Abstract
To assess the clinical utility of genome-wide oligonucleotide arrays in diagnosis of mental retardation and to address issues relating to interpretation of copy number changes (CNCs), we collected results on a total of 1499 proband patients from five academic diagnostic laboratories where the same 44K array platform has been used. Three of the five laboratories achieved a diagnostic yield of 14% and the other two had a yield of 11 and 7%, respectively. Approximately 80% of the abnormal cases had a single segment deletion or duplication, whereas the remaining 20% had a compound genomic imbalance involving two or more DNA segments. Deletion of 16p11.2 is a common microdeletion syndrome associated with mental retardation. We classified pathogenic CNCs into six groups according to the structural changes. Our data have demonstrated that the 44K platform provides a reasonable resolution for clinical use and a size of 300 kb can be used as a practical cutoff for further investigations of the clinical relevance of a CNC detected with this platform. We have discussed in depth the issues associated with the clinical use of array CGH and provided guidance for interpretation, reporting, and counseling of test results based on our experience.
Mental retardation (MR) and developmental delay (DD) occurs in ∼1% of the general population.1,2 Chromosomal abnormalities are detected in about 3.7% children with global DD using conventional G-banding analysis.3 Fluorescence in situ hybridization (FISH) has enhanced the detection rate but can only target well-characterized microdeletion syndromes and subtelelomeric regions. Genome-wide analysis using array-based comparative genomic hybridization (aCGH) has shown an ∼10% of additional diagnostic yield in patients referred for unexplained MR or DD.4,5
A number of array platforms are commercially available, and each of them has some unique features in terms of coverage, resolution, and utility.6 It is now well-known that copy number variants (CNVs) occur with a high frequency in normal human population.7,8,9 A higher resolution of an array platform may lead to a higher diagnostic yield but is also associated with a higher detection rate of benign CNVs in patients,10,11,12 and this can make interpretation of the aCGH results challenging at a clinical laboratory setting. Currently available databases have been built with information generated from different aCGH platforms, and each laboratory may have developed its own standard for confirmation and reporting.
The platform of Agilent 44K oligonucleotide array has been validated for clinical use in our early studies.11,13 To further assess the utility of this platform for clinical testing on developmental disorders, we have collected results of 1499 consecutive cases tested from five laboratories. Here we summarize the overall detection rate and the spectrum of pathogenic copy number changes with further discussions on the issues relating to the clinical applications of aCGH.
Materials and Methods
Patients
Clinical information and aCGH results of 1499 patients were collected from five clinical laboratories at University of Miami Miller School of Medicine (M), Children's Hospital Boston and Harvard Medical School (H), Yale University School of Medicine (Y), Tulane University School of Medicine (T), and University of South Alabama (A). All patients were referred by physicians as part of clinical assessment of unexplained MR or DD with or without dysmorphic features. Conventional chromosome analysis was performed in most of these cases with a normal karyotype reported. Patients with an indication of autism, pervasive developmental disorder or Asperger syndrome have not been included and will be dealt with separately.
The aCGH Analysis
The standard Agilent 44K platform (Agilent Technologies, Santa Clara, CA) has been used by all five laboratories. This platform is composed of 44,290 60-mer oligonucleotide probes for the mapped genes or unique DNA sequences with an average spatial resolution of ∼30–35 kb. It was noted that this platform lacks coverage for the pseudoautosomal regions of the X and the Y chromosomes. An aCGH test was performed using the standardized protocol as recommended by the manufacturer as described previously.11 Briefly, patient and reference DNA samples were differentially labeled and cohybridized to Agilent 4 × 44K arrays. Reference DNA samples were prepared either from apparently healthy individual (mixture of four to eight same gender individuals) or purchased (Promega, Madison, WI). Hybridized slides were scanned with microarray scanner (Agilent G2505B or Axon Genepix 4200A), and the image data were extracted and converted to text file with Feature Extraction (Agilent Technologies). DNA Analytics 4.0 (Agilent Technologies) was used to plot the log2 ratio of signal intensity of each probe across the whole genome. The copy number data were visualized along each chromosome with correspondent National Center for Biotechnology Information annotated gene information for each probe. For copy number change (CNC) identification, we used DNA Analytics build-in aberration detection algorithm followed by visual inspection. Each lab performed technical validation of the platform before using it as a diagnostic tool. A consensus cutoff for recording an alteration was a CNC involving ≥3 consecutive probes based on our previous validation studies.11
Criteria for Result Interpretation
We used the term “CNC” to describe a DNA copy alteration observed, the term “pathogenic CNC” to describe a CNC associated with abnormal phenotypes and the term “CNV” for a benign CNC. These labs have used the following consensus criteria for interpretation of pathogenic CNCs versus benign variants: A CNC is considered likely to be pathogenic if 1) it involves a region known to be associated with a microdeletion or microduplication syndrome, 2) it is inherited from a similarly affected parent, 3) it involves known dosage sensitive gene(s), or 4) it is a multigene imbalance, either de novo or inherited from a parent as a product of segregation of a balanced translocation/insertion or recombination of an inversion. In contrast, a CNC is considered likely to be benign if 1) it is a known CNV in normal populations, 2) it is inherited from a normal parent, or 3) it does not involve the regions associated with known microdeletion or microduplication syndromes or dosage-sensitive genes. Possible exceptions are described in Discussion. We have used Database of Genome Variants (http://projects.tcag.ca/variation/, July 24, 2009) as a reference for currently known CNVs in normal individuals, DECIPHER (DatabasE of Chromosomal Imbalances and PHenotypes using Ensembl Resources) (www.sanger.ac.uk/PostGenomics/decipher/, July 24, 2009) for currently known microdeletion and microduplication syndromes and Online Medelian Inheritance in Man (www.ncbi.nlm.nih.gov/Omim/getmorbid.cgi, July 24, 2009) for currently known disease-causing genes, gene functions, and inheritance patterns. Literature search on a particular CNC was also an important step to determine its clinical significance. A final result was reported as abnormal when a CNC was interpreted as pathogenic or normal when no alteration was detected or observed alterations were judged as benign. The term “variant of unknown clinical significance” was used when an imbalance was >300 kb involving multiple genes, but the significance of the imbalance could not be determined based on available knowledge and family studies.
Results
Results on a total of 1499 consecutive proband patients including previously published 150 cases11,13 were collected (Table 1). Pathogenic CNCs were reported in 177 patients representing an overall diagnostic yield of 12% with a variation from 7 to 14%. variant of unknown clinical significance was reported ∼5% of cases (Supplemental Table S1, see http://jmd.amjpathol.org). One or more benign CNVs have been reported in ∼48% of the patients. The sizes of the pathogenic imbalances varied from 166 Kb to152 Mb (Tables 234). For the compound imbalances, this size represented the total size of two or more DNA segments. The imbalances were >500 kb in the vast majority of abnormal cases (95.5%). Pathogenic CNCs <500 kb but >300 kb were detected in a small number of patients (4.5%). It is very important to find out the functions of the genes involved in a small CNC, particularly when it is not a known CNV and parental studies cannot be completed. For example, we reported a CNC <300 kb as pathogenic in a patient because of deletion of the TSC2 gene. Pathogenic imbalances >10 Mb were detected in 28% of patients, and most of them did not have a conventional chromosome study before aCGH. We noticed that deletions or duplications >10 Mb were missed by chromosome karyotyping in several patients. A significant proportion (19%) of pathogenic CNCs had a size of 5 to ∼10 Mb (Table 2). However, we could not determine the percentage of the cases missed by chromosome analysis, because in many cases, the initial karyotyping was performed outside of the five laboratories. A few cases with a chromosomal aneuploidy were observed but were not included in this article.
Table 1.
Array CGH Results from Five Labs
| Lab | No. of patients | No. patients with pathogenic CNCs | Diagnostic yield (%) |
|---|---|---|---|
| M | 545 | 74 | 14 |
| Y | 275 | 38 | 14 |
| H | 319 | 21 | 7 |
| T | 227 | 25 | 11 |
| A | 133 | 19 | 14 |
| Total | 1499 | 177 | 12 |
M, University of Miami Miller School of Medicine; H, Children's Hospital Boston and Harvard Medical School; Y, Yale University School of Medicine; T, Tulane University School of Medicine; and A, University of South Alabama.
Table 2.
Size Distribution of Pathogenic Imbalances
| Size of imbalance | No. cases with imbalances | Percentage |
|---|---|---|
| >10 Mb | 50 | 28.25 |
| 5 to ∼10 Mb | 33 | 18.64 |
| 1 to ∼5 Mb | 71 | 40.11 |
| 0.5 to ∼1 Mb | 15 | 8.47 |
| 0.3 to ∼0.5 Mb | 7 | 3.95 |
| <0.3 Mb | 1 | 0.56 |
| Total | 177 | 100 |
Table 3.
Pathogenic CNCs Involving a Single DNA Segment
| Patient | Imbalance | Size (kb) | ISCN description | NCBI build |
|---|---|---|---|---|
| A0024 | del | 1,060 | 1q21.1(143789559-144849923)x1 | hg18 |
| M0894 | del | 1,170 | 1q21.1(145031367-146201635)x1 | hg18 |
| A0021 | dup | 14,762 | 1q42.13q43(225784048-240546387)x3 | hg18 |
| M0516 | dup | 1,883 | 1 q42.2q43(233617930-235500555)x3 | hg18 |
| M1006 | del | 837 | 2p14p14(67241932-68078842)x1 | hg18 |
| M0099 | del | 13,769 | 2q14.1q14.3(115776113-129545173)x1 | hg17 |
| Y0008 | del | 13,698 | 2q24.3q31.1(162106534-175804618)x1 | hg17 |
| M1060 | del | 17,759 | 2q31.12q32.2(171993989-189752539)x1 | hg18 |
| T0074 | del | 12,456 | 3p26.3p25.1(224527-12680213)x1 | hg17 |
| Y0295 | dup | 40,927 | 3p22.1(224727-41152166)x3 | hg17 |
| H0243 | del | 2,090 | 3p21.31(45998019-48087642)x1 | hg18 |
| A0005 | del | 10,147 | 3q22.3q24(138095979-148242751)x1 | hg18 |
| M0773 | dup | 51,470 | 4q31.1q35.2(139651136-191121344)x3 | hg18 |
| Y0068 | dup | 6,457 | 4q35.1q35.2(184033371-190490075)x3 | hg17 |
| Y0173 | del | 3,781 | 5p15.33(148243-3929457)x1 | hg17 |
| M0717 | dup | 964 | 5p13.3(31244617-32209107)x3 | hg18 |
| A0027 | dup | 5,069 | 5q13.1q13.3(68203607-73272937)x3 | hg18 |
| M0254 | dup | 5,025 | 5q14.3q15(87529305-92554625)x3 | hg18 |
| H0181 | del | 8,761 | 5q23.1q23.3(119295079-128055733)x1 | hg18 |
| M0381 | del | 5,299 | 6q14.3q15(86137153-91436233)x1 | hg18 |
| T0018 | del | 10,481 | 6q16.1q21(95641152-106121873)x1 | hg17 |
| M0109 | del | 19,086 | 6q22.1q22.32(107058287-126144058)x1 | hg17 |
| M0840 | dup | 20,097 | 6q23.3q25.3(135720981-155817943)x3 | hg18 |
| Y0078 | dup | 14,601 | 6q24.1q25.3(142510586-157111635)x3 | hg17 |
| Y0004 | del | 9,452 | 6q26q27(161357458-170809875)x1 | hg17 |
| Y0014 | del | 7,953 | 6q26q27(162903508-170856715)x1 | hg17 |
| M0866 | del | 903 | 6q27(169830830-170734227)x1 | hg18 |
| Y0033 | del | 7,880 | 7p21.1p15.2(18809907-26689559)x1 | hg17 |
| M0975 | del | 3,048 | 7p21.1p15.3(19151860-22199856)x1 | hg18 |
| A0008 | del | 19,584 | 7p21.3p15.2(6275894-25859565)x1 | hg17 |
| Y0519 | del | 946 | 7p15.1(29959761-30905557)x1 | hg17 |
| M0531 | del | 11,444 | 7p15.3p14.3(20567604-32011280)x1 | hg18 |
| T0009 | del | 3,976 | 7q11.22q11.23(69801305-73777467)x1 | hg17 |
| Y0148 | dup | 457 | 7q11.23(72171229-72628014)x3 | hg17 |
| Y0431 | dup | 392 | 7q11.23(72723421-73115544)x3 | hg17 |
| Y0298 | del | 5,058 | 7q22.1q31.1(103693910-108751427)x1 | hg17 |
| M0979 | dup | 1,397 | 7q36.3(157384149-158781397)x3 | hg18 |
| M0300 | del | 2,995 | 8p23.3p23.2(181530-3176742)x1 | hg18 |
| M0798 | del | 4,602 | 8q22.3q23.1(102281230-106883502)x1 | hg18 |
| A0016 | del | 7,565 | 9p24.3p24.1(204367-7769314)x1 | hg18 |
| M0112 | del | 3,598 | 9q33.3q34.11(124897698-128495943)x1 | hg17 |
| Y0059 | del | 506 | 9q34.3(137723586-138229984)x1 | hg17 |
| M0266 | del | 5,773 | 10q25.3q25.2(112249547-118022746)x1 | hg18 |
| Y0534 | del | 13,168 | 10q26.12q26.3(122125789-135293404)x1 | hg17 |
| M0650 | dup | 1,574 | 10q26.3(132780691-134355172)x3 | hg18 |
| M0681 | dup | 376 | 11p15.4(6239388-6615806)x3 | hg18 |
| M1015 | dup | 1,823 | 11p15.5p15.4(1515185-3338575)x3 | hg18 |
| M0522 | del | 6,836 | 11p15.4p15.1(9967639-16803929)x1 | hg18 |
| M1003 | dup | 2,591 | 11q22.1q22.3(100828925-103420043)x3 | hg18 |
| Y0207 | del | 5,708 | 12p13.2p12.3(11678312-17386429)x1 | hg17 |
| M0702 | del | 12,758 | 13q33.1q34(101319059-114077122)x1 | hg18 |
| Y0039 | del | 13,239 | 13q33.1q34(100838094-114077122)x1 | hg17 |
| A0006 | del | 1,091 | 14q21.1q21.2(41116021-42206812)x1 | hg18 |
| M0152 | del | 1,150 | 14q32.2q32.31(99330427-100480657)x1 | hg17 |
| Y0048 | dup | 5,791 | 15q11.2q13.1(21208344-26999801)x3 | hg17 |
| Y0528 | dup | 7,891 | 15q11.2q13.1(19109124-26999801)x3 | hg17 |
| M0028 | del | 3,042 | 15q24.1q24.2(70764625-73807021)x1 | hg17 |
| T0001 | del | 3,292 | 15q26.1q26.2(89197342-92489641)x1 | hg17 |
| T0059 | del | 2,234 | 16p13.12p13.11(13949643-16183616)x1 | hg17 |
| H0221 | del | 166 | 16p13.3(2005613-2171887)x1 | hg18 |
| M0742 | dup | 3,392 | 16p13.11p12.3(15062188-18454260)x3 | hg18 |
| H0195 | del | 446 | 16p11.2(29581455-30027260)x1 | hg18 |
| H0264 | del | 446 | 16p11.2(29581455-30027260)x1 | hg18 |
| M0961 | del | 523 | 16p11.2(29581455-30104842)x1 | hg18 |
| M0480 | del | 525 | 16p11.2(29581455-30106101)x1 | hg18 |
| M0611 | del | 525 | 16p11.2(29581455-30106101)x1 | hg18 |
| H0338 | dup | 544 | 16p11.2(29560500-30104842)x3 | hg18 |
| H0367 | del | 546 | 16p11.2(29560500-30106852)x1 | hg18 |
| H0240 | dup | 659 | 16p11.2(29581455-30240082)x3 | hg18 |
| T0008 | dup | 16,113 | 16q22.3q24.3(72538455-88651780)x3 | hg17 |
| H0301 | del | 5,077 | 16q23.1q23.3(76508445-81585846)x1 | hg18 |
| H0376 | del | 1,207 | 17p13.3(48539-1255546)x1 | hg18 |
| T0017 | del | 4,132 | 17p13.3p13.2(116343-4248317)x1 | hg17 |
| M0481 | dup | 547 | 17p13.3(183662-730351)x3 | hg18 |
| Y0313 | dup | 2,493 | 17p13.2p13.1(6277211-8769730)x3 | hg17 |
| M0280 | dup | 319 | 17p13.1(9989893-10308758)x3 | hg18 |
| H0302 | del | 1,383 | 17p12(14040843-15423597)x1 | hg18 |
| M0855 | del | 1,380 | 17p12(14052497-15432473)x1 | hg18 |
| Y0273 | del | 3,590 | 17p11.2(16543855-20133761)x1 | hg17 |
| H0113 | del | 3,618 | 17p11.2(16543855-20162287)x1 | hg18 |
| H0194 | dup | 3,590 | 17p11.2(16644378-20234630)x3 | hg18 |
| H0074 | dup | 3,590 | 17p11.2(16543855-20133761)x3 | hg18 |
| H0371 | del | 1,288 | 17p11.2(16723271-18010992)x1 | hg18 |
| Y0494 | dup | 1,351 | 17q12(31891535-33242217)x3 | hg17 |
| M0761 | del | 627 | 17q21.3(41073486-41700815)x1 | hg18 |
| A0028 | dup | 14,749 | 18 p11.32p11.2(170229-14918854)x3 | hg18 |
| M0673 | del | 7,267 | 18q12.3q21.1(37556785-44824255)x1 | hg18 |
| M0482 | del | 4,391 | 18q12.3q21.2(38169137-42560421)x1 | hg18 |
| Y0489 | del | 2,719 | 18q21.2(48686656-51405390)x1 | hg17 |
| M0523 | del | 4,818 | 18q22.3q23(71264994-76083117)x1 | hg18 |
| M0262 | del | 511 | 19p13.3(737550-1248499)x1 | hg18 |
| M0015 | del | 3,459 | 19p13.2(10291150-13749674)x1 | hg17 |
| M0308 | dup | 733 | 19p13.2(10128082-10861364)x3 | hg18 |
| M0680 | del | 8,231 | 19q12q13.13(34854071-43085470)x1 | hg18 |
| Y0005 | del | 7,284 | 21q22.3(39608300-46892294)x1 | hg17 |
| M0054 | del | 6,182 | 21q21.3q22.12(28802339-34984201)x1 | hg17 |
| T0062 | dup | 14,789 | 21q22.11q22.3(32103127-46892494)x3 | hg17 |
| T0050 | dup | 2,268 | 22q11.21(17021209-19289184)x3 | hg17 |
| M0541 | del | 1,118 | 22q11.21q11.22(20128705-21246612)x1 | hg18 |
| A0029 | dup | 1,994 | 22q11.22q11.23(21322838-23316556)x3 | hg18 |
| T0014 | del | 8,003 | 22q13.2q13.33(40993242-48996488)x1 | hg17 |
| M0491 | dup | 3,144 | Xp22.33(000000-3144100)x3* | hg18 |
| M0958 | dup | 151,784 | Xp22.33q28(2710316-154494649)x3 | hg18 |
| T0049 | del | 1,431 | Xp22.31(6560955-7992261)x1 | hg17 |
| Y0268 | del | 1,525 | Xp22.31(6317139-7841856)x1 | hg17 |
| Y0497 | del | 1,431 | Xp22.31(6410891-7841856)x1 | hg17 |
| T0038 | del | 1,431 | Xp22.31(6560955-7992261)x1 | hg17 |
| M0849 | dup | 4,705 | Xp11.23p11.22(48005240-52710691)x3 | hg18 |
| Y0357 | del | 5,224 | Xp11.2(43850835-49074718)x1 | hg17 |
| A0013 | dup | 9,370 | Xp22.11p21.1(22924079-32294279)x3 | hg18 |
| H0121 | del | 2,734 | Yp11.2(6457810-9191638)x1 | hg18 |
NCBI, National Center for Biotechnology Information; ISCN, International System for Human Cytogenetic Nomenclature (2009).
The cases with common microdeletion syndromes involving 1p36, 4p16, 7q11.23, 15q11.2, and 22q11.2 have been excluded from this table.
Duplication of the terminal region. Upper breakpoint could not be assigned due to lack of probe coverage for the region.
Table 4.
Pathogenic CNCs Involving Two or More DNA Segments
| Patient | Imbalance | Size (kb) | ISCN description | NCBI build | Classification |
|---|---|---|---|---|---|
| M099 | del/dup | 4,134/2,164 | 1p36.33p36.32(799622-4933181)x1/Xq28(152241308-154405159)x3 | hg18 | 2 |
| A011 | del/dup | 9,275/5,885 | 1q43q44(236147009-245422419)x1/9p24.3p24.1(204367-6089101)x3 | hg17 | 2 |
| M183 | del/dup | 1,496/1,718 | 2p25.3(29193-1525513)x1/4q13.2q13.3(69802468-71520579)x3 | hg18 | 3 |
| M127 | del/dup | 3,471/6,495 | 4p16.3p16.2(62447-3533180)x1/7p22.3p22.1(149268-6644183)x3 | hg18 | 2 |
| M564 | del/dup | 4,349/1,397 | 4p16.3p16.2(62447-4411346)x1/7q36.3(157384149-158781397)x3 | hg18 | 2 |
| M192 | del/del | 2,132/1,431 | 4q31.21q31.22(144878812-147011076)x1/Xp22.31(6561155-7992120)x1 | hg18 | 6 |
| T013 | del/dup | 28,400/11,947 | 4q32.2q35.2(162914399-191314689)x1/3q27.2q29(187378521-199325500)x3 | hg17 | 2 |
| M470 | del/dup | 5,739/9,993 | 5p15.33p15.32(148243-5887664)x1/16q23.2q24.3(78658713-88651780)x3 | hg18 | 2 |
| T037 | del/dup | 1,872/27,999 | 6p25.3(204328-2076297)x1/9p24.3p21.1(204167-28203565)x3 | hg17 | 2 |
| M411 | del/dup | 9,362/2,984 | 6p25.2p24.1(4014025-13376010)x1/6p24.1p22.3(13419730-16403770)x3 | hg18 | 4 |
| M347 | del/dup | 3,181/5,277 | 7p22.3p22.2(149268-3330301)x1/9q34.13q34.3(134852111-140128736)x3 | hg18 | 3 |
| Y451 | del/dup | 14,193/3,283 | 7p21.3p15.3(7407526-21600331)x1/7p22.3p22.2(149268-3431999)x3 | hg17 | 4 |
| M119 | del/dup | 7,702/6,748 | 7q36.1q36.3(150706898-158409214)x1/22q13.31q13.33(42720092-49468408)x3 | hg18 | 2 |
| M449 | del/dup | 3,918/5,822 | 7q36.2q36.3(154684956-158602499)x1/9q34.13q34.3(134251761-140073968)x3 | hg18 | 2 |
| M479 | del/dup | 6,720/17,530 | 8p23.1p23.3(181530-6901486)x1/8p12p23.1(12627630-30157579)x3 | hg18 | 4 |
| Y222 | dup/dup | 42,685/40,034 | 8p23.31p11.2(181530-42866112)x3/9p24.3p12(204367-40238102)x3 | hg17 | 2 |
| Y006 | del/dup | 7,109/10,669 | 8p23.3p23.1(181530-7290597)x1/8p21.3p23.1(12285464-22954412)x3 | hg17 | 4 |
| M323 | del/dup | 4,366/38,156 | 9p24.2p24.1(4142060-8508353)x1/9q13q31.2(70225166-108381595)x3 | hg18 | 5 |
| T019 | del/del | 14,426/470 | 9p22.3p24.3(204367-14629971)x1/9q34.11(129926621-130396376)x1 | hg17 | 6 |
| A018 | del/dup | 2,683/58,847 | 10q26.3(132610756-135293404)x1/4q28.3q35.2(132273948-191121344)x3 | hg18 | 2 |
| A002 | del/dup | 1,422 /5,589 | 10q26.3(133871004-135293404)x1/17q25.3(73034631-78623230)x3 | hg17 | 2 |
| Y279 | dup/dup | 17,723/4,253 | 11q23.3q25(116228648-133951370)x3/22q11.1q11.2(14433473-18686317)x3 | hg17 | 6 |
| M613 | del/dup | 5,607/18,843 | 12p13.33p13.31(179323-5786793)x1 | hg18 | 2 |
| 15q25.2q26.3(81325482-100168718)x3 | |||||
| Y017 | del/dup | 10,269/3,107 | 13q33.2q34(103855141-114123908)x1/1p36.3(807922-3914685)x3 | hg17 | 2 |
| M160 | del/dup | 1,300/1,178 | 14q13.2q13.3(35053035-36352972)x1/9p22.1p22.2(17492885-18671089)x3 | hg18 | 3 |
| A014 | dup/dup | 9,791/11,936 | 15q11.2q13.3(18362555-28153357)x3/15q11.2q13.3(18362555-30298096)x3 | hg17 | 6 |
| T058 | del/dup | 86/49,036 | 18p11.32(170029-255680)x1/18q12.11q23(27047211-76083258)x3 | hg17 | 5 |
| H177 | del/dup | 13,535/1,714 | 18p11.32p11.21(121700-13656290)x1/18p11.21(13656231-153706830)x3 | hg18 | 4 |
| T074 | del/dup | 2,943/8,234 | 18q23(73139833-76083258)x1/7p22.3p21.3(1490689-9724287)x3 | hg17 | 3 |
| H158 | del/dup | 5,545/20,681 | 18q22.3q23(70565916-76110964)x1/13q31.3q34(93433835-114114568)x3 | hg18 | 2 |
| Y199 | del/dup | 2,477/44,675 | 21q21.1(13926078-16402867)x1/13q11.1q21.31(18650699-63325724)x3 | hg17 | 3 |
| M483 | del/dup | 11,797/1,121 | 21q22.12q22.3(35084120-46880878)x1/21q22.11q22.12(33918888-35040082)x3 | hg18 | 4 |
| M295 | del/dup | 7,830/1,137 | 22q13.2q13.33(41219395-49049035)x1/22q13.2(40003772-41140907)x3 | hg18 | 4 |
| Y001 | del/dup/dup | 35,770/3,767/2,165 | Xp22.33p11.4(2693677-38463601)x1/9q34.13q34.3(134517453-138284752)x3/22q11.2(17539511-19704512)x3 | hg17 | 6 |
| M111 | del/dup | 3,735/61,522 | Xq28(150669991-154405159)x1/7q21.3q36.3(96887100-158409214)x3 | hg18 | 2 |
| T040 | del/dup | 4,020/25,202 | Yq11.222q11.223(19263493-23283748)x1/7q33q36.3(133400864-158602499)x3 | hg17 | 2 |
NCBI, National Center for Biotechnology Information; ISCN, International System for Human Cytogenetic Nomenclature (2009).
We classified the 177 cases with pathogenic CNCs into two groups. The first group had a gain or loss of a single DNA segment (141 of 177, 80%; Table 3). Among them, 46 patients had one of the previously well-known microdeletion/duplication syndromes involving 1p36, 4p16, 7q11, 15q11, 17p11, 17p13, 22q11, and Xp22 region. The remaining cases of the group had recently recognized syndromes or novel pathogenic imbalances, including the imbalances of the16p11.2 region in eight patients (six deletions and two duplications, 0.5% of the total patients) with a size ranging from ∼450 to 660 kb. The second group (36 of 177, 20%) had an imbalance involving two DNA segments except for a single case with more complex alterations (Table 4). These two groups were further divided into six classes: 1) deletion or duplication of a single DNA segment; 2) a deletion of the terminal region of one chromosome and a duplication of the terminal region of another chromosome; 3) a deletion in one chromosome and a duplication in another with at least one interstitial alteration; 4) a deletion and a duplication involving a contiguous genomic region within the same chromosome arm; 5) a deletion and a duplication on different arms of the same chromosome; and 6) others including more complex structural changes and mosaicism (Figure 1). We noted that four of these patients had both a chromosomally visible alteration and a submicroscopic imbalance <4 Mb (cases T036, Y451, T019, and A018; Table 4).
Figure 1.
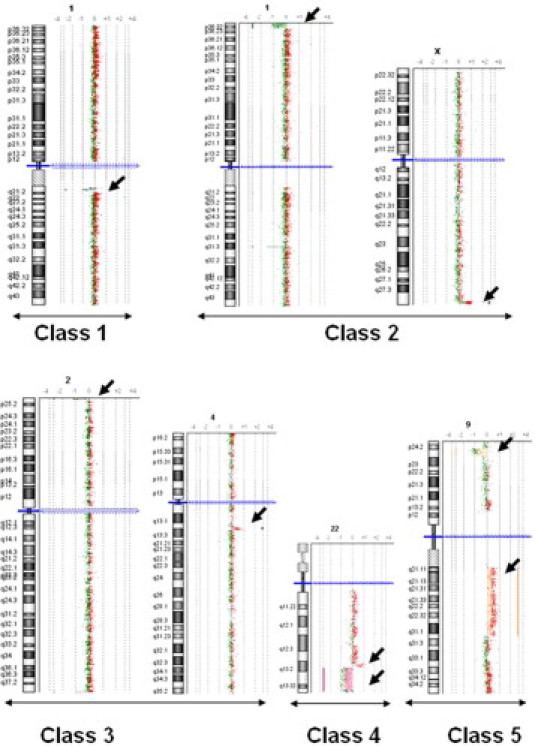
Exemplary array CGH plots of pathogenic CNCs. Class 1: a deletion or duplication of a single DNA segment; class 2: a deletion of the terminal region of one chromosome and a duplication of the terminal region of another chromosome; class 3: a deletion in one chromosome and a duplication in another with at least one interstitial alteration; class 4: a deletion and a duplication involving a contiguous genomic region within the same chromosome arm; class 5: a deletion and a duplication on different arms of the same chromosome; and class 6: represents a heterogeneous group of compound imbalances, not included in this figure.
Discussion
Our results derived from a cohort of 1499 proband patients with MR showed an overall diagnostic yield of 12% (Table 1). With use of the same platform and consensus analytical criteria, a diagnostic yield of 14% was achieved in three of five laboratories. One lab had a diagnostic yield of 11%. A molecular genetics lab had a significantly lower detection rate (7%), most likely due to less stringent referring criteria and exclusion of chromosomal abnormalities. A review5 on the results of a total of 1364 patients collected from 17 published reports, including 973 patients studied with genome-wide BAC arrays (11 reports) and 391 patients studied with genome-wide oligonucleotide arrays (6 reports) showed that oligonucleotide-based arrays appear to give a higher diagnostic yield than the BAC-based arrays (14.83 versus 9.76%). Our results are consistent with the reported data on genome-wide oligonucleotide arrays.
We have categorized the pathogenic imbalances detected in 177 patients into two major groups: one (141 patients) had a simple single DNA segment deletion or duplication that can be terminal or interstitial (Table 3); the second group (36 patients) had compound imbalances involving two or more DNA segments (Table 4). Forty-six patients in the first group had imbalances associated with common microdeletion or microduplication syndromes, and 33 patients in the second group had imbalances involving the terminal regions. All these genomic imbalances (79 of 177, 45%) are detectable by FISH using commercially available probes. In a report on 1500 consecutive patients tested with an array targeting the common microdeletion syndromes and the subtelomeric regions, 5.6% showed clinically relevant genomic alterations.14 A detection rate of 9.8% was achieved when a targeted array was used in conjunction with a low-resolution BAC clone-based genome-wide array in a cohort of 1176 patients.15 Our collective data have clearly demonstrated the diagnostic efficacy of the 44K genome-wide olionucleotide array. Recently, the 44K oligonucleotide array has been customized with enriched probes for the genes or regions known to be associated with phenotypic anomalies.16 The customized array may reveal subtle copy number changes in the targeted regions and therefore enhances detection rate. This platform has been used in one of the five laboratories recently, and deletion of a single gene, UBE3A, has been detected in 1 of 16 patients studied (data not included). A larger study population to assess the clinical utility of the customized 44K platform is desirable.
The diagnostic yield is also determined by other variables such as patient selection, array resolution, and the genomic coverage of the array used. As aCGH is used widely in genetics laboratories, the criteria for patient selection will be less stringent as we have experienced in subtelomere FISH studies in the past years. Arrays have been designed with spatial resolutions ranging from 1 Mb to <1 kb,6 and the latest oligonucleotide array platforms made by major commercial manufacturers have ranged from 13 kb to 700 bp with multiple sets and formats.17,18 The Agilent 44K array used by our laboratories has a spatial resolution of 30 to 35 kb. Previous studies have used oligonucleotide arrays with a resolution of 30 kb, and the vast majority of reported pathogenic CNCs were >500 kb.11,12,19,20 Similarly, our current large cohort has shown that 95.5% of confirmed pathogenic CNCs are >500 kb; 3.95% are 300 to ∼500 kb and a single case (0.56%) with a size <300 kb (Table 2). Our data and the previous reports provide good evidence that a size of 300 kb can be used as a cutoff for further investigations of the clinical relevance of a CNC observed with this platform although a smaller imbalance can be pathogenic. The use of a genome-wide array with an overall spatial resolution <30 kb may increases diagnostic yield but also reveals many more benign CNVs. A reliable database for benign CNVs and software for screening these CNVs appear to be prerequisite for clinically implementing genome-wide arrays with a resolution >30 kb.
The pathogenic CNCs detected in this cohort were unevenly distributed throughout the genome with clusters in 1p36, 4p16, 7pter-q11, 15q12-13, 16p, 17p, 18q, 22q11, and Xp. Approximately 45% of the pathogenic CNCs involved the regions associated with the previously known common microdeletion or microduplication syndromes and the subtelomeric regions. The remaining pathogenic CNCs may represent emerging recurrent syndromes or novel pathogenic genomic imbalances. Of particular interests, we observed two cases (A0024 and M0894; Table 3) with a 1q21.1deletion that has been recently described as a recurrent genomic aberration associated with variable phenotypes.21 We had one case (M0761; Table 3) with a 17q21.3 deletion that was reported as a recurrent microdeletion associated with a common inversion polymorphism.22,23 One case had a deletion of 15q24 (M0028; Table 3) that was previously detected by FISH in several cases.24 Most significantly, we have detected imbalances of the16p11.2 region in eight patients (0.5%) with a size ranging from ∼450 to 660 kb. Deletion of 16p11.2 was reported to be associated with variable phenotypes including MR or DD and autism spectrum disorders.25,26,27 Our results showed that the imbalance of 16p11.2 represents one of the most frequent microdeletion/microduplication syndromes associated with MR or DD.
Approximately 80% of the pathogenic CNCs involved a deletion or duplication of a single DNA segment (Table 3). The remaining 20% had alterations of two DNA segments or occasionally more than two DNA segments or chromosomes (Figure 1). Similar to conventional cytogenetics, a single deletion or duplication could be the result of the segregation of a balanced insertion. A deletion and a duplication in the same patient could be derived from a balanced translocation. The finding of both deletion and duplication on the same chromosome can be caused by an inversion in a parent. For genetic counseling purposes, we have recommended a conventional karyotyping for the parents to rule out a balanced translocation, inversion, or insertion when an alteration was likely to be chromosomally visible or FISH when an alteration involved subtelometic regions or known microdeletion regions. Parental studies are critical in determining the clinical significance of novel CNCs. Classification of the pathogenic CNCs and discussion of the possible mechanism in the report can provide useful information for genetic counseling. On the other hand, our results also shown that a submicroscopic deletion or duplication may exist and contribute to abnormal phenotypes in the cases with structural abnormalities detected with G-banding analysis. Further characterization of the submicroscopic genomic variants may help us to better understand the architecture of the genome such as inversions or presents of low copy number repeats.28
All five laboratories have used the same criteria and process, which are similar to those previously described11,29 for interpreting the clinical relevance of CNCs. However, clinical correlation can be challenges in some patients, such as 1) the inheritance of a CNC may not be determined due to lack of parental samples; 2) a CNC inherited from a normal parent can be pathogenic due to incomplete penetrance or variable expressivity; 3) a small deletion or duplication can be pathogenic when a critical gene is involved; 4) a variant deletion in normal individuals can be pathogenic by unmasking a recessive gene mutation in a patient; and 5) a CNC involving an autosomal dominant gene may not be pathogenic depending on the nature of the function of the gene. For example, one of our patients had a 4.8 Mb of deletion in 18q22.2q23 (M0523; Table 3) with global DD, growth retardation, MR, hypotonia, genitourinary anomalies, and dysmorphic features. The same deletion was found in her mother who had a graduate degree in teaching and music and had no dysmorphic features except for bifid uvula. We reported this deletion as pathogenic on the bases of the gene content and patient's phenotypes, which were consistent with the 18q deletion syndrome. We hypothesized that a mosaicism might be the explanation for the normal mother but could not confirm it. This case has illustrated the complex nature in genotype-phenotypes correlation. The phenotype variability was also described in the patients with 1q21 deletion21,30 and 16p11.2 deletions.26
In summary, our collective data has shown that genome-wide oligonucleotide array Agilent 44K has provided reasonable resolution for clinical use while an array with a higher resolution particularly for the regions containing known disease genes is desirable. Using 300 kb as a cutoff for further investigation of the clinical relevance appears to be a practical approach when a CNC is observed using this platform. We have confirmed that 16p11.2 deletion is a frequent cause of MR or DD. We stress that appropriate interpretation of array CGH results is critical and a discussion on the structural type of a pathogenic CNC is useful for counseling and patient management. Understanding of the complex genotype-phenotype relationship and reviewing currently available literatures on particular genomic alterations are necessary for appropriate clinical correlation of genomic alterations observed.
Footnotes
B.-L.W., P.L., M.M.L., T.-J.C., and Y.-S.F. are senior authors for each group of this collaboration study.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Web Extra Material
JMDI Data Supplement File Sizes
References
- 1.Roeleveld N, Zielhuis GA, Gabreels F. The prevalence of mental retardation: a critical review of recent literature. Dev Med Child Neurol. 1997;39:125–132. doi: 10.1111/j.1469-8749.1997.tb07395.x. [DOI] [PubMed] [Google Scholar]
- 2.Pope A, Tarlov A. Disability in America: Toward a National Agenda for Prevention. Institute of Medicine; Washington, DC: 1991. [Google Scholar]
- 3.Shevell MI, Majnemer A, Morin I. Etiologic yield of cerebral palsy: a contemporary case series. Pediatr Neurol. 2003;28:352–359. doi: 10.1016/s0887-8994(03)00006-7. [DOI] [PubMed] [Google Scholar]
- 4.Shaffer LG, Theisen A, Bejjani BA, Ballif BC, Aylsworth AS, Lim C, McDonald M, Ellison JW, Kostiner D, Saitta S, Shaikh T. The discovery of microdeletion syndromes in the post-genomic era: review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet Med. 2007;9:607–616. doi: 10.1097/gim.0b013e3181484b49. [DOI] [PubMed] [Google Scholar]
- 5.Koolen DA, Pfundt R, de Leeuw N, Hehir-Kwa JY, Nillesen WM, Neefs I, Scheltinga I, Sistermans E, Smeets D, Brunner HG, van Kessel AG, Veltman JA, de Vries BB. Genomic microarrays in mental retardation: a practical workflow for diagnostic applications. Hum Mutat. 2009;30:283–292. doi: 10.1002/humu.20883. [DOI] [PubMed] [Google Scholar]
- 6.Aradhya S, Cherry AM. Array-based comparative genomic hybridization: clinical contexts for targeted and whole-genome designs. Genet Med. 2007;9:553–559. doi: 10.1097/gim.0b013e318149e354. [DOI] [PubMed] [Google Scholar]
- 7.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 8.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 10.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, Reijmersdal S, Nillesen WM, Huys EH, Leeuw N, Smeets D, Sistermans EA, Feuth T, van Ravenswaaij-Arts CM, van Kessel AG, Schoenmakers EF, Brunner HG, Veltman JA. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan YS, Jayakar P, Zhu H, Barbouth D, Sacharow S, Morales A, Carver V, Benke P, Mundy P, Elsas LJ. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007;28:1124–1132. doi: 10.1002/humu.20581. [DOI] [PubMed] [Google Scholar]
- 12.Friedman JM, Baross A, Delaney AD, Ally A, Arbour L, Armstrong L, Asano J, Bailey DK, Barber S, Birch P, Brown-John M, Cao M, Chan S, Charest DL, Farnoud N, Fernandes N, Flibotte S, Go A, Gibson WT, Holt RA, Jones SJ, Kennedy GC, Krzywinski M, Langlois S, Li HI, McGillivray BC, Nayar T, Pugh TJ, Rajcan-Separovic E, Schein JE, Schnerch A, Siddiqui A, Van Allen MI, Wilson G, Yong SL, Zahir F, Eydoux P, Marra MA. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang B, Li A, Valentin D, Nowak NJ, Zhao H, Li P. Analytical and clinical validity of whole-genome oligonucleotide array comparative genomic hybridization for pediatric patients with mental retardation and developmental delay. Am J Med Genet A. 2008;146A:1942–1954. doi: 10.1002/ajmg.a.32411. [DOI] [PubMed] [Google Scholar]
- 14.Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, Bejjani BA. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Pickering DL, Eudy JD, Olney AH, Dave BJ, Golden D, Stevens J, Sanger WG. Array-based comparative genomic hybridization analysis of 1176 consecutive clinical genetics investigations. Genet Med. 2008;10:262–266. doi: 10.1097/GIM.0b013e31816b64ad. [DOI] [PubMed] [Google Scholar]
- 16.Baldwin EL, Lee JY, Blake DM, Bunke BP, Alexander CR, Kogan AL, Ledbetter DH, Martin CL. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genet Med. 2008;10:415–429. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Irons M, Miller DT, Cheung SW, Lip V, Sheng X, Tomaszewicz K, Shao H, Fang H, Tang HS, Irons M, Walsh CA, Platt O, Gusella JF, Wu BL. Development of a focused oligonucleotide-array comparative genomic hybridization chip for clinical diagnosis of genomic imbalance. Clin Chem. 2007;53:2051–2059. doi: 10.1373/clinchem.2007.090290. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Wu BL. Microarray-based genomic DNA profiling technologies in clinical molecular diagnostics. Clin Chem. 2009;55:659–669. doi: 10.1373/clinchem.2008.112821. [DOI] [PubMed] [Google Scholar]
- 19.Wagenstaller J, Spranger S, Lorenz-Depiereux B, Kazmierczak B, Nathrath M, Wahl D, Heye B, Glaser D, Liebscher V, Meitinger T, Strom TM. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am J Hum Genet. 2007;81:768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoyer J, Dreweke A, Becker C, Gohring I, Thiel CT, Peippo MM, Rauch R, Hofbeck M, Trautmann U, Zweier C, Zenker M, Huffmeier U, Kraus C, Ekici AB, Ruschendorf F, Nurnberg P, Reis A, Rauch A. Molecular karyotyping in patients with mental retardation using 100K single-nucleotide polymorphism arrays. J Med Genet. 2007;44:629–636. doi: 10.1136/jmg.2007.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Raber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJ, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BB, Vermeesch JR, Barber JC, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, Porter K, Prigmore E, Krepischi-Santos AC, Varela MC, Koiffmann CP, Lees AJ, Rosenberg C, Firth HV, de Silva R, Carter NP. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 23.Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, Schinzel A, Baumer A, Anderlid BM, Schoumans J, Knoers NV, van Kessel AG, Sistermans EA, Veltman JA, Brunner HG, de Vries BB. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 24.Cushman LJ, Torres-Martinez W, Cherry AM, Manning MA, Abdul-Rahman O, Anderson CE, Punnett HH, Thurston VC, Sweeney D, Vance GH. A report of three patients with an interstitial deletion of chromosome 15q24. Am J Med Genet A. 2005;137:65–71. doi: 10.1002/ajmg.a.30836. [DOI] [PubMed] [Google Scholar]
- 25.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 27.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 28.Lupski JR. Genome structural variation and sporadic disease traits. Nat Genet. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- 29.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39(7 Suppl):S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 30.Ledbetter DH. Cytogenetic technology—genotype and phenotype. N Engl J Med. 2008;359:1728–1730. doi: 10.1056/NEJMe0806570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
JMDI Data Supplement File Sizes


