Abstract
Recent evidence indicates that the presence of epidermal growth factor receptor (EGFR) or KRAS mutations in non-small cell lung cancer (NSCLC) can predict the response of the tumor to gefinitib. However, it is difficult to detect these mutations using formalin-fixed, paraffin-embedded (FFPE) tissues because the fixation process and aging can damage the DNA. In this study, we describe our work in adapting the Smart Amplification Process version 2 (SmartAmp2) to detect EGFR or KRAS mutations in DNA extracted from FFPE tissues. We were able to detect these mutations in 37 (97%) of 38 FFPE lung cancer tissue samples within 60 minutes with the SmartAmp2 assay and to confirm the correlation between EGFR mutations in FFPE tissues and gefitinib responsiveness. All mutations had previously been confirmed in the 38 samples using DNA extracted from frozen tissues. Electrophoresis results indicated that PCR analysis was not reliable for DNA extracted from FFPE tissue when primers with a long amplicon (>300 bp) were used. This study confirms that the SmartAmp2 assay is suitable for use with DNA extracted from FFPE as well as frozen tissues.
Lung cancer, which is responsible for 1.18 million deaths annually worldwide, is the most common cause of cancer mortality in men and the second most common cause in women.1 Treatment involves a combination of surgery, chemotherapy, and radiation therapy determined based on histological results obtained in biopsy of cancer cells from the individual patient. Despite the greater availability of treatment and substantial research efforts, the prognosis for lung cancer remains poor, and the development of more effective treatments is one of the most important topics in oncology today.
Recent studies have indicated that mutations in the epidermal growth factor receptor (EGFR) gene and KRAS gene help physicians decide the course of chemotherapy in patients with non-small cell lung cancer (NSCLC). EGFR mutations2,3,4,5 and KRAS mutations6,7,8,9,10 occur in 8 to 10% and 33% of NSCLC patients, respectively, and in 27 to 56% and 5 to 15% of East Asian NSCLC patients, respectively. They are negatively correlated in NSCLC such that patients who have a mutation in the tyrosine kinase domain of the EGFR respond to tyrosine kinase inhibitors such as gefitinib and erlotinib, whereas patients with mutations of the KRAS gene do not respond to this treatment.3,9,11 Consequently, NSCLC patients with EGFR mutations have a favorable prognosis,12,13 whereas the prognosis for those with KRAS mutations is poor.14,15,16,17 Therefore, to provide the optimal therapy, physicians must be able to determine whether patients have EGFR or KRAS gene mutations.
Many methods used to detect EGFR or KRAS mutations in clinical samples include restriction fragment length polymorphism,18 single-strand conformation polymorphism,19 PCR sequencing,20 high-resolution melting analysis,21,22 and Scorpions Amplified Refractory Mutation System.23 All of these methods require careful DNA extraction and purification, involve many steps, and must be performed by skilled technicians. Some of these methods are more sensitive than simple sequencing but are unsuitable for routine clinical use because of their complexity and long turnaround times.
Recently, Mitani et al24 developed a rapid, simple, and sensitive mutation detection assay called the Smart Amplification Process version 2 (SmartAmp2). This assay has shown the ability to detect mutations in samples containing as little as 1% mutant allele.25,26 This assay can be used in the clinical setting, and it allows for the detection of EGFR and KRAS gene mutations within 60 minutes (including sample preparation) and enables high-throughput screening.
There are a number of archival formalin-fixed, paraffin-embedded (FFPE) tissue banks worldwide. FFPE tissue is relatively cheap, is easy to ship and handle, provides superior morphological quality, and is compatible with nearly all relevant immunohistochemical antibodies. Consequently, most surgical specimens are stored in FFPE tissue for later analysis of gene mutations if necessary. However, it is time-consuming to extract DNA from FFPE tissue and often difficult to detect mutations because the fixation process and aging can damage DNA.20,27 The present study demonstrates a technique for adapting the SmartAmp2 method to detect mutations from FFPE tissue. The procedure can detect mutations with high accuracy, and unlike any other method, it gives a reliable diagnostic result based exclusively on amplification. The SmartAmp2 assay provides reliable information from old specimens and marks a major advance in cancer diagnostics.
Materials and Methods
Clinical Samples
Tumor samples surgically resected from NSCLC patients at the Gunma University Hospital (Gunma, Japan) between 2003 and 2007 were used. Institutional approval and informed consent from all patients was obtained in writing. All tumor tissue was diagnosed for lung cancer by H&E stain. After surgical removal, a portion of each sample was immediately frozen and stored at −80°C until DNA extraction; the remainder was preserved in paraffin blocks after formalin fixation.
Sample Selection
A total of 43 samples that were available in both frozen and FFPE form were selected for the study. The SmartAmp2 assay was used to determine that both EGFR and KRAS gene mutations were present in the frozen tissue. Previous examination with a microscope confirmed that each FFPE tissue sample contained a sufficient number of tumor cells for analysis.
DNA Extraction
To suppress the tumor heterogeneity and to obtain a sufficient number of tumor cells, thin sections sliced from the tumor at the surface with maximum diameter were selected and cut into small pieces. DNA was extracted from a 3- to 5-mm cube collected from the small pieces using a DNA Mini Kit (Qiagen, Hilden, Germany), and the solution was serially diluted to a concentration of 20 ng/μl. For each tumor, the FFPE block with the maximum number of tumor-rich areas was selected and sliced into three 5-μm-thick sections. The tumor area of the section was macrodissected, and DNA was extracted using a QIAamp DNA FFPE Tissue Kit (Qiagen). The extracted solution was not diluted. We added RNase during the DNA extraction, although this was an optional step according to the protocol included with the kit. We obtained concentrations of at least 40 ng/μl in all of the extracted solutions. Although the concentration was lower for the first extraction of some specimens, we extracted again using more sections until the concentration reached 40 ng/μl. After extraction, all DNA templates were stored at −20°C until use.
SmartAmp2 Assay
The SmartAmp2 assay has been described previously.25,26 It is the first one-step mutation–detection technology to enable the precise amplification of only target sequences. Using a new DNA polymerase (Aac pol) and a unique five-set primer design, we performed rapid and sensitive assays under isothermal conditions (Figure 1A).24 The assays were performed in parallel sets using the same template, with one assay detecting the wild-type sequence, and the other detecting the mutant sequence. Detection of the wild-type sequence served as a positive control, allowing us to distinguish between assay failure and a true negative result.
Figure 1.
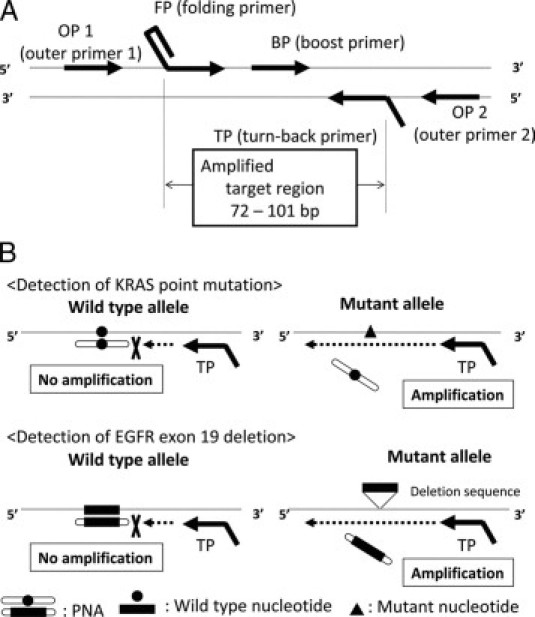
A: Amplification by the SmartAmp2 assay requires five unique primers: turn-back primer (TP), folding primer (FP), boost primer (BP), and two outer primers (OP1 and OP2). The genomic sequence between and including the TP and FP primers is the target region to be amplified in the SmartAmp2 assay. B: SmartAmp2 assay PNA clamp. The PNA clamp competitive probe is designed for the wild-type allele sequence. The greater stability of the PNA probe in hybridization inhibits SmartAmp2 amplification and suppresses wild-type allele amplification. Amplification of the mutant allele is not inhibited by PNA regardless of the point mutation (KRAS) or deletion (EGFR exon 19).
In the present study, we used two versions of the SmartAmp2 assay, conventional and peptide nucleic acid (PNA) clamp. In the conventional SmartAmp2 assay, the target sequence was detected by primer genotyping. When the primer sequence was completely complementary to the target sequence, an amplified product was produced. Therefore, we needed two different primer sets to detect wild-type alleles and mutant alleles using this method. For the PNA clamp SmartAmp2 assay, we used PNA and a single primer set which was designed to amplify the target sequence in both the wild-type and mutant allele. The PNA was exactly homologous to the wild-type allele. However, because PNA is not recognized by the polymerase as natural DNA, it cannot serve as a primer for polymerization or as a substrate for Taq polymerase exonuclease activity. In addition, the melting temperature of a perfectly matched PNA-DNA duplex is higher than that of DNA-DNA of the same length, although a single mismatch destabilizes the PNA-DNA hybrids, causing a melting temperature shift of 10 to 18°C. The greater stability of the hybridized PNA probe inhibits the SmartAmp2 amplification; thus, amplification of the wild-type allele is suppressed (Figure 1B). When the assay is performed with both the primer set and PNA, any mutant sequences in the template are amplified. All template sequences are amplified when using the same primer set without PNA, which served as a control. Conventional SmartAmp2 was used to detect some EGFR deletions and the L858R mutation (sample numbers 1–13 and 19–30) and PNA clamp SmartAmp2 was used to detect other EGFR deletions and KRAS mutations (sample numbers 14–18 and 31–38). In this article, “SmartAmp2” refers to both assay methods. The mutation detection kits were from K.K. DNAFORM (Yokohama, Japan). The SmartAmp2 assay was used to detect mutations in both frozen and FFPE tissues.
PCR and Electrophoresis
PCR was performed on DNA extracted from both frozen and FFPE tissues. We used several sets of primers to amplify the EGFR and KRAS gene mutations. Each primer was designed to produce various amplicon lengths, including that of the mutation site (Table 1). Although the PCR primer sequences for EGFR and KRAS amplification were used previously,20,21,27,28,29,30 the other experimental conditions were unique to this study. The PCRs were performed in a total volume of 25 μl containing 10× PCR Gold Buffer, 1.5 mmol/L MgCl2, 200 μmol/L deoxyribonucleotide triphosphates, 500 nmol/L each primer, 1 U of TaqDNA Gold Polymerase (Applied Biosystems Japan, Tokyo, Japan), and 2 μl of extracted genomic DNA. The thermal cycling regimen was as follows: 5 minutes at 94°C, followed by 35 cycles at 94°C for 15 s, annealing for 30 s, 72°C for 1 minute, and 1 cycle of 72°C for 5 minutes. Annealing temperatures were based on the results of the PCR gradient in a pilot study (see Table 1).
Table 1.
PCR and Sequencing Primers for the EGFR and KRAS Genes
| Exon | Primer name | Forward primer | Reverse primer | Amplicon length (bp) | Annealing temperature (°C) | Refs. |
|---|---|---|---|---|---|---|
| EGFR | 19-1 | 5′-ACCATCTCACAATTGCCAGTTAAC-3′ | 5′-GAGGTTCAGAGCCATGGACC-3′ | 192 | 60 | 28 |
| Exon19 | 19-2 | 5′-CCAGATCACTGGGCAGCATGTGGCACC-3′ | 5′-AGCAGGGTCTAGAGCAGAGCAGCTGCC-3′ | 265 | 60 | 27 |
| 19-3 | 5′-CCTTAGGTGCGGCTCCACAGC-3′ | 5′-CATTTAGGATGTGGAGATGAGC-3′ | 349 | 62 | 20 | |
| EGFR | 21-1 | 5′-TCACAGCAGGGTCTTCTCTGTTT-3′ | 5′-ATGCTGGCTGACCTAAAGCC-3′ | 212 | 61 | 28 |
| Exon21 | 21-2 | 5′-TCAGAGCCTGGCATGAACATGACCCTG-3′ | 5′-GGTCCCTGGTGTCAGGAAAATGCTGG-3′ | 297 | 62 | 27 |
| 21-3 | 5′-CAGCCATAAGTCCTCGACGTGG-3′ | 5′-CATCCTCCCCTGCATGTGTTAAAC-3′ | 374 | 60 | 20 | |
| KRAS | KRAS-1 | 5′-TCATTATTTTTATTATAAGGCCTGCTGAA-3′ | 5′-CAAAGACTGGTCCTGCACCAGTA-3′ | 189 | 59 | 21 |
| KRAS-2 | 5′-ACTGGTGGAGTATTTGATAG-3′ | 5′-ACTCATGAAAATGGTCAGAG-3′ | 288 | 59 | 29 | |
| KRAS-3 | 5′-TGAAGTACAGTTCATTACGATACACG-3′ | 5′-GGAAAGTAAAGTTCCCATATTAATGGT-3′ | 499 | 58 | 30 |
Annealing temperature in PCR thermal cycle is indicated as above.
The amplicon length and annealing temperature used in the PCR thermal cycle are indicated.
The PCR products of all primers were confirmed by electrophoresis in 2% agarose gel containing ethidium bromide.
Sequencing Analysis
The PCR products generated from FFPE tissues were purified using the QIAquick PCR Purification Kit (Qiagen) and processed for DNA sequencing reaction using ABI PRISM BigDye Terminator version 3.1 (Applied Biosystems Japan) with a forward primer of EGFR 19-1, EGFR 21-1, and KRAS-1 (Table 1). Sequence data were generated using the ABI PRISM 3100 DNA Analyzer (Applied Biosystems Japan).
The presence of mutations in the frozen tissue was confirmed by direct sequencing and PNA-enriched sequencing in our previous study. The PNA-enriched sequencing method has been described previously.31,32,33,34
Results
EGFR and KRAS Mutation Detection Using the SmartAmp2 Assay and Direct Sequencing
We performed the SmartAmp2 assay on 43 DNA samples extracted from both frozen and FFPE tissues. In the frozen tissue, 38 mutations were detected: 18 samples had EGFR mutations in exon 19, 12 samples had EGFR mutations in exon 21, and 8 samples had KRAS mutations in exon 2. No mutations were detected in five samples (Table 2). The presence of these mutations was previously examined by PNA-enriched sequencing, and the results were completely concordant with SmartAmp2 (data not shown). In DNA extracted from the FFPE tissue, we used the SmartAmp2 assay to detect 37 of the 38 mutations (97%): 18 samples had EGFR mutations in exon 19, 11 had EGFR mutations in exon 21, and 8 had KRAS mutations in exon 2. No mutations were detected in six samples (Table 2). The L858R mutation could not be detected in one FFPE tissue (sample 25). A typical SmartAmp2 assay is shown in Figure 2. The results from all of the SmartAmp2 assays were obtained within 60 minutes. Several samples of wild-type lung cancer DNA were extracted from FFPE tissue and assayed in concentrations ranging from 20 to 200 ng/μl; no false-positive results were observed.
Table 2.
Results of SmartAmp2 Assay, Electrophoresis, and Direct Sequencing
| DNA extracted from frozen sample as templates |
DNA extracted from FFPE sample as templates |
|||||||
|---|---|---|---|---|---|---|---|---|
| Sample no. | SmartAmp2 mutation site and type | Visual band by any primers | Direct sequencing | SmartAmp2 | Visual band | Direct sequencing | ||
| Short | Medium | Long | ||||||
| 1 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Mut | Mut | + | − | − | Mut |
| 2 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Wt | Mut | + | − | − | Mut |
| 3 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Mut | Mut | + | + | + | Mut |
| 4 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Mut | Mut | + | − | − | Wt |
| 5 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Mut | Mut | + | − | − | Mut |
| 6 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Mut | Mut | + | + | − | Mut |
| 7 | EGFR Ex 19 E746-A750 del (c.2235-2249del15) | + | Mut | Mut | + | + | − | Wt |
| 8 | EGFR Ex 19 E746-A750 del (c.2236-2250del15) | + | Wt | Mut | + | − | − | Wt |
| 9 | EGFR Ex 19 E746-A750 del (c.2236-2250del15) | + | Mut | Mut | + | − | − | Mut |
| 10 | EGFR Ex 19 E746-A750 del (c.2236-2250del15) | + | Mut | Mut | + | − | − | Mut |
| 11 | EGFR Ex 19 E746-A750 del (c.2236-2250del15) | + | Wt | Mut | + | − | − | Wt |
| 12 | EGFR Ex 19 E746-A750 del (c.2236-2250del15) | + | Wt | Mut | + | − | − | Mut |
| 13 | EGFR Ex 19 E746-A750 del (c.2236-2250del15) | + | Mut | Mut | + | − | − | Mut |
| 14 | EGFR Ex 19 T747-T751 del | + | Wt | Mut | + | − | − | Mut |
| 15 | EGFR Ex 19 L747-E749 del, A750P | + | Mut | Mut | − | − | − | Wt |
| 16 | EGFR Ex 19 L747-E749 del, A750P | + | Mut | Mut | + | − | − | Wt |
| 17 | EGFR Ex 19 E746-E749, S752-P753 del | + | Wt | Mut | + | − | − | Mut |
| 18 | EGFR Ex 19 L747-K754 del, K754N | + | Mut | Mut | + | + | − | Mut |
| 19 | EGFR Ex 21L858R | + | Wt | Mut | + | − | − | Wt |
| 20 | EGFR Ex 21L858R | + | Wt | Mut | + | − | − | Wt |
| 21 | EGFR Ex 21L858R | + | Mut | Mut | + | − | − | Mut |
| 22 | EGFR Ex 21L858R | + | Wt | Mut | + | − | − | Mut |
| 23 | EGFR Ex 21L858R | + | Mut | Mut | + | − | − | Wt |
| 24 | EGFR Ex 21L858R | + | Mut | Mut | + | + | − | Wt |
| 25 | EGFR Ex 21L858R | + | Wt | Wt | + | + | + | Wt |
| 26 | EGFR Ex 21L858R | + | Wt | Mut | + | − | − | Mut |
| 27 | EGFR Ex 21L858R | + | Mut | Mut | + | − | − | Wt |
| 28 | EGFR Ex 21L858R | + | Mut | Mut | + | − | − | Wt |
| 29 | EGFR Ex 21L858R | + | Wt | Mut | + | − | − | Wt |
| 30 | EGFR Ex 21L858R | + | Wt | Mut | + | − | − | Wt |
| 31 | KRAS Ex2 G12V | + | Wt | Mut | + | − | − | Wt |
| 32 | KRAS Ex2 G12C | + | Wt | Mut | + | − | − | Wt |
| 33 | KRAS Ex2 G12A | + | Wt | Mut | + | − | − | Wt |
| 34 | KRAS Ex2 G12V | + | Mut | Mut | + | − | − | Wt |
| 35 | KRAS Ex2 G12V | + | Mut | Mut | + | + | − | Wt |
| 36 | KRAS Ex2 G12V | + | Mut | Mut | + | − | − | Mut |
| 37 | KRAS Ex2 G12F | + | Mut | Mut | + | − | − | Wt |
| 38 | KRAS Ex2 G12V | + | Mut | Mut | + | − | − | Mut |
| 39 | Wt | + | Wt | Wt | + | − | − | Wt |
| 40 | Wt | + | Wt | Wt | + | + | − | Wt |
| 41 | Wt | + | Wt | Wt | + | − | − | Wt |
| 42 | Wt | + | Wt | Wt | + | + | − | Wt |
| 43 | Wt | + | Wt | Wt | + | − | − | Wt |
Wt, wild type; Mut, mutant type.
PCR was performed using primers that produced each length of amplicon to amplify the sequence, including that of the suspected mutations (see Table 1). We defined three primers for each exon as “short,” “medium,” or “long” in ascending order of amplicon size. PCR was performed using all primers for samples 39–43.
+, PCR product of a primer produced a visual band in the electrophoresis gel; −, no visual band was detected.
Figure 2.
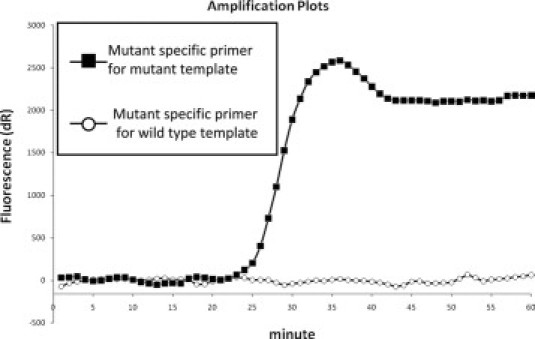
A typical SmartAmp2 assay result. The graphs represent amplification curves of the L858R mutation detection in wild-type and mutant templates. If the amplification started within 40 minutes, it was defined as a positive reaction.
Direct sequencing was also performed on 43 DNA samples extracted from frozen and FFPE tissues, and the results were compared with those obtained with SmartAmp2 from frozen tissue. The mutation in EGFR exon 19 was detected by direct sequencing in 12 of 18 samples (67%) from frozen tissue. Similarly, the mutations in EGFR exon 21 and KRAS exon 2 were detected in 5 of 12 samples (42%) and in 5 of 8 samples (63%), respectively, from frozen tissue. Meanwhile, the mutation in EGFR exon 19 was detected by direct sequencing in 12 of 18 samples (67%) from FFPE tissue. Similarly, the mutations in EGFR exon 21 and KRAS exon 2 were detected in 3 of 12 samples (25%) and in 2 of 8 samples (25%), respectively, from FFPE tissue. Taken together, the mutations for EGFR exon 19, EGFR exon 21, or KRAS exon 2 were detected in 22 of 38 samples (58%) from frozen tissue and in 17 of 38 samples (45%) from FFPE tissue (Table 2).
As described, with direct sequencing we could not detect mutations well, especially when FFPE tissue samples were used. However, with SmartAmp2 detection of mutations was almost 100% using both FFPE tissue and frozen tissue samples.
Agarose Gel Electrophoresis of PCR Products
Agarose gel electrophoresis of PCR products was performed on DNA extracted from the 43 frozen and FFPE tissues to elucidate the reason for SmartAmp2′s superior performance. A visible band was apparent in all samples for all primers in the DNA extracted from the frozen tissue. In contrast, in the FFPE tissue, it was difficult to amplify DNA using primers with a long amplicon. The DNA showed visible bands in 42 samples in which the primers had a short amplicon; however, bands were visible in only 9 samples of primers with a medium amplicon and in only 2 samples with a long amplicon (Figure 3, Table 2). This indicates that DNA extracted from FFPE tissue was severely fragmented.
Figure 3.
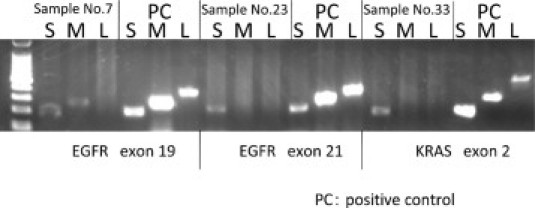
Electrophoresis results of samples 7, 23, and 33. The PCR products of DNA extracted from blood samples were used as a positive control. S, short; M, medium; L, long amplicon of each primer. For example, S lane of EGFR exon 19 represents the result of electrophoresis using the PCR product of the 192-bp amplicon of EGFR exon 19. The left lane is used as the 100-bp ladder marker.
Correlation between EGFR or KRAS Mutations and Gefitinib Responsiveness
To investigate whether the mutations of EGFR detected in FFPE tissue are associated with sensitivity to gefitinib, we followed the 43 patients and checked the history of gefitinib therapy. The response to gefitinib was assessed for the best response to therapy according to the use of Response Evaluation Criteria in Solid Tumors guidelines.35 The clinical features and results of the mutational analyses in FFPE tissues are summarized in Table 3. By August 2009, 13 patients had been treated with gefitinib, and all had a recurrence of lung cancer. Of the 13 patients, 11 patients had EGFR mutations, 1 had a KRAS mutation, and 1 had wild-type alleles. Ten of the 13 patients had a partial response to gefitinib, all of whom had EGFR mutations. Therefore, the response rate to gefitinib in patients with EGFR mutations detectable in FFPE tissue was 91% (10 of 11). The patient with a KRAS mutation and the patient with wild-type alleles detectable in FFPE tissue were classified as having progressive disease and stable disease, respectively.
Table 3.
Characteristics of 13 Patients Treated with Gefitinib Therapy
| Sample no. | Age | Sex | SmartAmp2 mutation site and type | Direct sequencing | Response to gefitinib |
|---|---|---|---|---|---|
| 2 | 65 | F | EGFR Ex 19 E746-A750 del | Mut | PR |
| 3 | 69 | F | EGFR Ex 19 E746-A750 del | Mut | SD |
| 9 | 71 | F | EGFR Ex 19 E746-A750 del | Mut | PR |
| 10 | 50 | F | EGFR Ex 19 E746-A750 del | Mut | PR |
| 12 | 40 | M | EGFR Ex 19 E746-A750 del | Mut | PR |
| 13 | 66 | F | EGFR Ex 19 E746-A750 del | Mut | PR |
| 15 | 55 | M | EGFR Ex 19 L747-E749 del, A750P | Wt | PR |
| 16 | 63 | F | EGFR Ex 19 L747-E749 del, A750P | Wt | RP |
| 17 | 62 | F | EGFR Ex 19 E746-E749, S752-P753 del | Mut | PR |
| 21 | 74 | F | EGFR Ex 21L858R | Mut | PR |
| 27 | 66 | M | EGFR Ex 21L858R | Wt | PR |
| 37 | 71 | F | KRAS Ex2 G12F | Wt | PD |
| 42 | 74 | F | Wt | Wt | SD |
Wt, wild type; Mut, mutant type; F, female; M, male; PR, partial response; SD, stable disease; PD, progressive disease.
Age, gender, and the results of mutational analysis in FFPE tissue are indicated. Among the treated patients, 10 were PR and all had EGFR mutations detected in FFPE tissue.
Discussion
In general, frozen tissue is preferable to FFPE tissue for genetic analysis because the DNA is not as degraded. However, it is difficult to keep frozen tissue in the general hospital because of space limitations or absence of equipments, and therefore, most clinical materials are preserved as FFPE tissues. There are several methods for detecting mutations in DNA from FFPE tissues, but they are time-consuming and complicated. Only a few studies have compared the detection of EGFR or KRAS mutations in DNA extracted from samples obtained from the same patient and prepared as both frozen and FFPE tissues, and the results may differ significantly.36 This has led to the belief that it is difficult to obtain concordant results from both frozen and FFPE tissues.
The recent developments in SmartAmp2 have made rapid and sensitive detection of EGFR and KRAS mutations possible. The assay produces good results even with crude samples such as those taken directly from lysed blood and purification of DNA is not necessary.24 In this study, we investigated the performance of the SmartAmp2 assay for detecting EGFR and KRAS mutations when we extracted DNA from FFPE tissues.
We used the QIAamp DNA FFPE Tissue kit for DNA extraction and the SmartAmp2 assay for mutation detection. These are both easy, simple techniques that require no special skills and can be performed quickly. We initially extracted DNA without RNase, but mutation detection was not as good as expected, and the results were improved after we added the RNase treatment step. We needed more FFPE tissue sections to obtain a sufficient DNA concentration in the extracted solution with RNase than without it. Without RNase treatment, the RNA remaining in the extraction solution likely led to an overestimation of the DNA concentration, and consequently, the actual amount of DNA used for the assay of some specimens might have been insufficient to allow the detection of mutations. The RNase treatment step was essential in our protocol.
With SmartAmp2, only 1 of 43 FFPE tissue samples gave a different result compared with the frozen tissue samples. This is a significant improvement over previous comparative analyses.36 Additionally, the results of SmartAmp2 assay can be obtained within 60 minutes, considerably faster than other procedures. Unfortunately, we could not detect a mutation in sample number 25, despite using several FFPE blocks from different areas of the same tumor. We could find no reasonable explanation for our inability to detect the mutation in only FFPE blocks from sample number 25.
Direct sequencing was less effective at detecting mutations in DNA extracted from FFPE tissue than from frozen tissue, because mutations were detected in only 17 of the 38 FFPE tissue samples (45%) shown by SmartAmp2 to have mutations compared with 22 of 38 frozen tissue samples (58%).
To clarify the advantage of SmartAmp2 using FFPE tissue, we performed electrophoresis on the PCR products and demonstrated the difficulty of amplifying DNA in FFPE samples using PCR, especially with longer amplicon primers. This finding agrees with previous reports indicating that DNA from FFPE tissue degenerates into fragments, making longer amplicons (>300 bp) difficult to amplify.27,36,37 The SmartAmp2 assay was able to detect amplify template and detect mutations in almost all FFPE tissues. This may be because SmartAmp2 primers are designed for amplification of a short target region (Figure 1A: EGFR exon 19, 101 bp; EGFR exon 21, 77 bp; KRAS exon 2, 72 bp), and thus, DNA fragmentation had little influence in the SmartAmp2 assay.24,25,26 Certainly, with a short amplicon primer, most FFPE tissues (42 of 43; 98%) could be amplified by PCR.
As noted previously, most mutations in FFPE tissue could not be detected by direct sequencing, which requires the mutation be present in at least 20% of the sample.38 This implies that most samples had a lower mutant to wild-type ratio, which allowed detection with the SmartAmp2 assay, but not with direct sequencing. On histological examination, these samples, as represented by sample number 31 (Figure 4), tended to have considerable inflammatory cell infiltrate or fibrotic changes, which might have provided a greater source of nonmutated DNA in these samples. Thus, although a sufficient DNA concentration was extracted from the tumor, it might have contained a higher percentage of nonmutated DNA and, consequently, a lower percentage of the mutant allele. In contrast to direct sequencing methods, the SmartAmp2 assay, owing to its high sensitivity, can detect a mutation present as <1% of sample DNA, as may be the case for severe tumor conditions like those in sample number 31.25,26
Figure 4.
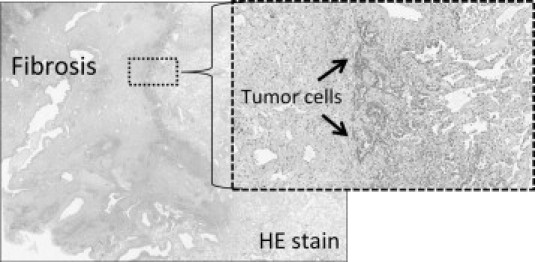
Microscopic examination of sample number 31. A typical case of a tumor with severe fibrotic changes. Most of the tumor had undergone fibrotic changes, and true tumor cells were found in only a small portion of the tumor, as indicated by the arrows. In such cases, mutations were difficult to detect by direct sequencing.
It is worth noting that detection in FFPE tissues may have benefited from the Aac DNA polymerase used in the SmartAmp2 assay. It has very strong strand displacement activity, and because DNA in FFPE tissues is cross-linked to surrounding proteins, strand displacement activity may be necessary for amplifying DNA extracted from this tissue.37
In this study, a comparison between the use of the SmartAmp2 assay and direct sequencing for the detection of mutations in DNA extracted from FFPE tissue preparations demonstrated the superiority of SmartAmp2. However, extremely sensitive PCR-based detection methods continue to be developed. For example, pyrosequencing, an advanced direct sequencing technology, is more sensitive than conventional direct sequencing and has been reported to be applicable for FFPE tissue analysis.39,40,41 Although we might have obtained similar results with pyrosequencing, SmartAmp2 has advantages compared with sequencing-based methods, including pyrosequencing. First, because SmartAmp2 does not require a separate amplification process, because amplification itself is the signal for detection, it can be performed in fewer steps and within a shorter time than other methods. Second, the results of the SmartAmp2 assay are clear and very easy to read. In sequencing-based methods, the data must be carefully examined for the presence of signals, which may be difficult to discern for samples with a low percentage of mutation. Thus, these methods may not provide definitive results for guiding the development of therapeutic strategies. In contrast, with SmartAmp2, only the amplification of the sequence needs to be observable. The unambiguous results with SmartAmp2 can improve the therapeutic approach in patients. These two advantages are valuable, especially in clinical practice.
As shown above, we can detect very low levels of EGFR or KRAS mutations using SmartAmp2; however, the association between the effects of tyrosine kinase inhibitors such as gefitinib and the presence of extremely low levels of the mutant alleles has not been determined. In a recent study, patients with an EGFR mutation that was detected with SmartAmp2, but not by direct sequencing, were reported to respond to gefitinib.25 Our results showing a high response rate to gefitinib in patients with EGFR mutations (Table 3) confirm earlier similar findings.3,11 Moreover, tumors such as sample numbers 15, 16, and 27 with EGFR mutations detectable by SmartAmp2 assay, but not by direct sequencing, responded to gefitinib. Therefore, direct sequencing by itself may not provide enough information to determine therapeutic strategies. A more sensitive EGFR mutation detection method such as SmartAmp2 is essential to accurately predict a response to gefitinib. Only one tumor with a KRAS mutation, which was detected by SmartAmp2 but not by direct sequencing, was treated with gefitinib, and it did not respond to gefitinib. This suggests that low levels of KRAS mutations may correlate with resistance to gefitinib, further study with many samples will be needed to establish this.
In conclusion, we have compared the results of SmartAmp2 and direct sequencing using both FFPE and frozen tissue samples taken from the same tumor and have demonstrated that with SmartAmp2 we could detect mutations effectively in DNA extracted from the FFPE tissue. To our knowledge, this is the first report of applying SmartAmp2 to the analysis of FFPE tissue. The protocol for accurately identifying EGFR and KRAS mutations in DNA from FFPE tissue is quick, easy, and reliable. This new method will allow physicians to identify NSCLC patients who are the most likely to respond to tyrosine kinase inhibitors and ultimately provide better diagnostic options for these patients.
Footnotes
Supported by grant-in-aid for scientific research KAKENHI 19390359 from the Japan Society for the Promotion of Science.
The present study was performed at the Department of Thoracic and Visceral Organ Surgery, Gunma University Graduate School of Medicine.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 5.Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Shibata T, Sakiyama T, Yoshida T, Tamura T. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 6.Toyooka S, Tsukuda K, Ouchida M, Tanino M, Inaki Y, Kobayashi K, Yano M, Soh J, Kobatake T, Shimizu N, Shimizu K. Detection of codon 61 point mutations of the K-ras gene in lung and colorectal cancers by enriched PCR. Oncol Rep. 2003;10:1455–1459. doi: 10.3892/or.10.5.1455. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 8.Shepherd FA, Rosell R. Weighing tumor biology in treatment decisions for patients with non-small cell lung cancer. J Thorac Oncol. 2007;2(Suppl 2):S68–S76. doi: 10.1097/01.JTO.0000269737.05962.a0. [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 11.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki H, Endo K, Mizuno K, Yano M, Fukai I, Yamakawa Y, Fujii Y. EGFR mutation status and prognosis for gefitinib treatment in Japanese lung cancer. Lung Cancer. 2006;51:135–136. doi: 10.1016/j.lungcan.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, da Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 14.Nelson HH, Christiani DC, Mark EJ, Wiencke JK, Wain JC, Kelsey KT. Implications and prognostic value of K-ras mutation for early-stage lung cancer in women. J Natl Cancer Inst. 1999;91:2032–2038. doi: 10.1093/jnci/91.23.2032. [DOI] [PubMed] [Google Scholar]
- 15.Gandara DR, Lara PN, Lau DH, Mack P, Gumerlock PH. Molecular-clinical correlative studies in non-small cell lung cancer: application of a three-tiered approach. Lung Cancer. 2001;34(Suppl 3):S75–S80. doi: 10.1016/s0169-5002(01)00368-3. [DOI] [PubMed] [Google Scholar]
- 16.Pajkos G, Kiss I, Sandor J, Ember I, Kishazi P. The prognostic value of the presence of mutations at the codons 12, 13, 61 of K-ras oncogene in colorectal cancer. Anticancer Res. 2000;20:1695–1701. [PubMed] [Google Scholar]
- 17.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–418. [PubMed] [Google Scholar]
- 18.Hatzaki A, Razi E, Anagnostopoulou K, Iliadis K, Kodaxis A, Papaioannou D, Labropoulos S, Vasilaki M, Kosmidis P, Saetta A, Mihalatos M, Nasioulas G. A modified mutagenic PCR-RFLP method for K-ras codon 12 and 13 mutations detection in NSCLC patients. Mol Cell Probes. 2001;15:243–247. doi: 10.1006/mcpr.2001.0367. [DOI] [PubMed] [Google Scholar]
- 19.Huang CL, Taki T, Adachi M, Konishi T, Higashiyama M, Kinoshita M, Hadama T, Miyake M. Mutations of p53 and K-ras genes as prognostic factors for non-small cell lung cancer. Int J Oncol. 1998;12:553–563. doi: 10.3892/ijo.12.3.553. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Nakano J, Ueno M, Masuya D, Nakashima T, Yokomise H, Yube K, Huang CL. A useful protocol for analyses of mutations of the epidermal growth factor receptor gene. Oncol Rep. 2006;15:1503–1505. [PubMed] [Google Scholar]
- 21.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: kRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008;8:142. doi: 10.1186/1471-2407-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horiike A, Kimura H, Nishio K, Ohyanagi F, Satoh Y, Okumura S, Ishikawa Y, Nakagawa K, Horai T, Nishio M. Detection of epidermal growth factor receptor mutation in transbronchial needle aspirates of non-small cell lung cancer. Chest. 2007;131:1628–1634. doi: 10.1378/chest.06-1673. [DOI] [PubMed] [Google Scholar]
- 24.Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y, Itoh M, Miyagi T, Takakura H, Hoshi K, Kato C, Arakawa T, Shibata K, Fukui K, Masui R, Kuramitsu S, Kiyotani K, Chalk A, Tsunekawa K, Murakami M, Kamataki T, Oka T, Shimada H, Cizdziel PE, Hayashizaki Y. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 25.Hoshi K, Takakura H, Mitani Y, Tatsumi K, Momiyama N, Ichikawa Y, Togo S, Miyagi T, Kawai Y, Kogo Y, Kikuchi T, Kato C, Arakawa T, Uno S, Cizdziel PE, Lezhava A, Ogawa N, Hayashizaki Y, Shimada H. Rapid detection of epidermal growth factor receptor mutations in lung cancer by the SMart-Amplification Process. Clin Cancer Res. 2007;13:4974–4983. doi: 10.1158/1078-0432.CCR-07-0509. [DOI] [PubMed] [Google Scholar]
- 26.Tatsumi K, Mitani Y, Watanabe J, Takakura H, Hoshi K, Kawai Y, Kikuchi T, Kogo Y, Oguchi-Katayama A, Tomaru Y, Kanamori H, Baba M, Ishidao T, Usui K, Itoh M, Cizdziel PE, Lezhava A, Ueda M, Ichikawa Y, Endo I, Togo S, Shimada H, Hayashizaki Y. Rapid screening assay for KRAS mutations by the modified smart amplification process. J Mol Diagn. 2008;10:520–526. doi: 10.2353/jmoldx.2008.080024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomizawa Y, Iijima H, Sunaga N, Sato K, Takise A, Otani Y, Tanaka S, Suga T, Saito R, Ishizuka T, Dobashi K, Minna JD, Nakajima T, Mori M. Clinicopathologic significance of the mutations of the epidermal growth factor receptor gene in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:6816–6822. doi: 10.1158/1078-0432.CCR-05-0441. [DOI] [PubMed] [Google Scholar]
- 28.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non-small cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 29.Hayes VM, Westra JL, Verlind E, Bleeker W, Plukker JT, Hofstra RM, Buys CH. New comprehensive denaturing-gradient-gel-electrophoresis assay for KRAS mutation detection applied to paraffin-embedded tumours. Genes Chromosomes Cancer. 2000;29:309–314. [PubMed] [Google Scholar]
- 30.Kim DH, Kim JS, Park JH, Lee SK, Ji YI, Kwon YM, Shim YM, Han J, Park J. Relationship of Ras association domain family 1 methylation and K-ras mutation in primary non-small cell lung cancer. Cancer Res. 2003;63:6206–6211. [PubMed] [Google Scholar]
- 31.Soh J, Toyooka S, Aoe K, Asano H, Ichihara S, Katayama H, Hiraki A, Kiura K, Aoe M, Sano Y, Sugi K, Shimizu N, Date H. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int J Cancer. 2006;119:2353–2358. doi: 10.1002/ijc.22190. [DOI] [PubMed] [Google Scholar]
- 32.Luo JD, Chan EC, Shih CL, Chen TL, Liang Y, Hwang TL, Chiou CC. Detection of rare mutant K-ras DNA in a single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe. Nucleic Acids Res. 2006;34:e12. doi: 10.1093/nar/gnj008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taback B, Bilchik AJ, Saha S, Nakayama T, Wiese DA, Turner RR, Kuo CT, Hoon DS. Peptide nucleic acid clamp PCR: a novel K-ras mutation detection assay for colorectal cancer micrometastases in lymph nodes. Int J Cancer. 2004;111:409–414. doi: 10.1002/ijc.20268. [DOI] [PubMed] [Google Scholar]
- 34.Sun X, Hung K, Wu L, Sidransky D, Guo B. Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat Biotechnol. 2002;20:186–189. doi: 10.1038/nbt0202-186. [DOI] [PubMed] [Google Scholar]
- 35.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 36.Gallegos Ruiz MI, Floor K, Rijmen F, Grunberg K, Rodriguez JA, Giaccone G. EGFR and K-ras mutation analysis in non-small cell lung cancer: comparison of paraffin embedded versus frozen specimens. Cell Oncol. 2007;29:257–264. doi: 10.1155/2007/568205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25:409–418. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 38.Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, Maher EA, Distel RJ, Meyerson M, Johnson BE. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–758. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 39.Poehlmann A, Kuester D, Meyer F, Lippert H, Roessner A, Schneider-Stock R. K-ras mutation detection in colorectal cancer using the pyrosequencing technique. Pathol Res Pract. 2007;203:489–497. doi: 10.1016/j.prp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Packham D, Ward RL, Ap Lin V, Hawkins NJ, Hitchins MP. Implementation of novel pyrosequencing assays to screen for common mutations of BRAF and KRAS in a cohort of sporadic colorectal cancers. Diagn Mol Pathol. 2009;18:62–71. doi: 10.1097/PDM.0b013e318182af52. [DOI] [PubMed] [Google Scholar]
- 41.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


