Abstract
Chronic lymphocytic leukemia (CLL) is the most common leukemia affecting adults in the western world. The clinical course of CLL is highly variable: cases that express mutated immunoglobulin heavy chain variable regions (IgVH) typically have a more indolent clinical course compared with those with unmutated IgVH. The use of the VH3-21 variable region has also been found to confer a poor prognosis, independent of mutation status. Here we describe an assay for the identification of the expressed VH segment and its mutation status in CLL. This test uses whole blood-derived RNA and PCR primers annealing to the leader regions and the joining region segments. This approach allows more accurate determination of the IgVH mutation status relative to using framework region specific VH primers. An additional primer specific for the leader region of the VH3-21 segment is described and is shown to be necessary to identify this diagnostically important variable region. We successfully analyzed 99 of 103 samples, including five expressing the VH3-21 variable region. Approximately 5% of cases had complement determining region 3 sequences similar to previously reported cases, and overrepresentation of the VH1-69 segment was observed among unmutated cases. These results confirm the proper functioning and high success rate of this valuable prognostic for CLL designed for the use in a clinical laboratory setting.
Chronic lymphocytic leukemia (CLL) is a neoplasm of small mature B-cells and the most common leukemia affecting adults in the United States and Europe.1 Almost all cases of CLL express CD5 along with pan-B-cell markers such as CD19 and show other characteristic immunophenotypic features that can be easily identified by flow cytometry analysis.1,2,3 Many patients are asymptomatic in early stage disease at diagnosis and are now often identified through routine blood testing.4 However, the clinical course of CLL is highly variable with many cases behaving indolently with little affect on survival and others behaving aggressively with patients succumbing to their disease after only a few years.2,4 Since traditional staging methods cannot predict the clinical course of disease in patients with early stage CLL, the identification of biological prognostic markers to potentially help guide treatment decisions has assumed increased importance.4,5
One of the earlier biological markers reported to correlate with CLL clinical course was the somatic mutation status of the expressed Ig heavy chain variable region gene segment (VH).6,7 Cases that express mutated VH gene segments (less than 98% homology to the germline counterparts) typically have a more favorable clinical course than those expressing unmutated VH segments (98% or greater homology to the germline segment). Subsequent studies confirmed these findings and also that usage of the VH3-21 gene segment confers a poor prognosis, regardless of the IgVH mutation status.8,9 The basis for the prognostic significance of VH mutational status and expression of VH3-21 is still unclear, but likely relates to signaling differences mediated by surface Ig, the CLL antigen receptor.2,9 The importance of direct antigen receptor stimulation in the development and growth of CLL is also supported by studies revealing preferential use of certain VH gene segments and cases from different patients with nearly identical VH and VL genes, including the highly variable complement determining region 3 (CDR3).9,10,11 Expression of ZAP-70 and CD38 have been found to correlate with the immunoglobulin gene mutation status,7,12,13 and measurement of these markers by flow cytometry is used as a surrogate for IgVH mutation status. Several recurrent cytogenetic abnormalities present in CLL typically identified by fluorescence in situ hybridization have also been shown to have prognostic significance such as presence of deletions in the long arms of chromosomes 13 [del(13q14.10)], 11 [del(11q)], and 6 [del(6q)] and deletions in the short arm of chromosome 17 [del(17p)].14,15,16 However, recent studies have shown that VH mutational status still has prognostic significance in CLL independent of cytogenetic findings.13,17
Here we describe a method for the accurate determination of the identity and mutation status of the IgVH segment expressed in CLL. Our assay has high sensitivity and several features that help with implementation in a routine clinical laboratory.
Materials and Methods
RNA Extraction and RT-PCR Analysis
A total of 103 cases with characteristic CLL immunophenotypes and other features consistent with CLL were obtained from the Associated Regional and University Pathologists Hematological Flow Cytometry laboratory. The research use of these left over specimens was approved by the University of Utah Institutional Review Board, number 11905. The average percentage of neoplastic cells in the patient samples was 88% (range 49% to 98%). Total RNA was prepared from whole blood or cryopreserved white blood cells (WBC) by using the Qiamp RNA Blood Mini Kit (Qiagen, Inc., Valencia, CA). Five microliters of RNA were used to generate random-primed cDNA by using the Superscript III First Strand cDNA Synthesis Kit for RT-PCR (Invitrogen Corp, Carlsbad, CA). For PCR amplification cDNAs were diluted in water to an equivalent of 2 to 10 ng of RNA per microliter. Previously described VH family-specific forward primers that anneal to the leader region and reverse primers that anneal to the JH region (Table 1) were used to amplify rearranged heavy chain variable regions.18,19 A unique primer was also used that perfectly matched the leader region of the VH3-21 gene segment. Twenty-microliter PCR reactions contained 2 μL of diluted cDNA, leader primer(s) (0.2 μmol/L), JH and JH-1 primer (0.2 μmol/L each), deoxynucleoside triphosphates (0.2 mmol/L each), MgCl2 (3 mmol/L), GoTaq Flexi DNA polymerase (1 unit; Promega Corp., Madison, WI), and GoTaq Flexi Green buffer (1X; Promega). The VH1, VH3, and VH4 primers were in separate reactions while VH2, VH5, VH6, and VH3-21 were multiplexed together. After 2 minutes at 94°C, samples were amplified in 30 cycles of 20 seconds at 94°C, 10 seconds at 55°C, and 30 seconds at 72°C, followed by an additional 2 minutes at 72°C and a cool down to 4°C. Five mircoliters of each PCR reaction were run on a 2% agarose gel and visualized with ethidium bromide (0.5 μg/ml) under UV light.
Table 1.
Primers
| Name | Sequence |
|---|---|
| VH1* | 5′-CACCATGGACTGGACCTGGA-3′ |
| VH2 | 5′-ATGGACACACTTTGCTCCAC-3′ |
| VH3 | 5′-CCATGGAGTTTGGGCTGAGC-3′ |
| VH3-21† | 5′-CCATGGAacTgGGGCTccGC-3′ |
| VH4 | 5′-ATGAAACACCTGTGGTTCTT-3′ |
| VH | 5′-ATGGGGTCAACCGCCATCCT-3′ |
| VH6 | 5′-ATGTCTGTCTCCTTCCTCAT-3′ |
| JH | 5′-ACCTGAGGAGACGGTGACCAGGGT-3′ |
| JH-1 | 5′-ACCTGAGGAGACGGTGACC-3′ |
†Mismatches to the VH3 consensus primer are shown in lower case.
Note that in reference 19 this primer has a typo.
DNA Sequence Analysis
The PCR product to be sequenced was cleaned up by incubating 10 μL of the PCR reaction with 2 μL ExoSAP-IT (USB Corp., Cleveland, OH) at 37°C for 45 minutes followed by heat inactivation for 15 minutes at 85°C and a cool down to 4°C. The DNA was then diluted in water and sequenced in both directions with the appropriate leader primer and the JH-1 primer by using BigDye terminator chemistry and the ABI3730 instrument (Applied Biosystems, Inc., Foster City, CA). Forward and reverse sequencing results were aligned, and a consensus sequence was searched against the Immuno Genetics (http://imgt.cines.fr, last accessed December 13, 2009) database of human immunoglobulin sequences with the V-QUEST program.20 The search returns the closest matching germline VH segment and the percentage of sequence identity to it. Insertions and deletions are not taken into consideration in the calculation and must be included manually (see below).
Dilutional Sensitivity Study
The percentages of CLL cells and normal peripheral blood B-cells before dilution were determined by using flow cytometry staining for CD19 and CD5, normal B-cells being CD19+CD5− and CLL cells being CD19+CD5+. Total RNA was prepared from both samples and brought to the same concentration (10 ng/uL). The CLL RNA was then serially twofold diluted in healthy donor RNA followed by cDNA synthesis. On the basis of the flow cytometry data, the hypothetical percentage of CLL cells among total B cells (CD19+) and total WBCs was calculated for each dilution. Rearranged variable gene segments were amplified in the diluted samples and analyzed by agarose gel electrophoresis, and the resultant bands were directly sequenced as described above.
Results
We were able to identify the full length VH segment and determine mutation status in 99 of 103 analyzed CLL cases. The remaining four cases did not yield a PCR band in any of the four reactions possibly related to primer mismatches due to excessive numbers of mutations. VH segments from all families were amplified including those from the VH7 family, which have leader sequences that match the VH1 consensus leader primer (Figure 1). In addition to the dominant bands typically present that were successfully directly sequenced, faint bands corresponding to background B-cells could also be identified in some cases. For several cases, the weak PCR bands were also subjected to DNA sequence analysis but always yielded polyclonal sequences (data not shown). The multiplex reaction for VH2, VH5, and VH6 segments and VH3-21 also worked well in that dominant bands, when present, could all be directly sequenced with multiplexed leader primers. To validate the use of the VH3-21-specific leader primer, four cases expressing VH3-21 and nine cases expressing other VH3 family segments were amplified with the VH3 consensus leader primer and the VH3-21 leader primer. As shown in Figure 2, the VH3-21 cases could only be effectively amplified with the VH3-21 primer and vice versa. Four cases yielded strong PCR bands of the correct size in two PCR reactions, which could both be directly sequenced and corresponded to productively rearranged VH genes. Amplification of VH genes expected to have minor mismatches of one to three nucleotides relative to the consensus leader primers was observed for 16 cases, including some with known mismatches near the 3′ end of the primer (VH2–26, VH3-48).
Figure 1.
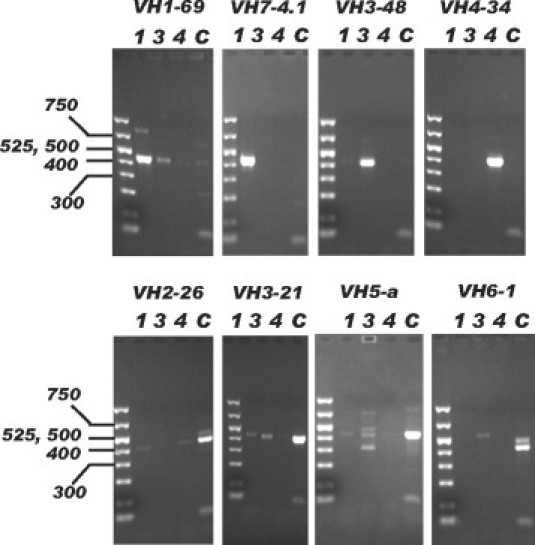
Gel electrophoresis of PCR products. IgVH fragments from eight CLL samples (each positive for a different VH gene family and the VH3-21 segment) were amplified. PCR reactions with the consensus leader primers for the VH1, VH3, or VH4 gene families are labeled 1, 3, and 4, respectively. The multiplexed PCR reaction combining the VH2, VH5, and VH6 consensus leader primers and the VH3-21 leader primer is labeled C. Fragment sizes are indicated in bp.
Figure 2.
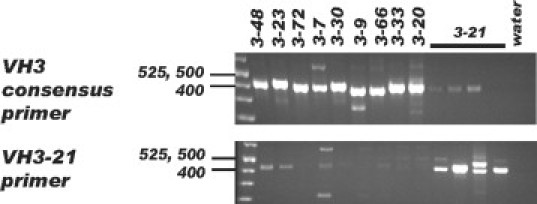
PCR amplification of the VH3-21 variable region. Four CLL cases expressing the VH3-21 segment and 9 cases expressing other VH3 variable regions were amplified with the VH3 consensus leader primer (top) or the VH3-21 specific primer (bottom). Fragment sizes are indicated in bp.
Among the 99 successfully sequenced samples, only 30 of approximately 40 possible functional variable region segments were represented (Figure 3). Ten percent of all cases expressed VH1-69, almost exclusively by those with unmutated VH genes in agreement with previously reported studies.21 Biases in VH segment usage were also observed for VH4-34 and VH4-39 as previously reported.6 In addition, two cases had nearly identical CDR3 sequences (Table 2) that closely resembled CDR3 sequences of other VH3-21 expressing CLL cases reported in the literature.8 Moreover, two of our CLL cases using VH1-69 and one using the VH4-34 gene segment were also found to closely resemble CDR3 sequences of unrelated previously reported CLL cases using the same VH gene segments.10,21
Figure 3.
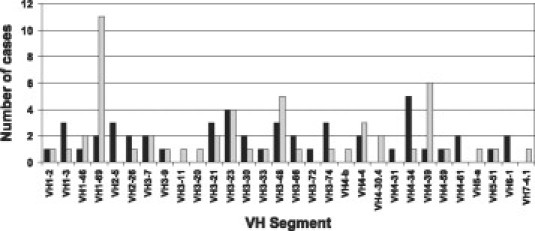
Distribution of VH segment usage. The number of times each VH segment was observed is indicated. The numbers are separated into mutated (black bars) and unmutated (gray bars) cases.
Table 2.
Recurring CDR3 Sequences
| Sample | VH segment | CDR3 sequence | Study |
|---|---|---|---|
| D-57 | VH3-21 | CAVDRNGMDV | |
| E-76 | VH3-21 | ..S....... | |
| 18G | VH3-21 | ..R....... | Thorselius et al8 |
| C-88 | VH1-69 | CATPGSVDIVVVPAAMSYYYYGMDV | |
| FS41 | VH1-69 | ..RG.––.................. | Messmer et al10 |
| C-74 | VH1-69 | CARGGLYDYIWGSYRPNDAFDI | |
| CLL-F | VH1-69 | ...................... | Widhopf et al21 |
| MCLL258 | VH1-69 | .....I...V............ | Messmer et al10 |
| D-49 | VH4-34 | CARGFPDTAVVRRYYYYGMDV | |
| CLL4B | VH4-34 | ...AY...PM........... | Messmer et al10 |
D-57, E-76, C-88, C-74, and C-49 are cases from this study. The references for the other cases are indicated. Identical residues are indicated by dots, missing residues are indicated by dashes.
Approximately half (50) of the successfully analyzed cases showed sequence identities of 98% or greater to the closest matching germline VH segment and were, therefore, considered unmutated. Analysis of the distribution of the number of mutations found among the samples showed a biphasic distribution peaking around 93% to 94% germline sequence identity for the mutated cases and a smaller number of cases with 97% to 98% homology values, close to the 98% cut off separating mutated from unmutated cases (Figure 4). In addition to single point mutations, two cases were identified with in-frame insertions/deletions with respect to the closest germline sequence match. One case expressing VH3-7 had a 3 bp deletion in the CDR2 region, and a case expressing VH3-9 had a 9 bp insertion in the CDR1 region. These in-frame insertion/deletions were counted as one single mutation when calculating the mutation status of the variable region. We estimated that about 5% of cases expressed VH genes with occasional mixed bases of nearly equal intensity in the sequence electropherograms but otherwise unambiguous sequence in the CDR3 portion, consistent with ongoing somatic hypermutation.22 Since these cases were highly mutated, the mixed positions in the electropherogram would not have affected the mutation status. Evidence of ongoing mutation was also observed for a case expressing VH3-21, where a portion of the PCR product had a 3-bp insertion in the CDR2 region.
Figure 4.

Distribution of the observed percentage of germline sequence identity. The calculated percentages of sequence identity to the closest matching germline sequence from 99 CLL samples were combined into bins of 2% ranges. The number of cases falling into each bin is indicated.
The PCR portion of this and other VH mutation tests can amplify expressed variable gene segments not only from the clonal CLL cells but also from other B cells that may be present that may limit the sensitivity of detecting the CLL VH gene by direct sequencing of the PCR product. To examine this issue, dilutional studies using normal polyclonal peripheral blood B-cells were performed by using a CLL case that expressed a VH3-48 segment from the largest VH family with 99.7% identity to the germline sequence. With increasing dilution of the CLL sample, PCR bands also appeared in the VH1, VH4, and multiplexed reactions as expected (Figure 5). Successful identification of the VH gene segment by direct sequence analysis of the PCR product was still possible in the 16-fold dilution where the CLL cells comprised 50% of total B cells. Since the VH3 family is the largest, containing approximately half of the functional VH segments, these results represent a lower limit of our test's sensitivity.
Figure 5.
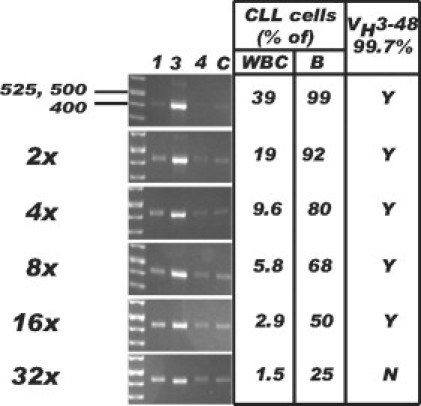
Detection limit by direct sequencing. Gel electrophoresis of PCR products from serial twofold dilutions of a CLL sample into a normal sample (see text for more details) is shown. The fold dilution is indicated on the left. The calculated theoretical concentration of CLL cells in percent among WBC and total B cells (B) is shown on the right. The ability to identify the expected VH3-48 variable region with 99.7% identity to the germline sequence is indicated in the far right column (Y or N). The gel lanes are labeled according to the leader primer used in the PCR reaction: 1, VH1; 3, VH3; 4, VH4; C, VH2, VH3-21, VH5, and VH6 multiplex. DNA fragment length is indicated in bp.
Discussion
We described a method for analysis of immunoglobulin heavy chain variable region genes expressed by CLL cases that gives an accurate determination of the mutational status and is also well suited for use in a clinical laboratory. We used RNA as a starting material because immunoglobulin transcripts are highly expressed in B cells, which enriches the target sequences and enhances RNA-based VH amplification, which may be important in problematic cases. Although RNA-based methods may be more susceptible than DNA-based methods to degradation artifacts, our high rate of success in identifying the expressed VH genes in 99 of 103 tested cases (96%) attests to our method's robustness and high reliability, where the majority of specimens were overnight shipped. Amplification of VH genes with leader primers as we have done has several advantages over using framework region-specific primers, which has also been reported.23,24 First, the complete variable region is obtained allowing the determination of the mutation status with highest accuracy, as pointed out by the European Research Initiative on CLL.25 Second, leader primers are mostly family specific and amplify only a subset of possible variable gene segments, thereby lowering possible polyclonal background signals and increasing the sensitivity of the test if not excessively multiplexed. Third, somatic hypermutation may occur less frequently in the 5′ leader region26 ensuring a higher success rate when amplifying heavily mutated cases. Studies suggest, however, that identifying CLL VH gene segments through amplification of cDNA with leader primers shows a high degree of concordance relative to amplification of DNA with the BIOMED-2 framework 1 region primers.23
Several features of our assay were introduced to specifically help facilitate performance and implementation in a clinical lab. First, a consensus reverse primer annealing to all six JH segments was used in all reactions along with an additional JH primer with extended homology to four of the six segments (JH1, JH2, JH4, and JH5) to better handle somatic mutations that could be present in the JH segment. The annealing temperature was also kept low at 55°C to prevent PCR failure due to the presence of mutations. An extra leader primer designed to be specific for the VH3-21 gene was used. This was done because the VH3-21 gene is mismatched at five locations relative to the standard VH3 leader primer. In addition, we showed that this additional primer is necessary even with our low annealing temperature of 55°C because the standard VH3 leader primer failed to effectively amplify any of four VH3-21 expressing cases tested. Moreover, the VH3-21 leader primer is sufficiently different from other VH3 family segments, so that they were not amplified with this primer. It is interesting that the VH3-21 gene has been identified by using the standard VH3 leader primer in other studies of CLL cases,6,23 but, due to the primer mismatches we have described, may be underdetected relative to its true frequency of use. Expression of VH3-21 is particularly important to identify because cases of CLL using this VH segment have a very poor prognosis regardless if mutated or unmutated.8,9 Based on the leader region sequences published in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov, last accessed December 13, 2009), only minor mismatches exist between the other standard consensus leader primers and their respective target sequences making more additions to the original set unnecessary. As such, all variable gene segments should be analyzable by our assay, which is further supported by our finding 16 cases expected to have primer mismatches and other studies using leader primers.6,7
The VH1, VH3, and VH4 families, which contain approximately 90% of the functional VH gene segments, were amplified in separate reactions with individual leader primers. This was done to help minimize co-amplification of background non-CLL B-cells that may be present if these primers were multiplexed together, which could complicate direct sequence analysis of the PCR product in some cases. However, to help keep the number of separate reactions more manageable, primers for the VH2, VH5, and VH6 families, along with the VH3-21 leader primer, were multiplexed in a single reaction. Doing this did not interfere with our ability to directly sequence any of the 16 PCR products generated with this combination because these genes are only infrequently used by normal B-cells.
For testing of clinical samples, we also ran a control reaction with three forward FW1 primers that cover the VH1, VH3, and VH4 families to ensure rearranged VH genes can be identified, as well as a no template control reaction in addition to the four leader primer reactions described above. Running six separate reactions that we have outlined above can be easily performed, and also insures high initial sensitivity to minimize the necessity of running additional follow-up or repeat reactions. Typically, only one leader primer reaction generates a predominant band that is then subjected to direct sequencing. The occasional faint bands that were also present in some cases could not be directly sequenced indicating they were nonclonal. However, four cases yielded two predominant PCR bands that generated easily readable sequence on direct sequencing, corresponding to two different clones expressing different VH, DH, and JH gene segments. These cases could represent examples of bi-allelic VH gene expression and lack of allelic exclusion, reported to occur in approximately 5% of CLL cases.27 However, we cannot exclude the possibility that one of the two VH genes in these cases has stop codons located in areas we did not analyze, eg, the end of the JH region. Biclonal CLL cases, which have also been found to occur, could be an alternative possibility.28 However, the possibility that one of the two VH genes represents a small reactive clone is unlikely because flow cytometry studies demonstrated that 80% of the leukocytes or 90% of the lymphocytes were B-cells with characteristic monotypic CLL phenotypes without other phenotypically distinct CD5−, CD19+ B-cell populations identified. Southern blot heavy chain analysis (not shown) further supported two of these cases having biallelic rearrangements as the two nongermline bands identified were of comparable intensity for all three enzymes used.
Approximately half of our cases were mutated, with an average mutation load close to 94%, similar to other studies.6 It has recently been suggested that the cut-off value of 98% germline sequence identity for discrimination between mutated and unmutated cases may not be adequate because there may be a continuum of worsening prognoses for cases showing between 97% and 100% germline sequence identity.29 We, therefore, flag those cases in the 97% sequence identity range as potentially borderline with poorer prognosis. Some of the few mutations observed in the 98% to 99% range may be due to allelic differences between the patient and sequences in the database queried. It is, therefore, of importance to use a comprehensive database of germline VH sequences for this test. In our hands the Immuno Genetics database returned a larger number of cases with 100% germline sequence identity than other databases, as also reported by others.30
The overrepresentation of specific gene segments among unmutated and mutated cases we and others have observed, in addition to recurrent CDR3 sequences found in this and other studies, highlights the importance of immunoglobulin mediated stimulation in the development of CLL. However, the restricted VH gene repertoire of CLL does not simplify clinical laboratory analysis of VH gene segment usage and mutational status from what we have described in this study.
In summary, we described a sensitive method for VH gene analysis of CLL cases that can be easily implemented in a clinical laboratory and provides important prognostic information. Although more expensive and involved than flow cytometric analysis of ZAP-70, flow analysis of ZAP-70 is problematic and nonstandardized, and therefore not necessarily reliable or accurate.31 Moreover, the relative ease of sequencing short PCR products in modern clinical laboratories and lower cost compared with several years ago have removed much of the perceived necessity for using surrogate markers of VH mutational status.
Footnotes
Supported by the Associated Regional and University Pathologists Institute for Clinical and Experimental Pathology, LLC, and NIH grant 5R21DE017136 (to D.W.B.).
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. [Google Scholar]
- 2.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 3.Ho AK, Hill S, Preobrazhensky SN, Miller ME, Chen Z, Bahler DW. Small B-cell neoplasms with typical mantle cell lymphoma immunophenotypes often include chronic lymphocytic leukemias. Am J Clin Pathol. 2009;131:27–32. doi: 10.1309/AJCPPAG4VR4IPGHZ. [DOI] [PubMed] [Google Scholar]
- 4.Shanafelt TD, Geyer SM, Kay NE. Prognosis at diagnosis: integrating molecular biologic insights into clinical practice for patients with CLL. Blood. 2004;103:1202–1210. doi: 10.1182/blood-2003-07-2281. [DOI] [PubMed] [Google Scholar]
- 5.Binet JL, Caligaris-Cappio F, Catovsky D, Cheson B, Davis T, Dighiero G, Dohner H, Hallek M, Hillmen P, Keating M, Montserrat E, Kipps TJ, Rai K. Perspectives on the use of new diagnostic tools in the treatment of chronic lymphocytic leukemia. Blood. 2006;107:859–861. doi: 10.1182/blood-2005-04-1677. [DOI] [PubMed] [Google Scholar]
- 6.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 7.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 8.Thorselius M, Krober A, Murray F, Thunberg U, Tobin G, Buhler A, Kienle D, Albesiano E, Maffei R, Dao-Ung LP, Wiley J, Vilpo J, Laurell A, Merup M, Roos G, Karlsson K, Chiorazzi N, Marasca R, Dohner H, Stilgenbauer S, Rosenquist R. Strikingly homologous immunoglobulin gene rearrangements and poor outcome in VH3-21-using chronic lymphocytic leukemia patients independent of geographic origin and mutational status. Blood. 2006;107:2889–2894. doi: 10.1182/blood-2005-06-2227. [DOI] [PubMed] [Google Scholar]
- 9.Ghia EM, Jain S, Widhopf GF, 2nd, Rassenti LZ, Keating MJ, Wierda WG, Gribben JG, Brown JR, Rai KR, Byrd JC, Kay NE, Greaves AW, Kipps TJ. Use of IGHV3-21 in chronic lymphocytic leukemia is associated with high-risk disease and reflects antigen-driven, post-germinal center leukemogenic selection. Blood. 2008;111:5101–5108. doi: 10.1182/blood-2007-12-130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messmer BT, Albesiano E, Efremov DG, Ghiotto F, Allen SL, Kolitz J, Foa R, Damle RN, Fais F, Messmer D, Rai KR, Ferrarini M, Chiorazzi N. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200:519–525. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T, Belhoul L, Stella S, Stavroyianni N, Crespo M, Hadzidimitriou A, Sutton L, Bosch F, Laoutaris N, Anagnostopoulos A, Montserrat E, Fassas A, Dighiero G, Caligaris-Cappio F, Merle-Beral H, Ghia P, Davi F. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood. 2007;109:259–270. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 12.Orchard JA, Ibbotson RE, Davis Z, Wiestner A, Rosenwald A, Thomas PW, Hamblin TJ, Staudt LM, Oscier DG. ZAP-70 expression and prognosis in chronic lymphocytic leukaemia. Lancet. 2004;363:105–111. doi: 10.1016/S0140-6736(03)15260-9. [DOI] [PubMed] [Google Scholar]
- 13.Rassenti LZ, Jain S, Keating MJ, Wierda WG, Grever MR, Byrd JC, Kay NE, Brown JR, Gribben JG, Neuberg DS, He F, Greaves AW, Rai KR, Kipps TJ. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 15.Mayr C, Speicher MR, Kofler DM, Buhmann R, Strehl J, Busch R, Hallek M, Wendtner CM. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107:742–751. doi: 10.1182/blood-2005-05-2093. [DOI] [PubMed] [Google Scholar]
- 16.Shanafelt TD, Witzig TE, Fink SR, Jenkins RB, Paternoster SF, Smoley SA, Stockero KJ, Nast DM, Flynn HC, Tschumper RC, Geyer S, Zent CS, Call TG, Jelinek DF, Kay NE, Dewald GW. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 17.Lin KI, Tam CS, Keating MJ, Wierda WG, O'Brien S, Lerner S, Coombes KR, Schlette E, Ferrajoli A, Barron LL, Kipps TJ, Rassenti L, Faderl S, Kantarjian H, Abruzzo LV. Relevance of the immunoglobulin VH somatic mutation status in patients with chronic lymphocytic leukemia treated with fludarabine, cyclophosphamide, and rituximab (FCR) or related chemoimmunotherapy regimens. Blood. 2009;113:3168–3171. doi: 10.1182/blood-2008-10-184853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell MJ, Zelenetz AD, Levy S, Levy R. Use of family specific leader region primers for PCR amplification of the human heavy chain variable region gene repertoire. Mol Immunol. 1992;29:193–203. doi: 10.1016/0161-5890(92)90100-c. [DOI] [PubMed] [Google Scholar]
- 19.Bahler DW, Miklos JA, Swerdlow SH. Ongoing Ig gene hypermutation in salivary gland mucosa-associated lymphoid tissue-type lymphomas. Blood. 1997;89:3335–3344. [PubMed] [Google Scholar]
- 20.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widhopf GF, 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 2004;104:2499–2504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 22.Gurrieri C, McGuire P, Zan H, Yan XJ, Cerutti A, Albesiano E, Allen SL, Vinciguerra V, Rai KR, Ferrarini M, Casali P, Chiorazzi N. Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification. J Exp Med. 2002;196:629–639. doi: 10.1084/jem.20011693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marasca R, Maffei R, Morselli M, Zucchini P, Castelli I, Martinelli S, Fontana M, Ravanetti S, Curotti M, Leonardi G, Cagossi K, Partesotti G, Torelli G. Immunoglobulin mutational status detected through single-round amplification of partial V(H) region represents a good prognostic marker for clinical outcome in chronic lymphocytic leukemia. J Mol Diagn. 2005;7:566–574. doi: 10.1016/S1525-1578(10)60589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews C, Catherwood M, Morris TC, Alexander HD. Routine analysis of IgVH mutational status in CLL patients using BIOMED-2 standardized primers and protocols. Leuk Lymphoma. 2004;45:1899–1904. doi: 10.1080/10428190410001710812. [DOI] [PubMed] [Google Scholar]
- 25.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer S, Stevenson F, Davi F, Rosenquist R. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia. 2007;21:1–3. doi: 10.1038/sj.leu.2404457. [DOI] [PubMed] [Google Scholar]
- 26.Longerich S, Tanaka A, Bozek G, Nicolae D, Storb U. The very 5′ end and the constant region of Ig genes are spared from somatic mutation because AID does not access these regions. J Exp Med. 2005;202:1443–1454. doi: 10.1084/jem.20051604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rassenti LZ, Kipps TJ. Lack of allelic exclusion in B cell chronic lymphocytic leukemia. J Exp Med. 1997;185:1435–1445. doi: 10.1084/jem.185.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsi ED, Hoeltge G, Tubbs RR. Biclonal chronic lymphocytic leukemia. Am J Clin Pathol. 2000;113:798–804. doi: 10.1309/V8AN-A2XP-7TDV-HR0T. [DOI] [PubMed] [Google Scholar]
- 29.Hamblin TJ, Davis ZA, Oscier DG. Determination of how many immunoglobulin variable region heavy chain mutations are allowable in unmutated chronic lymphocytic leukaemia: long-term follow up of patients with different percentages of mutations. Br J Haematol. 2008;140:320–323. doi: 10.1111/j.1365-2141.2007.06928.x. [DOI] [PubMed] [Google Scholar]
- 30.Pekova S, Baran-Marszak F, Schwarz J, Matoska V. Mutated or non-mutated? Which database to choose when determining the IgVH hypermutation status in chronic lymphocytic leukemia? Haematologica. 2006;91:ELT01. [PubMed] [Google Scholar]
- 31.Chen YH, Peterson L, Dittmann D, Evens A, Rosen S, Khoong A, Shankey TV, Forman M. Comparative analysis of flow cytometric techniques in assessment of ZAP-70 expression in relation to IgVH mutational status in chronic lymphocytic leukemia. Am J Clin Pathol. 2007;127:182–191. doi: 10.1309/230199FLE32ATUB0. [DOI] [PubMed] [Google Scholar]


