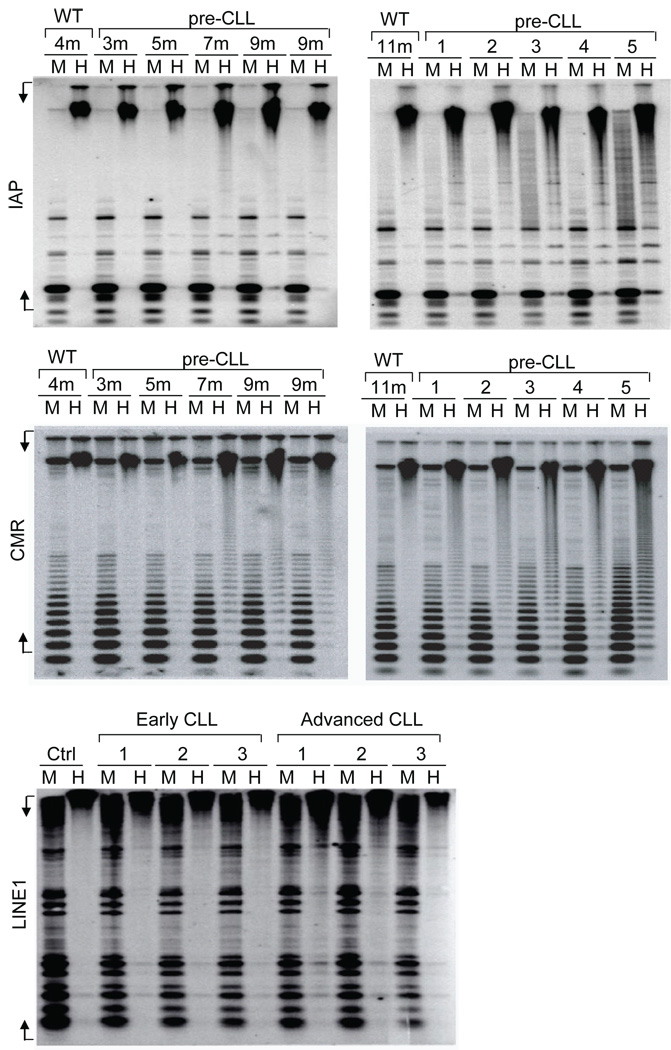
In addition to hypermethylation, previous tumor studies reported an overall decrease in 5-methylcytosine levels arising from hypomethylation of normally methylated repetitive elements might also contribute to tumorigenesis. Thus we analyzed whether global hypomethylation is occurring in the CLL cells of Eµ-TCL1 mice. The proviral sequences related to the intracisternal A particle (IAP) and centromeric repeat sequences were used as the probe for methylation analysis on the repetitive sequences by Southern blot. After the comparison of Hpall (methylation sensitive) and MspI (methylation insensitive) digests of DNA, we found that IAP (Figure 1A, top) and centromeric repeat sequences (Figure 1A, middle) were heavily methylated in 4 and 11 month old wild type C3H/B6 mice; but hypomethylated in Eµ-TCL1 mice from 7 and 9 month old as well as CLL mice. Moreover, hypomethylation of LINE1 repeat elements is also increased in comparing of primary untreated and relapsed CLL patients (Figure 1A, bottom).
Figure 1. Elevated global hypomethylation of repetitive sequences in CLL.
Hypomethylation profiles on mouse and human CLL samples from different stages were studied by Southern blotting as previous described34. Genomic DNAs of spleen cells from wild type and Eµ-TCL1 mice at indicated time points, or Eµ-TCL1 mice with symptomatic disease (n=5), were digested with MspI (M) or HpaII (H) restriction enzymes. The blots were hybridized with intracisternal A particle probe (IAP) (Top) or centromeric repeat sequences (CMR) (Middle). For CLL patient samples, the analysis was done using human LINE1 probes on the peripheral blood B cells from patients required no treatment (early CLL) or that underwent treatment (advanced CLL) (Bottom). MspI digested DNA was used as control. Undigested product of HpaII digestion represents methylated DNA, while only unmethylated DNA is HpaII digestible. Mouse IAP, CMS and human LINE1 probes were prepared by PCR amplification using forward (F) and reverse (R) primers: IAP-F: 5’CGTCATTGTTCAGAGCCAGA3’, IAP-R: 5’TCCCGGAAACTTTTGTTCAC3’; CMS-F: 5’GATAAAAACCTACACTGTAG3’, CMS-R: 5’GTTTCTAATTGTAACTCATTG3’; LINE1-F: CGGGTGATTTCTGCATTTCC and LINE1-R: GACATTTAAGTCTGCAGAGG.
The elevated methylation in CLL is favored by the de novo DNA methyl-transferases
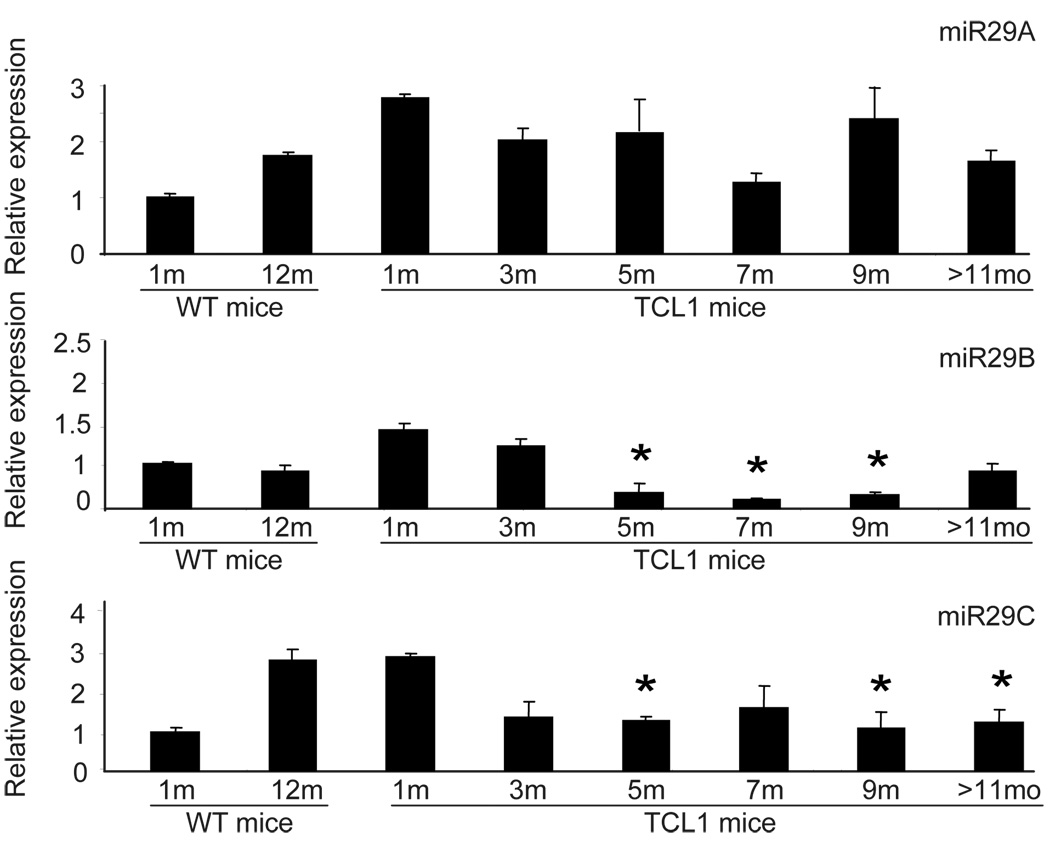
DNA methylation is mediated by transferring a methyl group from methyl donor S-adenosine methionine, to position 5 of the cytosine ring in the DNA. This reaction is catalyzed by DNA methyltransferases (DNMT)20. De novo methyltransferases (i.e. DNMT3A, 3B and 3L) establish the initial DNA methylation pattern within the genetic sequence 21, whereas the maintenance enzyme (DNMT1) maintains the methylation pattern during DNA replication 22. In our mice we have observed the lack of initiating DNA methyltransferases (DNMT3A and DNMT3B) in the genesis of transformation but increasing protein levels of DNMT3A and 3B in later stages. In contrast to protein expression, no significant changes were noted in the mRNA expression levels of these enzymes in CD19+ splenocytes from Eµ-TCL1 at any stage. These data suggests a posttranscriptional regulation of DNMT3A/3B possibly due to microRNAs, similar to recent findings in lung cancer, where one of the targets of miR29s is de novo DNA methyltransferase, DNMT3A and 3B 23. Indeed, diminished expression of miR29B and miR29C but not miR29A was noted in TCL1 mice at 5 and 7 months (Figure 2), corresponding to the increased DNMT3A and DNMT3B protein expression in line with the assumption of a direct regulation by the low-expressed miR29s in CLL B-cells 24. The increase in DNMT3A and 3B may be one of the factors needed for the increase in CpG island hypermethylation described above.
Figure 2. Decreased miR29s and increased DNMTs in Eµ-TCL1 mice.
Expression of miR29s by TaqMan PCR was done on TCL1 mouse spleen B cells. Each bar represents the average results from three mice at the indicated age. Results were normalized by the data obtained from untransformed B cells from 1 month old mice. Standard deviation was calculated by using mean ± SEM of respective data. Two tail unequal variant student T test was applied, star represents significant results with P value less than 0.05.
Silencing of Foxd3 a key event in progression to CLL
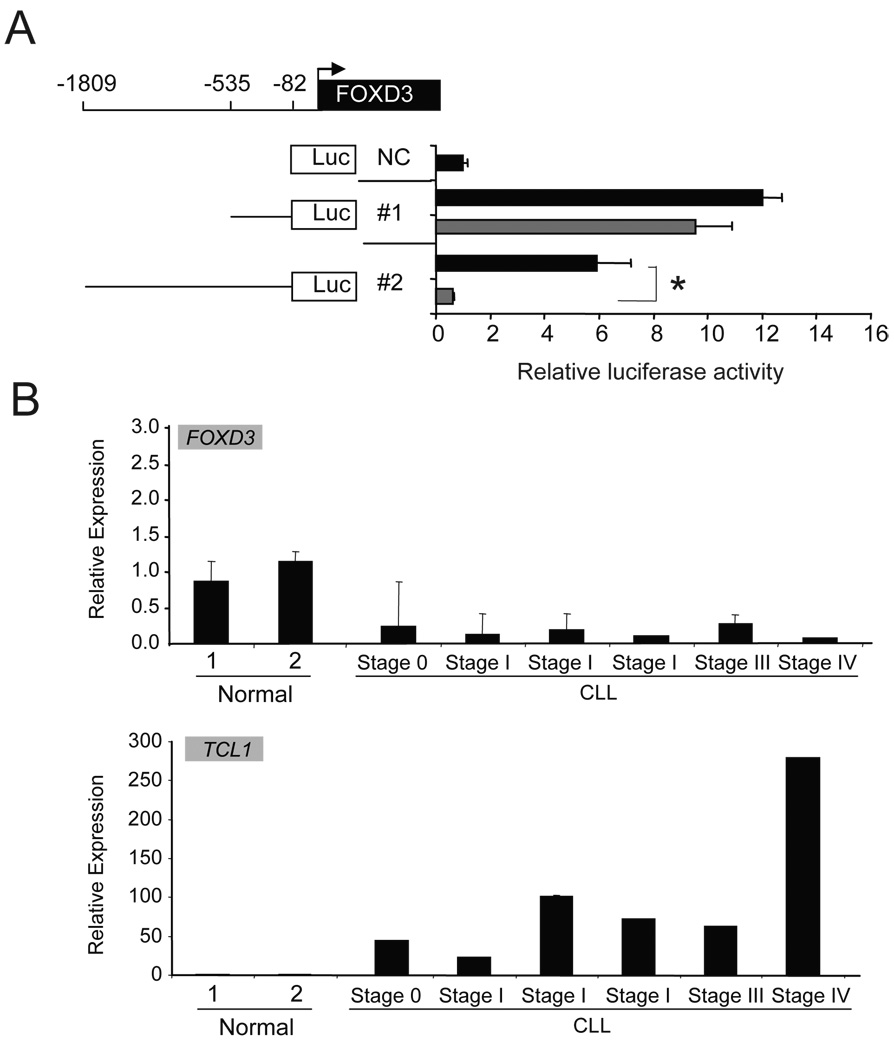
The fact that accumulated epigenetic alteration in CpG islands start from a small number of changes renders a thought of finding the initiating factor. For this purpose, we have searched the conserved domain that's specifically located within methylated promoters 19. Surprisingly, the results show that 70% methylated genes in Eµ-TCL1 mice are the putative targets of transcription activator FOXD3, a gene that was found methylated in TCL1 mouse B-cells prior to the transformation. FOXD3 is a member of fork head-box (FOX) family transcription factors characterized by a 100 amino acid monomeric DNA binding domain, which is highly conserved among all FOX proteins for nuclear localization and transcriptional regulation 25–27. FOXD3 plays a crucial role in gene regulation and is involved in a tight regulatory feedback loop with OCT4 and NANOG. The interaction and balanced expressions in this negative feedback loop formed by FOXD3, OCT4 and NANOG have been found essential in maintaining the multipotent properties of stem cells 28–31. In B lymphocytes, FOXD3 promoter activity was found negatively regulated by TCL1 in the CLL WAC3CD5 cell line (Figure 3A). Silenced FOXD3 were also noted in CLL samples with high TCL1 expression (Figure 3B).
Figure 3. Negative regulated FOXD3 by TCL1 in CLL cells.
(a) Luciferase assay was done on WAC3CD5 cell line transfected with 1µg pCMV-TCL1 (gray bar) or control vector (black bar). The diagram shows human FOXD3 5’ promoter region cloned into pGL3 vector for the analysis. Results from each cell line transfected with pGL3-basic vector (NC) were set as 1. Each bar represents the average result from the triplicate experiments using mean ± SEM of respective data. (b) FOXD3 and TCL1 expression were analyzed in two normal B cell samples and 6 CLL B cell samples from patients with different diagnosis. The error bars using mean ± SEM of respective data of triplicate experiments.
We then tried to unravel the mechanisms of gene deregulation involving transcription factor Foxd3. For the first time we were able to identify very early events in transformation by demonstrating TCL1 mediated transcriptional silencing of Foxd3 through a novel NF-κB p50/p50, Histone deacetylase 1 (HDAC1) co-repressor complex. The evidences suggest that silencing of transcription factors such as Foxd3 at early time point may be responsible for early transcriptional and later epigenetic silencing of multiple genes in the transformation of murine CLL 19.
Pre-leukemic stages in human CLL
Recent studies identified small B-cell clones using a six-colour flow cytometry with antibodies against CD45, CD19, CD5, CD10, kappa and lambda light chains in addition to immunoglobulin heavy chain gene rearrangement in healthy individuals with no signs of a lymphoproliferative disorder. The number of monoclonal B-cells in these conditions are below 5000 per cubic millimeter 32. This condition was termed monoclonal B-cell lymphocytosis, or MBL, and has now been shown to be a precursor for CLL. The frequency of MBL ranges from 3–5% in the general population. Interestingly, the majority of CLL patients (44 out of 45 individuals studied by Landgren et al.) showed this phenotype 33. The study samples were collected up to 77 months before the diagnosis of CLL thus arguing that MBL is a precursor stage of CLL. In the future it might be possible to utilize these samples for the discovery of early events in the progression to CLL. However due to the small number of individuals presenting with MBL (1% of the population) and the infrequent progression to CLL the mouse model presents a more robust study system at this time.
Acknowledgments
The authors thank Dr. Carlo Croce for kindly offered TCL1 transgenic mice, and Dr. Yuri Perkasky for pCMV-TCL1 plasmid. The authors would also like to thank all members of the Plass and Byrd labs for critical discussions; This publication was supported by National Cancer Institute grants CA110496 (J.C.B., C.P), A101956 (C.P. and J.C.B.) CA81534 to the CLL Research Consortium (J.C.B), P30 CA16058 (C.P. and J.C.B.), the Leukemia and Lymphoma Society (J.C.B. and C.P.), The D. Warren Brown Foundation (J.C.B), and the Thompson family. C.P. and J.C.B were Leukemia and Lymphoma Society scholar or clinical scholar, respectively.
References
- 1.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 2.Ripolles L, Ortega M, Ortuno F, Gonzalez A, Losada J, Ojanguren J, Soler JA, Bergua J, Coll MD, Caballin MR. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;171:57–64. doi: 10.1016/j.cancergencyto.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Stilgenbauer S, Sander S, Bullinger L, Benner A, Leupolt E, Winkler D, Krober A, Kienle D, Lichter P, Dohner H. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 4.Shanafelt TD, Witzig TE, Fink SR, Jenkins RB, Paternoster SF, Smoley SA, Stockero KJ, Nast DM, Flynn HC, Tschumper RC, Geyer S, Zent CS, Call TG, Jelinek DF, Kay NE, Dewald GW. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 6.Rush LJ, Plass C. Alterations of DNA methylation in hematologic malignancies. Cancer Lett. 2002;185:1–12. doi: 10.1016/s0304-3835(02)00288-4. [DOI] [PubMed] [Google Scholar]
- 7.Rush LJ, Raval A, Funchain P, Johnson AJ, Smith L, Lucas DM, Bembea M, Liu TH, Heerema NA, Rassenti L, Liyanarachchi S, Davuluri R, Byrd JC, Plass C. Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res. 2004;64:2424–2433. doi: 10.1158/0008-5472.can-03-2870. [DOI] [PubMed] [Google Scholar]
- 8.Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen SS, Hackanson B, Grever MR, Lucas DM, Matkovic JJ, Lin TS, Kipps TJ, Murray F, Weisenburger D, Sanger W, Lynch J, Watson P, Jansen M, Yoshinaga Y, Rosenquist R, de Jong PJ, Coggill P, Beck S, Lynch H, de la Chapelle A, Plass C. Downregulation of Death-Associated Protein Kinase 1 (DAPK1) in Chronic Lymphocytic Leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66:653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 10.Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, Liu S, Smith LT, Lee S, Rassenti L, Marcucci G, Byrd J, Caligiuri MA, Plass C. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- 11.Chim CS, Fung TK, Wong KF, Lau JS, Liang R. Infrequent Wnt inhibitory factor-1 (Wif-1) methylation in chronic lymphocytic leukemia. Leuk Res. 2006;30:1135–1139. doi: 10.1016/j.leukres.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Chim CS, Fung TK, Wong KF, Lau JS, Law M, Liang R. Methylation of INK4 and CIP/KIP families of cyclin-dependent kinase inhibitor in chronic lymphocytic leukaemia in Chinese patients. J Clin Pathol. 2006;59:921–926. doi: 10.1136/jcp.2005.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsirigotis P, Pappa V, Labropoulos S, Papageorgiou S, Kontsioti F, Dervenoulas J, Papageorgiou E, Panani A, Mantzios G, Economopoulos T, Raptis S. Mutational and methylation analysis of the cyclin-dependent kinase 4 inhibitor (p16INK4A) gene in chronic lymphocytic leukemia. Eur J Haematol. 2006;76:230–236. doi: 10.1111/j.1600-0609.2005.00604.x. [DOI] [PubMed] [Google Scholar]
- 14.Pinyol M, Cobo F, Bea S, Jares P, Nayach I, Fernandez PL, Montserrat E, Cardesa A, Campo E. p16(INK4a) gene inactivation by deletions, mutations, and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin's lymphomas. Blood. 1998;91:2977–2984. [PubMed] [Google Scholar]
- 15.Raval A, Lucas DM, Matkovic JJ, Bennett KL, Liyanarachchi S, Young DC, Rassenti L, Kipps TJ, Grever MR, Byrd JC, Plass C. TWIST2 demonstrates differential methylation in immunoglobulin variable heavy chain mutated and unmutated chronic lymphocytic leukemia. J Clin Oncol. 2005;23:3877–3885. doi: 10.1200/JCO.2005.02.196. [DOI] [PubMed] [Google Scholar]
- 16.Corcoran M, Parker A, Orchard J, Davis Z, Wirtz M, Schmitz OJ, Oscier D. ZAP-70 methylation status is associated with ZAP-70 expression status in chronic lymphocytic leukemia. Haematologica. 2005;90:1078–1088. [PubMed] [Google Scholar]
- 17.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, Croce CM. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson AJ, Lucas DM, Muthusamy N, Smith LL, Edwards RB, De Lay MD, Croce CM, Grever MR, Byrd JC. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood. 2006;108:1334–1338. doi: 10.1182/blood-2005-12-011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SSRA, Johnson AJ, Hertleinc E, Liu TH, Jin VX, Sherman M, Liu SJ, Dawsone DW, Williams KE, Lanasag M, Liyanarachchia S, Lin TS, Marcucci G, Pekarskya Y, Davuluria R, Croce CM, Guttridge DS, Teitell MA, Byrd JC, Plass C. Epigenetic changes during disease progression in a murine model of human chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2009 Jul 28; doi: 10.1073/pnas.0906455106. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson AT, Vertino PM, Spitzner JR, Baylin SB, Muller MT, Davidson NE. Role of estrogen receptor gene demethylation and DNA methyltransferase.DNA adduct formation in 5-aza-2'deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem. 1997;272:32260–32266. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- 21.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 22.Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 23.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox's in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 26.Katoh M, Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 27.Berry FB, Saleem RA, Walter MA. FOXC1 transcriptional regulation is mediated by N- and C-terminal activation domains and contains a phosphorylated transcriptional inhibitory domain. J Biol Chem. 2002;277:10292–10297. doi: 10.1074/jbc.M110266200. [DOI] [PubMed] [Google Scholar]
- 28.Tompers DM, Foreman RK, Wang Q, Kumanova M, Labosky PA. Foxd3 is required in the trophoblast progenitor cell lineage of the mouse embryo. Dev Biol. 2005;285:126–137. doi: 10.1016/j.ydbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M, Hromas R. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271:23126–23133. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- 30.Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. Faseb J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci U S A. 2002;99:3663–3667. doi: 10.1073/pnas.062041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti GE, Rawstron AC, Ghia P, Hillmen P, Houlston RS, Kay N, Schleinitz TA, Caporaso N. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol. 2005;130:325–332. doi: 10.1111/j.1365-2141.2005.05550.x. [DOI] [PubMed] [Google Scholar]
- 33.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, Marti GE, Caporaso NE. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–667. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]