Abstract
Daily rhythms generated by the circadian clock regulate many life functions, including responses to xenobiotic compounds. In Drosophila melanogaster, the circadian clock consists of positive elements encoded by cycle (cyc) and Clock (Clk) and negative elements encoded by period (per) and timeless (tim) genes. The ϵ-isoform of the PAR-domain protein 1 (Pdp1ε) transcription factor is controlled by positive clock elements and regulates daily locomotor activity rhythms. Pdp1 target genes have not been identified, and its involvement in other clock output pathways is not known. Mammalian orthologs of Pdp1 have been implicated in the regulation of xenobiotic metabolism; therefore, we asked whether Pdp1 has a similar role in the fly. Using pesticides as model toxicants, we determined that disruption of Pdp1ε increased pesticide-induced mortality in flies. Flies deficient for cyc also showed increased mortality, while disruption of per and tim had no effect. Day/night and Pdp1-dependent differences in the expression of xenobiotic-metabolizing enzymes Cyp6a2, Cyp6g1, and α-Esterase-7 were observed and likely contribute to impaired detoxification. DHR96, a homolog of constitutive androstane receptor and pregnane X receptor, is involved in pesticide response, and DHR96 expression decreased when Pdp1 was suppressed. Taken together, our data uncover a pathway from the positive arm of the circadian clock through Pdp1 to detoxification effector genes, demonstrating a conserved role of the circadian system in modulating xenobiotic toxicity.
Keywords: circadian clock, Pdp1, permethrin, Cyp6g1, α-Esterase-7, DHR96
The circadian clock generates daily rhythmic patterns in sleep, activity, and other physiological variables, including daily fluctuations in metabolism, cellular function, and gene expression (Schibler, 2007). Circadian rhythms are entrained by exogenous zeitgebers (time givers), especially light/dark cycles. Remarkably, most clock genes, their network properties, and the basic multioscillatory organization of the circadian systems are highly conserved between flies and humans (Giebultowicz, 2001; Schibler, 2007; Stanewsky, 2003). The core circadian clock in Drosophila melanogaster consists of two interacting molecular feedback loops organized into 24-h feedback cycles. In the first loop, positive clock elements encoded by genes Clock (Clk) and cycle (cyc) activate the transcription of period (per) and timeless (tim) in the early night (Hardin, 2005). This leads to daily increases in the levels of per/tim messenger RNA (mRNA) and PER/TIM proteins. These negative clock elements accumulate in cell nuclei late at night and inhibit CLK-CYC–mediated transcription of per and tim (Glossop et al. 2003). The second feedback loop consists of the genes vrille (vri) and the PAR-domain protein 1 (Pdp1) (Cyran et al., 2003; Glossop et al., 2003; Zheng, et al., 2009). Pdp1 is a complicated gene because multiple promoters and alternative splicing events produce seven annotated isoforms of this transcription factor (Reddy et al., 2000). Two isoforms RD and RJ are regulated in a circadian manner and are collectively called Pdp1ϵ (Zheng et al., 2009). Pdp1ϵ is controlled by positive clock elements and regulates Clk expression through a feedback loop. In addition, Pdp1ϵ is necessary for daily rhythms in locomotor activity (Benito et al., 2007; Cyran et al., 2003; Lim et al., 2007; Zheng et al., 2009). It is not known whether this gene regulates other output rhythms from the circadian clock as specific target genes of Pdp1 have not been identified.
Several studies have suggested that the circadian clock plays a role in regulating the response to xenobiotics, including therapeutics and toxins (Hooven et al., 2009; Levi and Schibler, 2007). Microarray studies have shown daily molecular oscillations in expression of xenobiotic-metabolizing genes in mammals (Yan et al., 2008) and flies (Wijnen and Young, 2006). Furthermore, studies have shown that the nuclear receptor and xenosensors constitutive of androstane receptor (CAR) and pregnane X receptor (PXR also known as steroid and xenobiotic receptor [SXR]) are expressed rhythmically (Yang et al., 2006; Zhang et al., 2009). While these studies illustrate daily rhythms in expression of genes important for xenobiotic metabolism, they did not address functional significance of these rhythms. A recent study in Drosophila identified daily rhythms in activity of specific xenobiotic-metabolizing enzymes and daily variation in mortality after exposure to specific pesticides, suggesting that circadian clock genes may play an important functional role in responses to toxins (Hooven et al., 2009).
Another line of evidence for a role of circadian clock genes in xenobiotic metabolism comes from genetic studies in mammals. Disruption of the genes Clk and the cyc homolog (BMAL1) renders mice highly sensitive to the anticancer drug cyclophosphamide, while disruption of negative clock elements did not (Gorbacheva et al., 2005). Furthermore, mice lacking all three paralogs of Pdp1, the PAR bZip transcription factors dbp, tef, and hlf, are hypersensitive to pentobarbital and some anticancer drugs (Gachon et al., 2006). These triple knockout mice have altered expression of genes related to drug metabolism and detoxification processes like cytochrome P450 monooxygenases (P450s), carboxlyesterases, and the CAR (Gachon et al., 2006).
The present study sought to determine whether circadian clock genes regulate the response to toxicants. An additional goal of this study was to identify links between the central circadian mechanism and oscillations in output pathways related to detoxification. Using pesticides as a model for toxicant exposure, we identified a new output pathway from the circadian clock in Drosophila through cyc and Pdp1 to genes implicated in pesticide metabolism. This novel finding is functionally significant as flies lacking cyc or Pdp1 appear to have an impaired detoxification response and are thus more sensitive to pesticide-induced death. Taken together with mammalian studies, our study demonstrates a conserved pathway from positive clock elements through Pdp1 to xenobiotic metabolism effector genes. Given this conservation, studies in the fly may provide a fundamental understanding of the circadian system in modulating xenobiotic toxicity in humans and suggest that the fly may be an effective model for chronopharmacological and chronotoxicological studies.
MATERIALS AND METHODS
Fly rearing, strains, and activity.
Drosophila melanogaster were raised on yeast (35 g/l), cornmeal and molasses diet at 25 ± 1°C, at low density to attain uniform size, under a 12-h light/12-h dark regimen (where Zeitgeber time 0 [ZT 0] is time of lights on and ZT 12 is time of lights off). Flies were separated 1–2 days after emergence, and 5-day-old males were used for all experiments. cyc01 (Rutila et al., 1998), per01 (Konopka and Benzer, 1971), and tim01 (Sehgal et al., 1994) mutants were backcrossed to the Canton-S (CS) control line, allowing free recombination for at least six generations to isogenize the genetic background. The control flies for each mutant isogenized to the CS background were designated as CSC, CSP, CST, and CSD for cyc01, per01, tim01, and DHR960, respectively. Attenuated expression of Pdp1 was accomplished through RNA interference (RNAi) by crossing flies carrying timGal4 and UAS-Pdp1 RNAi constructs. Pdp1ia and Pdp1ib denote a UAS-Pdp1 RNAi line from the laboratory of Dr Hardin (Benito et al., 2007) or Dr Choe (Lim et al., 2007), respectively. Control flies for RNAi experiments were generated by crossing the timGal4 driver with w1118 flies, the genotype used to generate the transgenic lines. The fly lines used in this study were generously shared and included: Pdp1ia from P. Hardin (Benito et al., 2007), Pdp1ib from J. Choe (Lim et al., 2007), Pdp13135 and their respective isogenized control iso131 flies from X. Zheng (Zheng et al., 2009), and DHR960 and their respective isogenized control CSD from C. Thummel (King-Jones et al., 2006). Rhythmic activity was measured in a standard Trikinetics locomotor activity monitor (Waltham, MA). Data were collected in 15-min bins. A quantitative measure of the rhythmicity data was obtained using a fast Fourier Transform (FFT) (ClockLab, Actimetrics, Wilmette, IL), and individuals with FFT < 0.04 were deemed arrhythmic and those with FFT > 0.06 were rhythmic. Total daily activity (counts/24 h) was averaged for all individuals tested and served as an overall measure of daily activity.
Chemicals and insect treatment.
Pesticides were obtained from ChemService (Westchester, PA), handled under a chemical hood, and were of the highest purity available. For pesticide exposure, 0.2 μl of permethrin (0–640 μg/ml) or malathion (0–32 μg/ml) dissolved in acetone was administered topically via gastight microsyringe to each fly (10 flies/vial replicated thrice). Mortality was recorded 24 h after exposure. Each set of experiments was repeated three or more times. The concentration of pesticide that was lethal to 50% of flies (LC50) was calculated using PROBIT analysis (Finney, 1978) with a code written for and executed in SAS software (SAS 9.1.3; SAS Institute Inc., Cary, NC).
Quantitative real-time PCR.
Twenty-five whole males were collected, frozen, and homogenized in TriReagent following manufacturer’s protocol (Sigma) using a Kontes handheld motor. Samples were purified using the RNeasy Mini Kit (Qiagen) with on-column DNAse digestion (Qiagen) according to manufacturer’s protocol. Synthesis of complementary DNA was achieved with Sprint RT Complete kit (Clontech) or iScript cDNA synthesis kit (Bio-Rad) according to manufacturer’s protocol. Real-time PCR was performed with iTaq SYBR Green Supermix with Rox (Bio-Rad) on an ABI Prism 7300, 7500, or Step-One Plus real-time machine in triplicate under default thermal cycling conditions. Primers were designed for target gene sequences using standard practices (specific primer sequences are available upon request) and obtained from Integrated DNA Technologies. Data were analyzed using the standard 2−ΔΔCT method normalized to the gene rp49 and expressed relative to control samples at ZT 4.
Enzymatic activity assays.
Twenty-five male flies were homogenized in 250 μl of buffer (100mM K-PO4, pH 7.0, containing 2mM EDTA and 0.1% by volume Triton X-100), sonicated on ice, centrifuged at 3000 × g for 5 min at 4°C, and the supernatant recentrifuged at 10,000 × g for an additional 15 min. The protein concentration of the diluted supernatant (1:10 in assay buffer, 100mM K-PO4, pH 7.0) was quantified using the bicinchoninic acid assay. All assays were performed using a BioTek Synergy 2 plate reader (Winooski, VT).
Esterase activity was assayed using Van Asperen’s method, with α- or β-naphthyl acetate as substrates in microtitre plates with some modifications (Bracco et al., 1999). Fast Blue B was added following incubation at 30°C for 10 min, and absorbance was read at 595 nm for α-naphthyl acetate–cleaving esterase and 540 nm for β-naphthyl acetate–cleaving esterase. The concentration of products was determined at end point from standard curves of α- or β-naphthol. The activity was expressed as millimole of product formed per minute per milligram protein.
4-Nitrophenyl acetate (PNPA; Sigma) hydrolysis measurements were assayed according to Kim and Lee (2000). Briefly, generation of 4-nitrophenol was monitored as changes in absorbance at 405 nm adjusted against negative-control wells (without enzyme source). A standard curve of 4-nitrophenol ranging from 0 to 1mM was used to calculate activity as 1 μmol of 4-nitrophenol per minute per milligram protein.
Glutathione S-transferase (GST) activity assay was adapted from the procedure described (Habig et al., 1974) using 1-chloro-2,4-dinitrobenzene (CDNB) solution in acetonitrile as substrate. Absorbance at 340 nm was recorded at intervals for 5 min at 25°C. The conjugation of CDNB with reduced glutathione is accompanied by an increase in absorbance at 340 nm, directly proportional to the GST activity. Total enzyme activity (cytosolic and microsomal) was expressed as nanomole per minute per milligram of protein.
RESULTS
Disrupting cyc and Pdp1 Expression Increases Sensitivity of Drosophila to Pesticides
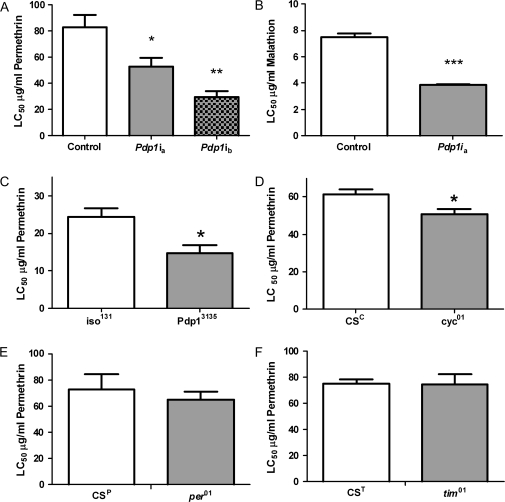
To identify components of the circadian system that modulate xenobiotic response, we exposed flies with disrupted expression of Pdp1, or null mutations in core clock genes, to pesticides. Flies were exposed to varying concentrations of each pesticide, and the lethal concentration to 50% of subjects was calculated (LC50). We used permethrin and malathion insecticides, which are used worldwide for the control of many insects, including the malarial vector Anopheles gambiae. These compounds represent two mechanistically distinct classes of insecticides (pyrethroids and organophosphates, respectively). No daily rhythm for permethrin-induced death was detected (Supplementary figs. 1A–C); thus, the pesticides were applied at ZT 4 according to a previous study by Hooven et al. (2009). We first examined the response to pesticides in flies where Pdp1 expression was suppressed in clock cells by RNAi. A 50–60% decrease in Pdp1ϵ and total Pdp1 mRNA expression levels was detected in both RNAi lines at ZT 4 and ZT 20 time points (Supplementary fig. 1D). The effectiveness of RNAi was confirmed by decreased daily rhythms in behavioral activity (Supplementary table 1). Suppression of Pdp1 in two independent fly lines resulted in a significant twofold and threefold reduction in the average LC50 for permethrin (Fig. 1A). Disruption of Pdp1 expression also resulted in a significant twofold reduction in the average LC50 for malathion (Fig. 1B). These data suggest that Pdp1 regulates the ability of flies to survive a toxic exposure. Given the similar mortality response observed with both pesticides when Pdp1 was disrupted, we focused the remainder of our survival experiments on permethrin exposure.
FIG. 1.
Disruption of Pdp1 or cyc expression results in increased sensitivity to pesticides. Flies were exposed to permethrin or malathion at various concentrations and data represent the average LC50 (+ SEM) for each mutant and their matched control flies in three independent experiments unless otherwise noted. (A) Data represent five independent experiments for control flies, six for Pdp1ia flies, and three for Pdp1ib. (D) Four independent experiments were completed for cyc01 and control flies. Statistical significance was determined by a one-way ANOVA and a Tukey's multiple comparison post-test for (A) and unpaired t-test for (B–F) where *p < 0.05, **p < 0.01, and ***p < 0.001. See also Supplementary figure 1 and Supplementary table 1.
To explore the specific role of clock-controlled Pdp1ϵ, we used the Pdp13135 mutant fly line where only the ϵ-isoforms are disrupted by a deletion in their second exon (Zheng et al., 2009). We confirmed that Pdp13135 flies were arrhythmic in locomotor activity (Supplementary table 1) as described (Zheng et al., 2009). Additionally, we showed that cycling of tim mRNA levels was significantly attenuated, which provides additional evidence that Pdp1ϵ is involved in modulation of the clock (Supplementary fig. 1E). Importantly, disruption of Pdp1ϵ alone resulted in a significant 1.7-fold reduction in the average LC50 for permethrin (Fig. 1C). These data suggest that clock-dependent Pdp1ϵ can regulate the ability of flies to survive a toxic exposure.
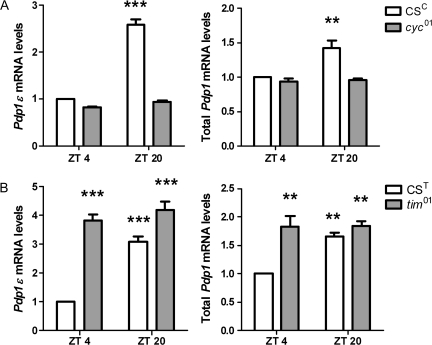
To determine whether clock elements upstream from Pdp1ϵ are essential for normal pesticide response, we exposed flies with null mutations in core clock genes to permethrin. Loss of cyc expression resulted in a significant decrease in the average LC50 for permethrin (Fig. 1D, p = 0.029), demonstrating that response to pesticides is regulated in part by the positive limb of the circadian clock. In contrast, loss of either per or tim function did not affect ability of flies to survive permethrin treatment (Figs. 1E and 1F), suggesting that repressive clock elements are not essential for a normal response to this compound. These data are consistent with Pdp1 being a key player in pesticide response because both total and Pdp1ϵ expression levels are suppressed and nonrhythmic in cyc01 flies but remain high and constitutively expressed in tim01 and per01 flies (Fig. 2; Benito et al., 2007; Cyran et al., 2003; Zheng et al., 2009). These data demonstrate that the circadian clock regulates the response of Drosophila to pesticides via the cyc-Pdp1ϵ axis.
FIG. 2.
Pdp1 mRNA expression in cyc and tim mutants. Pdp1ϵ (left) and total Pdp1 (right) mRNA levels are lower in (A) cyc01 flies with increased susceptibility to permethrin but not in (B) tim01 flies that do not show this sensitivity. Data represent mean mRNA levels + SEM for three independent experiments. Significant difference from control ZT 4 values, where values **p < 0.01 and ***p < 0.001, as calculated by ANOVA and Tukey's multiple comparison post-test.
DHR96 Affects Susceptibility to Permethrin and May Be Regulated in Part by Pdp1
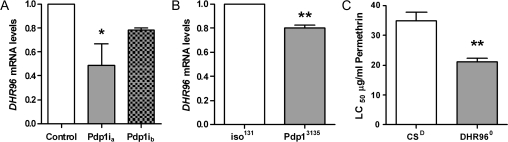
The Drosophila genome contains a single ortholog of both CAR and PXR, named DHR96 (Fisk and Thummel, 1995; King-Jones and Thummel, 2005; King-Jones et al., 2006). Given the regulation of CAR by PAR bZip transcription factors in mammals (Gachon et al., 2006) and its involvement in metabolism of xenobiotics (Goodwin and Moore, 2004), we asked whether DHR96 was regulated by Pdp1 in flies. Preliminary results using real-time PCR suggested that DHR96 mRNA levels did not change based on the circadian time of day (data not shown); therefore, we evaluated DHR96 mRNA levels in Pdp1 mutants at the time when flies were tested with pesticides. Disruption of Pdp1 via RNAi or mutation of Pdp1ϵ reduced DHR96 mRNA expression from 20 to 50% (Figs. 3A and 3B). These results suggest that Pdp1 may contribute to the regulation of DHR96 in the fly but also suggest that this multifunctional gene may be regulated by other mechanisms (Fisk and Thummel, 1995; King-Jones et al., 2006; Sieber and Thummel, 2009).
FIG. 3.
Influence of Pdp1 on DHR96 expression. (A–B) DHR96 mRNA expression levels in Pdp1 RNAi, Pdp13135, and their controls (control or iso131 flies, respectively) at ZT 4. Data represent mean relative expression (+ SEM) of three independent experiments. (C) Permethrin susceptibility is increased in the DHR96 mutant as compared to its respective control (CSD). Data are average LC50 (+ SEM) for four independent experiments. ANOVA and Tukey’s multiple comparison post-test (A) or an unpaired t-test (B–C) were used to determine statistical significance from control samples where *p < 0.05 and **p < 0.01.
PXR and CAR are known to bind a wide range of xenobiotic chemicals in mammals and activate many different target genes involved in detoxification (Goodwin and Moore, 2004; Maglich et al., 2002); thus, we asked in parallel experiments if flies with a mutation in DHR96 are more sensitive to an acute permethrin exposure. We show that loss of DHR96 expression significantly decreased the average LC50 for permethrin by 1.6-fold (Fig. 3C), demonstrating that DHR96 regulates the survival response of insects to a pyrethroid.
Disruption of Pdp1 Results in Altered Transcription of Xenobiotic-Metabolizing Genes and Esterase Activity
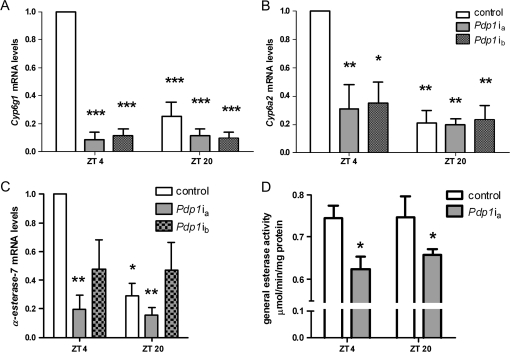
Recent work in our laboratory showed that both expression and activity of specific xenobiotic-metabolizing enzymes fluctuate during the day in Drosophila (Hooven et al., 2009). To determine the consequences of disrupted Pdp1 expression on xenobiotic-related genes, three effector-level genes, Cyp6a2, Cyp6g1, and α-Esterase-7, were selected. These genes were chosen because microarray studies suggested that they were rhythmically expressed and they are associated with pesticide metabolism and resistance (Daborn et al., 2002; Dunkov et al., 1997; Heidari et al., 2005; Hooven et al., 2009; Wijnen and Young, 2006). Real-time PCR revealed that mRNA levels of Cyp6g1 and Cyp6a2 are significantly higher at ZT 4 than ZT 20 in control flies, suggesting regulation by the circadian clock (Fig. 4A, Supplementary fig. 2A). Disruption of Pdp1 expression through RNAi resulted in significantly lower expression of both Cyp6g1 and Cyp6a2 (Figs. 4A and 4B). Disruption of Pdp1ϵ alone also resulted in a significant decline in the levels of Cyp6g1 mRNA and a similar trend in Cyp6a2 mRNA, suggesting regulation by the circadian clock (Supplementary fig. 2A).
FIG. 4.
Pdp1 regulates components of xenobiotic metabolism. (A–C) Time of day and Pdp1-dependent Cyp6g1, Cyp6a2, and α-Esterase-7 mRNA expression levels in control and Pdp1 RNAi flies. Data represent mean relative expression (+ SEM) for three independent experiments. ANOVA and Tukey's multiple comparison post-test were used to determine statistical significance from control ZT 4 samples where *p < 0.05, **p < 0.01, and ***p < 0.001. (D) Flies with disrupted Pdp1 expression have significantly lower general esterase activity for α-naphthyl acetate. Significant difference from control ZT 4 was determined by an ANOVA and Bonferonni post-test, p < 0.05. See also Supplementary figure 2.
Another group of phase I enzymes contributing to pesticide metabolism are esterases. Examination of α-Esterase-7 mRNA levels revealed a significantly higher amount of mRNA at ZT 4 than ZT 20 in control flies (Fig. 4C, Supplementary fig. 2A). Furthermore, both Pdp1ia and Pdp13135 flies had significantly lowered α-Esterase-7 mRNA at both time points (Fig. 4 and Supplementary fig. 2A). Pdp1ib flies also showed a trend of lowered α-Esterase-7 at both ZT 4 and ZT 20 (Fig. 4C). These data suggest that Pdp1 regulates α-Esterase-7 transcription placing this gene as an effector in a clock-controlled pathway. To test functional significance of the transcriptional rhythm in α-Esterase-7, we examined esterase activity and show that disruption of Pdp1 had substrate-specific effects. Activity toward α-naphthyl acetate, a substrate of α-Esterase-7 (Heidari et al., 2005), was significantly reduced in Pdp1 deficient flies (Fig. 4D). Thus, decreased esterase gene expression and enzymatic activity may contribute to the phenotype of increased sensitivity to pesticides. Additionally, esterase activity was significantly higher at ZT 20 than at ZT 4 for the substrates β-naphthyl acetate and PNPA, but this was not dependent on Pdp1 (Supplementary figs. 2B and 2C). We also examined GST activity, which plays an important role in phase II detoxification but found no Pdp1- or time-dependent change in GST activity levels (Supplementary fig. 2D).
DISCUSSION
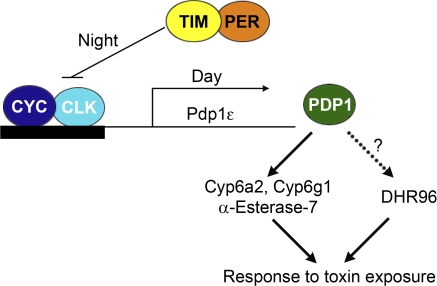
We identified a new circadian output pathway in Drosophila leading from the central clock gene cyc, to the transcription factor Pdp1, to xenobiotic-metabolizing genes Cyp6a2, Cyp6g1, and α-Esterase-7 (Fig. 5). Functional importance of a clock regulated detoxification pathway was demonstrated by showing that deficiency in cyc and Pdp1ϵ genes increases the sensitivity of insects to pesticide exposure. To our knowledge, this is the first demonstration that the circadian clock regulates the response of Drosophila to toxicants via the cyc-Pdp1ϵ axis. We observed a statistically significant difference between the cyc and Pdp1 mutants and their matched controls, e.g., a 1.7-fold reduction in the lethal dose for permethrin exposure was observed in Pdp1ϵ mutants. This signifies an important difference in the fly's ability to survive a toxic chemical; a 70% greater concentration of the toxicant was necessary to induce death when Pdp1ϵ is not mutated. Furthermore, a 1.7-fold reduction in the LC50 means that a farmer could cut the dose of pesticide by nearly half, which has important financial and environmental implications.
FIG. 5.
The positive arm of the circadian clock regulates xenobiotic-metabolizing genes and the response of flies to a toxic exposure.
Interestingly, abrogating expression of the Pdp1ϵ isoforms, or all Pdp1 isoforms, renders flies more sensitive to pesticide exposure than flies lacking cyc. This may be due to the fact that in addition to influencing the expression of core clock genes, Pdp1ϵ also functions as an output gene of the circadian clock (Supplementary fig. 1E and Benito et al., 2007; Cyran et al., 2003; Lim et al., 2007; Zheng et al., 2009). It is also noteworthy that this susceptibility is observed following permethrin treatment since there was no apparent circadian rhythm in response to this pesticide throughout the day. Thus, the functional circadian clock appears to significantly contribute to the response to a toxicant even when the end point response does not show daily fluctuations.
We show a significant decline in mRNA levels of the nuclear receptor DHR96 when Pdp1 expression was disrupted. This finding in flies is similar to the role observed in mammals for PAR bZip transcription factors in regulating CAR transcription. Specifically, PAR bZip triple knockout mice express only a low and noncircadian basal level of CAR mRNA (Gachon et al., 2006). The decline in DHR96 mRNA was more apparent when all isoforms of Pdp1 were suppressed by RNAi, suggesting that multiple isoforms of the gene may regulate DHR96. While our data support a role for Pdp1 regulation of DHR96, this multifunctional gene is also regulated by the insect hormone 20-hydroxyecdysone and is likely to be regulated by other additional mechanisms (Fisk and Thummel, 1995; Horner et al., 2009; Sieber and Thummel, 2009).
In murine liver, the xenosensors CAR, PXR (also known as SXR), aryl hydrocarbon receptor (AhR), and a number of other nuclear receptors are known to be expressed rhythmically (Gachon et al., 2006; Teboul et al., 2008; Zhang et al., 2009). In contrast, we did not detect a rhythm in expression of the orthologous DHR96 gene when whole flies were analyzed. While we cannot exclude that DHR96 mRNA may cycle in specific tissues, this is not the first time a nuclear receptor was shown to be differentially regulated in flies as compared to mammals. The Drosophila equivalent of AhR, spineless, is not rhythmically expressed in the fly and plays a diminished role in response to xenobiotics compared to mammalian species (Brown et al., 2005). These examples illustrate that while much of the pathway from the clock to xenobiotic metabolism is conserved between flies and mammals, the details may vary between species.
In this paper, we additionally identified that flies with a mutation in DHR96 are more susceptible to an acute exposure to permethrin, demonstrating that DHR96 regulates the survival response of insects to a pyrethroid. This is consistent with the report by King-Jones et al. (2006) who showed that flies lacking DHR96 are more sensitive to chronic exposure to the pesticide dichlorodiphenyltrichloroethane (DDT). At this time, it is not clear if DHR96 directly regulates the response to toxins or if this phenotype is at least in part indirectly regulated by altered triacylglycerol and cholesterol homeostasis, which has been observed in these animals (Horner et al., 2009; Sieber et al., 2009).
Our study uncovered novel links between the central circadian mechanism and rhythmic output pathways related to detoxification and xenobiotic metabolism. P450s and esterases are phase I enzymes that modify foreign compounds to decrease their hydrophobicity and promote conjugation by phase II enzymes, in preparation for excretion from the organism. We identified day/night and Pdp1-dependent differences in the expression of Cyp6a2, Cyp6g1, and α-Esterase-7. These results confirm previous microarray studies that suggested that these genes may be regulated by the circadian clock (Hooven et al., 2009; Wijnen and Young, 2006;). Rhythmic expression of α-Esterase-7 agrees well with the day/night and Pdp1-dependent differences in esterase activity. Differences in mRNA levels of Cyp6a2 and Cyp6g1 reported here are consistent with a previous report showing daily rhythm in P450 activity (Hooven et al., 2009). Upregulation of P450s may result in increased oxidative stress (Lewis, 2002). Coordinating the upregulation of P450s with the temporal window when the individual is active and ingesting food, and thus most likely to be exposed to toxins, would minimize oxidative stress. The timing of food intake is coordinated by the circadian clock; therefore, we speculate that clock genes also evolved to coordinate the expression of P450s when exposure to xenobiotics is most likely.
Taken together, the data suggest that circadian clock genes, and in particular Pdp1, play an important role in regulating xenobiotic metabolism across species and open a new avenue for understanding regulation of insect genes that have been associated with pesticide resistance. In particular, Cyp6a2 and Cyp6g1 are found to be overtranscribed in DDT-resistant isolates of D. melanogaster (Dunkov et al., 1997; Pedra et al., 2004), and cross-resistance to pyrethroids and organophosphates has been suggested to be mediated by insect orthologs of α-Esterase-7 (Heidari et al., 2005). Managing pesticide resistance is critical for protecting the food supply, environment, and human health.
While it has been known that both the core circadian clock and xenobiotic metabolism are evolutionary conserved between insects and mammals (King-Jones and Thummel, 2005; Schibler, 2007; Stanewsky, 2003), our work connects these two pathways in flies. Circadian clock genes and circadian rhythms have been implicated in the response to therapeutic compounds (Levi and Schibler, 2007). Similar to our results with insecticides, disruption of Clk and the cyc homolog BMAL1 renders mice highly sensitive to the anticancer drug cyclophosphamide, but disruption of a negative element of clock does not (Gorbacheva et al., 2005). Furthermore, high sensitivity of PAR bZip deficient mice to pentobarbital and the anticancer drugs mitoxantrone and cyclophosphamide were demonstrated (Gachon et al., 2006). Thus, a protective pathway involving positive clock elements and PAR bZip transcription factors is evolutionarily conserved between flies and mammals. Studies in the fly may help to provide a fundamental understanding of the functional significance of the circadian system in modulating xenobiotic toxicity and serve as an effective model for chronopharmacological and chronotoxicological studies.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of General Medical Sciences at the National Institute of Health (GM0737920 to J.M.G.); National Research Initiative at the United States Department of Agriculture (2007-04617 to J.M.G.); National Research Service Award at the National Institute of Health (5 F32 ES14994 to L.A.H.).
Supplementary Material
Acknowledgments
We thank Drs A. Sehgal and X. Zheng for sharing Pdp13135 flies and data prior to publication. We thank David Chin for assistance with fly rearing; Dr B. Pittendrigh for helpful discussions, and Drs P. Hardin, J. Choe, X. Zheng, and C. Thummel for flies.
References
- Benito J, Zheng H, Hardin PE. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J. Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracco JE, Barata JM, Marinotti O. Evaluation of insecticide resistance and biochemical mechanisms in a population of Culex quinquefasciatus (Diptera: Culicidae) from Sao Paulo. Brazil. Mem. Inst. Oswaldo Cruz. 1999;94:115–120. doi: 10.1590/s0074-02761999000100022. [DOI] [PubMed] [Google Scholar]
- Brown RP, McDonnell CM, Berenbaum MR, Schuler MA. Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene. 2005;358:39–52. doi: 10.1016/j.gene.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- Dunkov BC, Guzov VM, Mocelin G, Shotkoski F, Brun A, Amichot M, Ffrench-Constant RH, Feyereisen R. The Drosophila cytochrome P450 gene Cyp6a2: structure, localization, heterologous expression, and induction by phenobarbital. DNA Cell Biol. 1997;16:1345–1356. doi: 10.1089/dna.1997.16.1345. [DOI] [PubMed] [Google Scholar]
- Finney DJ. Statistical Method in Biological Assay. 3rd ed. London: Charles Griffin and Company Ltd; 1978. [Google Scholar]
- Fisk GJ, Thummel CS. Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Giebultowicz JM. Peripheral clocks and their role in circadian timing: insights from insects. Phil. Trans. R. Soc. B. 2001;356:1791–1799. doi: 10.1098/rstb.2001.0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Moore JT. CAR: detailing new models. Trends Pharmacol. Sci. 2004;25:437–441. doi: 10.1016/j.tips.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr. Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Heidari R, Devonshire AL, Campbell BE, Dorrian SJ, Oakeshott JG, Russell RJ. Hydrolysis of pyrethroids by carboxylesterases from Lucilia cuprina and Drosophila melanogaster with active sites modified by in vitro mutagenesis. Insect Biochem. Mol. Biol. 2005;35:597–609. doi: 10.1016/j.ibmb.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hooven LA, Sherman KA, Butcher S, Giebultowicz JM. Does the clock make the poison? Circadian variation in response to pesticides. PLoS One. 2009;4:e6469. doi: 10.1371/journal.pone.0006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DHDH, Lee SSSS. Origin of rate-acceleration in ester hydrolysis with metalloprotease mimics. Bioorg. Med. Chem. 2000;8:647–652. doi: 10.1016/s0968-0896(99)00310-7. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors—a perspective from Drosophila. Nat. Rev. Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- Lewis DFV. Oxidative stress: the role of cytochromes P450 in oxygen activation. J. Chem. Technol. Biotechnol. 2002;77:1095–1100. [Google Scholar]
- Lim C, Lee J, Koo E, Choe J. Targeted inhibition of Pdp1epsilon abolishes the circadian behavior of Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2007;364:294–300. doi: 10.1016/j.bbrc.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Pedra JH, McIntyre LM, Scharf ME, Pittendrigh BR. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7034–7039. doi: 10.1073/pnas.0400580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Wohlwill A, Dzitoeva S, Lin MH, Holbrook S, Storti RV. The Drosophila PAR domain protein 1 (Pdp1) gene encodes multiple differentially expressed mRNAs and proteins through the use of multiple enhancers and promoters. Dev. Biol. 2000;224:401–414. doi: 10.1006/dbio.2000.9797. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So V, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Schibler U. The daily timing of gene expression and physiology in mammals. Dialogues Clin. Neurosci. 2007;9:257–272. doi: 10.31887/DCNS.2007.9.3/uschibler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price J, Man B, Youngs M. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Teboul M, Guillaumond F, Grechez-Cassiau A, Delaunay F. The nuclear hormone receptors family round the clock. Mol. Endocrinol. 2008;12:2573–2582. doi: 10.1210/me.2007-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Zhang YK, Yeager RL, Klaassen CD. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab. Dispos. 2009;37:106–115. doi: 10.1124/dmd.108.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A. An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator. J. Neurosci. 2009;29:10920–10927. doi: 10.1523/JNEUROSCI.2133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.