Abstract
A notable success for evolutionary genetics during the past century was to generate a coherent, quantitative explanation for an apparent evolutionary paradox: the tendency for multicellular organisms to show declining fitness with age (senescence, often referred to simply as ‘ageing’). This general theory is now widely accepted and explains most of the features of senescence that are observed in natural and laboratory populations, but specific instantiations of that theory have been more controversial. To date, most of the empirical tests of these models have relied on data generated from biometric experiments. Modern population genetics and genomics provide new, and probably more powerful, ways to test ideas that are still controversial more than half a century after the original theory was developed. System-genetic experiments have the potential to address both evolutionary and mechanistic questions about ageing by identifying causal loci and the genetic networks with which they interact. Both the biometrical approaches and the newer approaches are reviewed here, with an emphasis on the challenges and limitations that each method faces.
Keywords: antagonistic pleiotropy, mutation accumulation, natural variation, QTL mapping, senescence
1. Introduction
Age is a very high price to pay for maturity.
(Tom Stoppard)
Declining vitality with age (senescence) is a nearly universal feature of multicellular organisms. Because senescence, by definition, is marked by progressively lower Darwinian fitness, it is not obviously an evolved trait. For example, one could argue that progressive deterioration is an inevitable property of complex systems (Comfort 1979; Gavrilov & Gavrilova 1991). Similar organisms sometimes senesce at very different rates, however, and some organisms appear able to postpone senescence nearly indefinitely (Comfort 1979; Finch 1990; Rose 1991). These observations indicate that senescence reflects species-specific biology and is not simply a property of complex systems.
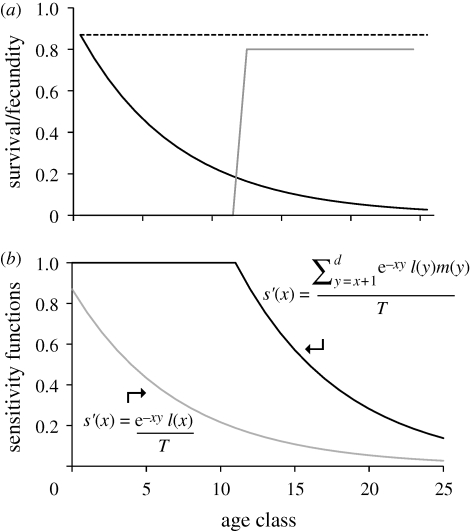
Indeed, evolutionary biologists proposed an explanation for senescence over 60 years ago, and this model accounts well for the general features of ageing (Rose 1991; Charlesworth 1994, 2000; Hughes & Reynolds 2005). The basic argument is simple: species have evolved senescent life histories because selection is weak against alleles that cause dysfunction only at old age (Medawar 1946, 1952; Williams 1957; Hamilton 1966). Hamilton (1966) was the first to demonstrate quantitatively that an originally non-senescent population will tend to evolve senescence through normal evolutionary processes of mutation and selection (figure 1). Hamilton's model was not based explicitly on population genetics, however. Subsequent work by Brian Charlesworth and others integrated Hamilton's model with standard models of population and quantitative genetics and demonstrated that Hamilton's equations are valid under many scenarios (reviewed by Charlesworth 1994, 2000). Hamilton's general model is now widely accepted (Hughes & Reynolds 2005; Partridge & Gems 2006), although a non-monotonic relationship between age and the strength of selection can occur under some parameterizations of the model (Baudisch 2005).
Figure 1.
(a) The demographic parameters for a hypothetical non-senescing population, as imagined by Hamilton (1966). Age-specific survival is constant, and cumulative survival declines geometrically. Age-specific reproduction rates are constant after the age of first reproduction, which in this case is age-class 12. (b) Hamilton's sensitivity functions, which reflect the strength of natural selection in the population described in (a). The symbols in the sensitivity functions represent terms from the discrete version of the Euler–Lotka equation, where r is the intrinsic rate of increase, l(x) is the probability of survival to age x, m(x) is the expected number of offspring produced by an individual of age x and T is a measure of generation time. (a) Dashed line, age-specific survival; black line, cumulative survival; grey line, age-specefic fecundity. (b) Black line, selection on survival; grey line, selection on fertility.
Modern experimental tests of this general theory of ageing began with a seminal paper by Rose & Charlesworth (1980). This was the first published work to quantitatively evaluate whether patterns of variation within populations conformed to predictions of Hamilton's model. This paper was also the first empirical attempt to differentiate between two different instantiations of the general model. These two scenarios have come to be known as the mutation accumulation (MA) and the antagonistic pleiotropy (AP) models.
Not coincidentally, discussion of these two models reflects classic evolutionary arguments about adaptive and non-adaptive genetic variation (Lewontin 1974) and more modern incarnations of the neutralist–selectionist debate. The non-adaptive MA scenario was first described by Medawar (1952). If mutations arise whose purely deleterious effects are confined to late ages, some of them will, because of weak selection and genetic drift, increase to high frequencies and will produce declines in individual health and performance at late ages. Mutations with deleterious effects on early ages will be subject to stronger purifying selection and so will accumulate in the population to a much lesser extent.
The adaptive AP scenario assumes the existence of alleles with beneficial effects at early ages but deleterious effects at later ages. Positive selection will cause such alleles to increase in frequency because their early-age effects are subject to stronger selection than their late-age effects. Alleles with the reverse pattern (deleterious early- and beneficial late-age effects) will be driven to low frequency by negative selection (Medawar 1952; Williams 1957).
The MA and AP processes are not mutually exclusive, and both could contribute to fixed differences in patterns of ageing between populations or to within-population variation. Although contributions of the AP process have been widely accepted, the idea that the non-adaptive MA process contributes substantially to variation in ageing within or among populations has been more contentious (Partridge & Gems 2002; Hughes & Reynolds 2005; Moorad & Promislow 2009). Below, I review several kinds of experiments that have been used to assess the relative importance of adaptive and non-adaptive processes in the evolution of ageing. New approaches using modern population genetics and genomics that could be deployed are also described, along with the initial results from some of the first attempts to use them.
2. Quantitative genetic approaches
Most attempts to disentangle MA and AP processes have, like the seminal Rose & Charlesworth (1980) experiment, used a quantitative genetic approach. Recent population- and quantitative-genetic theory predicts that the two processes should produce different patterns of age-specific genetic variance within populations (Charlesworth & Hughes 1996; Charlesworth 2001). Predictions include that MA should lead to an age-related increase in additive and non-additive genetic variance for fitness components and to corresponding increases in the inbreeding load. Subsequent empirical studies have generally supported these predictions (Hughes et al. 2002; Gong et al. 2006; Lesser et al. 2006; Swindell & Bouzat 2006; Borash et al. 2007; Reynolds et al. 2007; Escobar et al. 2008; Keller et al. 2008), but see Fox et al. (2006) and Fox & Stillwell (2009) for exceptions. Alternative formulations of these models have been proposed which predict that AP and MA will produce similar patterns of age-specific genetic variance (Moorad & Promislow 2009), but these models depend on assumptions about age-specific changes in allelic effects and dominance for which there is little evidence.
One issue that has not been widely discussed is that different species should be differentially susceptible to these two processes (Eyre-Walker 2006). Species with small effective population sizes (Ne), like humans, should be more susceptible to the accumulation of mutations with deleterious effects (MA). In contrast, species with large Ne, like Drosophila, should be more resistant to MA. A recent estimate indicates that the mean strength of selection acting on human polymorphisms is Nes < 1, that is, effective neutrality (Eyre-Walker et al. 2006). Most experiments that have attempted to differentiate AP and MA have been conducted in Drosophila or other invertebrates characterized by large population sizes. By the Ne argument, however, MA should be a more important contributor to ageing in humans and other species with small Ne.
The main prediction of the AP model for within-population variation is negative genetic correlation between early- and late-age fitness traits. Many studies, including some in natural populations, have documented these patterns, but others have not (Rose 1991; Hughes & Reynolds 2005; Charmantier et al. 2006; Hunt et al. 2006). Genetic correlations are notoriously difficult to estimate with precision, however; so failure to find them could simply reflect the limited statistical power in many studies.
Another prediction of the AP model is that genetic variation should be mainly due to alleles that segregate at intermediate frequencies rather than to rare alleles. Novel methods for testing this prediction have been developed recently (Kelly 1999, 2003, 2008; Macdonald & Long 2007). Applying these methods to the MA–AP debate has one limitation, however—deleterious alleles with delayed age of onset might behave as quasi-neutral alleles and therefore segregate at intermediate frequencies. This pattern is even more likely in species with small Ne. A solution to the ‘rare versus common-allele’ debate might therefore not resolve the question of whether MA, AP, or both contribute substantially to the evolution of senescence.
Differences between populations have also been used to test predictions of these models. Escobar et al. (2008) recently extended Charlesworth's earlier theoretical treatment of MA to include population structure. They argue that under MA, especially in species with low Ne, mutation–selection–drift balance should lead to independent fixation of deleterious alleles in different populations. Crosses between populations should therefore exhibit age-specific heterosis. Snails recently collected from nature confirmed this prediction (Escobar et al. 2008), and previous studies of long-term selection lines also documented this pattern (reviewed by Rose et al. 2007).
A concern with all quantitative genetic tests of these models is the sensitivity of parameter estimates to uncontrolled environmental and genetic effects. Empirical studies of wild populations cannot rule out genotype–environment interaction as a confounding effect (Wilson et al. 2008). These studies also suffer from generally high extrinsic mortality rates, which limit the number of individuals reaching truly advanced ages. Studies of recently established laboratory populations suffer from the ‘novel environment’ artefacts first described by Rose (1984): poor adaptation to the testing environment will lead to biased estimates of genetic variances and covariances among traits. Similar biases affect studies that extract chromosomes from one population and test them in a different, novel genetic background (cf. Promislow et al. 1996; Tatar et al 1996). Conversely, laboratory-adapted populations can suffer from inbreeding depression and from inadvertent selection for artificially short lifespan (Partridge & Gems 2007), although maintaining overlapping rather than discrete generations during laboratory culture should alleviate selection for artificially short lifespan (cf. Hughes 1995; Charlesworth & Hughes 1996; Hughes et al. 2002).
3. Gene-centric approaches
If quantitative genetics cannot resolve the question of whether variation in senescence is contributed mainly by adaptive (AP) or non-adaptive (MA) processes, then what can? The most direct test would be to identify alleles that cause senescence and to determine
— whether they have pleiotropic beneficial effects that are expressed at early ages and
— whether they exhibit molecular signatures of balancing selection (de Luca et al. 2003; Carbone et al. 2006).
Affirmative answers to these questions would support AP, whereas negative answers would support MA. Although these tests are simple to state, they are difficult to accomplish and rarely have been attempted even in a limited way, much less in a comprehensive, unbiased manner. Below, I describe some of the early attempts to gather the required data.
(a). Natural variants
Genes that contribute to senescence phenotypes can be identified essentially in two ways: screening of populations for naturally occurring variants and use of mutagenesis or other manipulations of gene expression to find genes with mutant phenotypes related to senescence. In one of the few studies to take the first approach and to successfully identify causal nucleotide variants, Carbone et al. (2006) found that a nucleotide substitution in the coding region of the Catecholamines up (Catsup) gene of Drosophila melanogaster was associated with longevity. Although the Catsup haplotypes sampled from the population had effects on several traits, including lifespan, locomotor behaviour and sensory bristle number, different sites within the gene were associated with different traits. These individual single-nucleotide polymorphisms (SNPs) were in linkage equilibrium. In other words, the nucleotide substitution that affected longevity did not affect any other measured trait, and it segregated independently of SNPs that did affect other traits. The minor allele frequency at the causal SNP was less than 5 per cent, and overall molecular diversity of the gene region was at the low end of values observed in this species. Patterns of variation in some regions of the gene were consistent with maintenance of variation by balancing selection, but the longevity SNP was not inside the region exhibiting this signature (Carbone et al. 2006; T. F. C. Mackay 2006, personal communication).
A related study by the same research group found that polymorphisms in the Dopa decarboxylase (Ddc) gene of D. melanogaster were also associated with longevity (de Luca et al. 2003). Although Ddc and Catsup function within the same biochemical pathway (catecholamine biosynthesis) and are located within 75 kb of each other, their patterns of association with longevity were dramatically different. Three variable sites in Ddc were in linkage disequilibrium and haplotypes had significant associations with longevity. One of these sites is within the promoter region of the gene, which exhibited a strong signature of balancing selection. Because of the strong linkage disequilibrium, effects of individual polymorphisms could not be determined, but the haplotype effects suggested strong epistatic interactions among the three SNPs within the gene. Also in contrast to Catsup, overall molecular diversity of this locus was high.
What is not obvious from this brief synopsis is the enormous effort that was required to generate these data. The chromosomal region containing Catsup and Ddc was first implicated as affecting longevity in a series of quantitative trait locus (QTL)-mapping experiments involving a cross between two inbred laboratory strains (Nuzhdin et al. 1997; Leips & Mackay 2000; Vieira et al. 2000). The QTL region was narrowed by crossing of the parental strains to stocks containing chromosomal deficiencies with well-defined borders (Pasyukova et al. 2000; de Luca et al. 2003; Mackay et al. 2006). Finally, association of the phenotype with individual polymorphisms was detected in a large sample of chromosome-extraction lines, in which the chromosomes were derived from a single natural population (de Luca et al. 2003; Carbone et al. 2006).
Despite this prodigious effort, interpreting the results in the context of MA and AP models of ageing is difficult. In these (and any other) mapping-based studies, choices had to be made to ensure efficient identification of causal variants. In this series of experiments, a gene had to be implicated both in differences between inbred laboratory strains and in a sample of chromosomes derived from nature and placed on an inbred genetic background. Genes identified in this way (and their allelic effects) might not be representative of all genes affecting senescence. In the Ddc study, the investigators sampled only 12 natural alleles to define the coding-region polymorphisms that would be scored in the large population sample, increasing the probability that only common variants would be detected. In the Catsup study, 169 natural alleles were completely sequenced for approximately 3000 bp, a scale that accounts for the ability of that study to detect association with rare variants. For both genes, lifespan and other phenotypes were measured in an environment that was novel to the genotypes being tested. Lifespan variation in a novel environment might reflect not the variation that would be observed in an environment to which the animal is well adapted but rather incidental variation in pre-adaptation to the new conditions (Rose 1984).
(b). Induced mutations
Mutagenesis and transgenic manipulation provide other ways to identify genes contributing to ageing. Analysis of induced mutations in model organisms has uncovered genes with large effects on lifespan and ageing-related traits (Kenyon 2001; Partridge & Gems 2002; Sinclair 2005). For example, manipulation of genes involved in an insulin signalling pathway can regulate maturation, reproduction and longevity in Caenorhabditis elegans, D. melanogaster and mammals (Bluher et al. 2003). Decreased signalling through this pathway leads to delayed and decreased reproduction and to increased longevity (Kenyon 2005; Piper et al. 2008). Other genes and pathways that apparently regulate lifespan have been indentified, and this literature is reviewed extensively elsewhere (Kenyon 2001; Partridge & Gems 2002; Hughes & Reynolds 2005; Sinclair 2005; Kuningas et al. 2008).
What do these mutations tell us about the evolution of ageing? Because many of these genetic lesions show the type of pleiotropic effects expected under the AP model (Partridge et al. 2005; Rose et al. 2007), we know that individual mutations can demonstrate antagonistic effects at young and old ages. A critical question is whether the same genes and pathways implicated in these molecular genetic studies also control natural variation in senescence, however (Partridge & Gems 2006). If the answer is yes, then testing the evolutionary models with sequence data from naturally occurring alleles should be a feasible approach to the problem.
At least one gene for which mutations strongly affect lifespan in D. melanogaster (Methuselah or Mth) also exhibits extensive natural polymorphism, a natural cline in allele frequencies that parallels a cline in lifespan, and a molecular signature of selection (Schmidt et al. 2000). Mutant analysis can therefore potentially reveal genes contributing to natural variation. Such genes can then be subjected to functional and evolutionary analyses that test model predictions. This path is not easy, however. Although the effects of Mth on lifespan have been recognized for more than a decade (Lin et al. 1998), natural allelic variation at this gene has still not been directly implicated in the clinal variation in lifespan (Paaby & Schmidt 2008). Of the many candidate genes nominated by mutagenic or transgenic manipulation, Mth is the only one that has been formally associated with natural variation in lifespan or ageing. More generally, loci contributing to natural variation in traits such as lifespan, starvation resistance and male mating behaviour show little overlap with those known to have large effects on the same phenotypes when mutated (Mackay et al. 2009). These results suggest that the candidate-gene approach is not an efficient one for documenting the genetic basis of natural variation in senescence.
4. Genomic approaches
Given how laborious it is to define causal polymorphisms for natural variation in ageing, perhaps genome-wide scans can resolve the debate about adaptive and non-adaptive causes of senescence without actually identifying the causal variants. For example, several QTL mapping studies in Drosophila and mice suggest that different alleles affect traits expressed at early and late ages (Nuzhdin et al. 1997; Curtsinger & Khazaeli 2002; Miller et al. 2005; Leips et al. 2006). Taken at face value, these results support the MA model, because it predicts that genes affecting late-life variation should be independent from those affecting early-life variation; under AP, the same alleles cause variation at both ages. QTL studies often have low power to detect alleles of small effect, which is thought to be one cause of failure to replicate results across studies (Shmookler Reis et al. 2006; Benfey & Mitchell-Olds 2008). A pattern of different QTLs detected at different ages within a single study might also be due to a high false-negative rate (Curtsinger & Khazaeli 2002). In any case, the pleiotropy cannot be confirmed until QTL regions are refined to the level of individual genes, a process that has been accomplished very rarely, even in Drosophila.
Large, well-designed whole-genome association studies provide another tool for addressing the same question: do different genomic regions control variation at early and late ages? To date, such studies have been applied mainly to humans with the goal of identifying risk factors for specific diseases, and none of these studies has included traits measured at different ages, presumably because of the high cost of phenotyping thousands of individual humans. In the future, long-term longitudinal studies could be used in this manner (Martin et al. 2007). Such studies, if they are large enough, might provide the best venue for determining whether different genes affect traits expressed at different ages.
Hybrid approaches that combine mapping experiments with high-throughput measures of phenotypic variation, gene expression and possibly other ‘intermediate’ phenotypes could also be brought to bear on questions about the evolution of ageing. Using this strategy, one can identify QTLs, and ultimately the causal molecular variant, for traditional phenotypes and for transcriptional, metabolic, protein or other molecular phenotypes. For example, QTL mapping of trait variation, genome-wide mRNA expression data and ‘expression-QTL’ mapping have been combined to generate models that relate causal polymorphisms to the downstream regulatory networks that regulate phenotypic variation (Chen et al. 2008; Emilsson et al. 2008; Ayroles et al. 2009; Harbison et al. 2009). A similar approach could be applied to well-designed whole-genome association-study mapping panels (Mackay et al. 2009). Research programmes that use these hybrid methods have been referred to as ‘genetical genomics’ (Jansen & Nap 2001) and ‘system genetics’ (Threadgill 2006).
A system-genetic approach has not yet been applied to age-specific trait variation. When it is, the results could address questions about the evolution of ageing and about the mechanisms of ageing simultaneously. System genetics will also provide a pathway to associate many unannotated genes with functions related to ageing (Mackay et al. 2009). Given the need to collect phenotypic and gene-expression data for multiple age classes and for many individuals, such an approach to ageing research is most practical currently for Drosophila and other model organisms that can be reared inexpensively and in large numbers.
One possibility is that community-based reference panels can be used. Panels of fully sequenced inbred lines, or of lines genotyped for many thousands of markers, are becoming available for several model organisms. These panels are intended to leverage the effort of an entire community of researchers to produce a rich body of phenotypic data (including transcriptional, metabolic and proteomic phenotypes) that can be related to genetic information. These reference panels thus provide a means to integrate age-specific data on a wide range of phenotypes, and should allow more comprehensive tests of the adaptive versus non-adaptive models of ageing than have been possible previously. Few of these community efforts are designed to survey natural variation derived from a single population, however. The Drosophila Genetic Reference Panel of inbred lines of D. melanogaster (DGRP; Mackay et al. 2008) was derived from a single natural population, but this panel is designed to detect alleles of moderate to large effect and alleles segregating at intermediate frequency. It will therefore be of limited use in testing the MA model. Using the sequenced lines of the DGRP as starting material for a series of crosses, in the tradition of classical biometrics, could provide the needed resolution. These same experiments are also likely to uncover novel molecular mechanisms of ageing and other life-history phenomena, in addition to allowing estimates of allelic effects in non-inbred individuals.
5. Concluding remarks
Like many fundamental questions in evolutionary genetics, determining whether senescence is primarily the result of adaptive or of non-adaptive processes has been a difficult challenge. Although some compelling evidence implies that both kinds of processes have contributed, at least in Drosophila, the issue is still not resolved. Although ascertainment bias will remain a concern, modern system-genetic approaches may provide a tool with which to uncover a relatively unbiased sample of polymorphisms that directly control variation in ageing within and between populations. Knowledge of the molecular pathways that these polymorphisms regulate, together with population-genetic tests of selection, should permit tests of the MA and AP models on a gene-by-gene basis. Fortuitously, the same methods will yield tremendous insight into the functional basis of senescence and the molecular mechanisms underlying life-history variation.
Acknowledgements
I thank Jessica Henrichs, Silvia Remolina and Anne B. Thistle for comments on drafts of this manuscript and Brian Charlesworth for encouraging and inspiring my interest in the evolution of ageing.
Footnotes
One contribution of 16 to a Theme Issue ‘The population genetics of mutations: good, bad and indifferent’ dedicated to Brian Charlesworth on his 65th birthday.
References
- Ayroles J. F., et al. 2009Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41, 299–307 (doi:10.1038/ng.332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudisch A.2005Hamilton's indicators of the force of selection. Proc. Natl Acad. Sci. USA 102, 8263–8268 (doi:10.1073/pnas.0502155102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P. N., Mitchell-Olds T.2008Perspective—from genotype to phenotype: systems biology meets natural variation. Science 320, 495–497 (doi:10.1126/science.1153716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M., Kahn B. B., Kahn C. R.2003Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574 (doi:10.1126/science.1078223) [DOI] [PubMed] [Google Scholar]
- Borash D. J., Rose M. R., Mueller L. D.2007Mutation accumulation affects male virility in Drosophila selected for later reproduction. Physiol. Biochem. Zool. 80, 461–472 (doi:10.1086/520127) [DOI] [PubMed] [Google Scholar]
- Carbone M. A., Jordan K. W., Lyman R. F., Harbison S. T., Leips J., Morgan T. J., de Luca M., Awadalla P., Mackay T. F. C.2006Phenotypic variation and natural selection at Catsup, a pleiotropic quantitative trait gene in Drosphila. Curr. Biol. 16, 912–919 (doi:10.1016/j.cub.2006.03.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B.1994Evolution in age-structured populations. Cambridge, UK: Cambridge University Press [Google Scholar]
- Charlesworth B.2000Fisher, Medawar, Hamilton and the evolution of aging. Genetics 156, 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B.2001Patterns of age-specific means and genetic variances of mortality rates predicted by the mutation-accumulation theory of ageing. J. Theor. Biol. 210, 47–65 (doi:10.1006/jtbi.2001.2296) [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Hughes K. A.1996Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl Acad. Sci. USA 93, 6140–6145 (doi:10.1073/pnas.93.12.6140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., McCleery R. H., Sheldon B. C.2006Quantitative genetics of age at reproduction in wild swans: support for antagonistic pleiotropy models of senescence. Proc. Natl Acad. Sci. USA 103, 6587–6592 (doi:10.1073/pnas.0511123103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Q., et al. 2008Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (doi:10.1038/nature06757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort A.1979The biology of senescence. Edinburgh, UK: Churchill Livingstone [Google Scholar]
- Curtsinger J. W., Khazaeli A. A.2002Lifespan, QTLs, age-specificity, and pleiotropy in Drosophila. Mech. Ageing Dev. 123, 81–93 (doi:10.1016/S0047-6374(01)00345-1) [DOI] [PubMed] [Google Scholar]
- de Luca M., Roshina N. V., Geiger-Thornsberry G. L., Lyman R. F., Pasyukova E. G., Mackay T. F. C.2003Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34, 429–433 (doi:10.1038/ng1218) [DOI] [PubMed] [Google Scholar]
- Emilsson V., et al. 2008Genetics of gene expression and its effect on disease. Nature 452, 423–428 (doi:10.1038/nature06758) [DOI] [PubMed] [Google Scholar]
- Escobar J. S., Jarne P., Charmantier A., David P.2008Outbreeding alleviates senescence in hermaphroditic snails as expected from the mutation-accumulation theory. Curr. Biol. 18, 906–910 (doi:10.1016/j.cub.2008.04.070) [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A.2006The genomic rate of adaptive evolution. Trends Ecol. Evol. 21, 569–575 (doi:10.1016/j.tree.2006.06.015) [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Woolfit M., Phelps T.2006The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics 173, 891–900 (doi:10.1534/genetics.106.057570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch C. E.1990Longevity, senescence, and the genome. Chicago, IL: University of Chicago Press [Google Scholar]
- Fox C. W., Stillwell R. C.2009Environmental effects on sex differences in the genetic load for adult lifespan in a seed-feeding beetle. Heredity 103, 62–72 (doi:10.1038/hdy.2009.31) [DOI] [PubMed] [Google Scholar]
- Fox C. W., Scheibly K. L., Wallin W. G., Hitchcock L. J., Stillwell R. C., Smith B. P.2006The genetic architecture of life span and mortality rates: gender and species differences in inbreeding load of two seed-feeding beetles. Genetics 174, 763–773 (doi:10.1534/genetics.106.060392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov L. A., Gavrilova N. S.1991The biology of life span: a quantitative approach. Chur, Switzerland: Harwood Academic [Google Scholar]
- Gong Y., Thompson J. N., Woodruff R. C.2006Effect of deleterious mutations on life span in Drosophila melanogaster. J.Gerontol. A Biol. Sci. Med. Sci. 61, 1246–1252 [DOI] [PubMed] [Google Scholar]
- Hamilton W. D.1966The moulding of senescence by natural selection. J. Theor. Biol. 12, 12–45 (doi:10.1016/0022-5193(66)90184-6) [DOI] [PubMed] [Google Scholar]
- Harbison S. T., Carbone M. A., Ayroles J. F., Stone E. A., Lyman R. F., Mackay T. F. C.2009Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat. Genet. 41, 371–375 (doi:10.1038/ng.330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. A.1995The evolutionary genetics of male life-history characters in Drosophila melanogaster. Evolution 49, 521–537 (doi:10.2307/2410276) [DOI] [PubMed] [Google Scholar]
- Hughes K. A., Reynolds R. M.2005Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 50, 421–445 (doi:10.1146/annurev.ento.50.071803.130409) [DOI] [PubMed] [Google Scholar]
- Hughes K. A., Alipaz J. A., Drnevich J. M., Reynolds R. M.2002A test of evolutionary theories of aging. Proc. Natl Acad. Sci. USA 99, 14 286–14 291 (doi:10.1073/pnas.222326199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt J., Jennions M. D., Spyrou N., Brooks R.2006Artificial selection on male longevity influences age-dependent reproductive effort in the black field cricket Teleogryllus commodus. Am. Nat. 168, E72–E86 (doi:10.1086/506918) [DOI] [PubMed] [Google Scholar]
- Jansen R. C., Nap J. P.2001Genetical genomics: the added value from segregation. Trends Genet. 17, 388–391 (doi:10.1016/S0168-9525(01)02310-1) [DOI] [PubMed] [Google Scholar]
- Keller L. F., Reid J. M., Arcese P.2008Testing evolutionary models of senescence in a natural population: age and inbreeding effects on fitness components in song sparrows. Proc. R. Soc. B 275, 597–604 (doi:10.1098/rspb.2007.0961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. K.1999An experimental method for evaluating the contribution of deleterious mutations to quantitative trait variation. Genet. Res 73, 263–273 (doi:10.1017/S0016672399003766) [DOI] [PubMed] [Google Scholar]
- Kelly J. K.2003Deleterious mutations and the genetic variance of male fitness components in Mimulus guttatus. Genetics 164, 1071–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.2008Testing the rare-alleles model of quantitative variation by artificial selection. Genetica 132, 187–198 (doi:10.1007/s10709-007-9163-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C.2001A conserved regulatory system for aging. Cell 105, 165–168 (doi:10.1016/S0092-8674(01)00306-3) [DOI] [PubMed] [Google Scholar]
- Kenyon C.2005The plasticity of aging: insights from long-lived mutants. Cell 120, 449–460 (doi:10.1016/j.cell.2005.02.002) [DOI] [PubMed] [Google Scholar]
- Kuningas M., Mooijaart S. P., van Heemst D., Zwaan B. J., Slagboom P. E., Westendorp R. G. J.2008Genes encoding longevity: from model organisms to humans. Aging Cell 7, 270–280 (doi:10.1111/j.1474-9726.2008.00366.x) [DOI] [PubMed] [Google Scholar]
- Leips J., Mackay T. F. C.2000Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155, 1773–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips J., Gilligan P., Mackay T. R. C.2006Quantitative trait loci with age-specific effects on fecundity in Drosophila melanogaster. Genetics 172, 1595–1605 (doi:10.1534/genetics.105.048520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser K. J., Paiusi I. C., Leips J.2006Genetic variation in age-specific immune response in Drosophila melanogaster. Aging Cell 5, 293–295 (doi:10.1111/j.1474-9726.2006.00219.x) [DOI] [PubMed] [Google Scholar]
- Lewontin R. C.1974The genetic basis of evolutionary change New York, NY: Columbia University Press [Google Scholar]
- Lin Y. J., Seroude L., Benzer S.1998Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282, 943–946 (doi:10.1126/science.282.5390.943) [DOI] [PubMed] [Google Scholar]
- Macdonald S. J., Long A. D.2007Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176, 1261–1281 (doi:10.1534/genetics.106.069641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Roshina N. V., Leips J. W., Pasyukova E. G.2006Complex genetic architecture of Drosophila longevity. In Handbook of the biology of aging (eds Masoro E. J., Austad S. N.), pp. 181–216 Boston, MA: Elsevier [Google Scholar]
- Mackay T. F. C., Richards S., Gibbs R.2008Proposal to sequence a Drosophila Genetic Reference Panel: a community resource for the study of genotypic and phenotypic variation. White Paper, NHGRI. See www.genome.gov/Pages/Research/Sequencing/SeqProposals/DrosophilaSeq.pdf [Google Scholar]
- Mackay T. F. C., Stone E. A., Ayroles J. F.2009The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10, 565–577 (doi:10.1038/nrg2612) [DOI] [PubMed] [Google Scholar]
- Martin G. M., Bergman A., Barzilai N.2007Genetic determinants of human health span and life span: progress and new opportunities. PLoS Genet. 3, e125 (doi:10.1371/journal.pgen.0030125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar P. B.1946Old age and natural death. Modern Quart. 1, 30–56 [Google Scholar]
- Medawar P. B.1952An unsolved problem of biology London, UK: H. K. Lewis [Google Scholar]
- Miller R. A., Berger S. B., Burke D. T., Galecki A., Garcia G. G., Harper J. M., Akha A. A. S.2005T cells in aging mice: genetic, developmental, and biochemical analyses. Immunol. Rev. 205, 94–103 (doi:10.1111/j.0105-2896.2005.00254.x) [DOI] [PubMed] [Google Scholar]
- Moorad J. A., Promislow D. E. L.2009What can genetic variation tell us about the evolution of senescence? Proc. R. Soc. B 276, 2271–2278 (doi:10.1098/rspb.2009.0183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin S. V., Pasyukova E. G., Dilda C. L., Zeng Z. B., Mackay T. F. C.1997Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 94, 9734–9739 (doi:10.1073/pnas.94.18.9734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Schmidt P. S.2008Functional significance of allelic variation at methuselah, an aging gene in Drosophila. PLoS ONE 3, e1987 (doi:10.1371/journal.pone.0001987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Gems D.2002Mechanisms of ageing: public or private? Nat. Rev. Genet. 3, 165–175 (doi:10.1038/nrg753) [DOI] [PubMed] [Google Scholar]
- Partridge L., Gems D.2006Beyond the evolutionary theory of ageing, from functional genomics to evo-gero. Trends Ecol. Evol. 21, 334–340 (doi:10.1016/j.tree.2006.02.008) [DOI] [PubMed] [Google Scholar]
- Partridge L., Gems D.2007Benchmarks for ageing studies. Nature 450, 165–167 (doi:10.1038/450165a) [DOI] [PubMed] [Google Scholar]
- Partridge L., Gems D., Withers D. J.2005Sex and death: what is the connection? Cell 120, 461–472 (doi:10.1016/j.cell.2005.01.026) [DOI] [PubMed] [Google Scholar]
- Pasyukova E. G., Vieira C., Mackay T. F. C.2000Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156, 1129–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. D. W., Selman C., McElwee J. J., Partridge L.2008Separating cause from effect: how does insulin/IGF signalling control lifespan in worms, flies and mice? J. Int. Med. 263, 179–191 [DOI] [PubMed] [Google Scholar]
- Promislow D. E. L., Tatar M., Khazaeli A. A., Curtsinger J. W.1996Age-specific patterns of genetic variance in Drosophila melanogaster. I. Mortality. Genetics 143, 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. M., Temiyasathit S., Reedy M. M., Ruedi E. A., Drnevich J. M., Leips J., Hughes K. A.2007Age specificity of inbreeding load in Drosophila melanogaster and implications for the evolution of late-life mortality plateaus. Genetics 177, 587–595 (doi:10.1534/genetics.106.070078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. R.1984Genetic covariation in Drosophila life history: untangling the data. Am. Nat. 123, 564–569 (doi:10.1086/284222) [Google Scholar]
- Rose M. R.1991The evolutionary biology of aging. Oxford, UK: Oxford University Press [Google Scholar]
- Rose M. R., Charlesworth B.1980A test of evolutionary theories of senescence. Nature 287, 141–142 (doi:10.1038/287141a0) [DOI] [PubMed] [Google Scholar]
- Rose M. R., Rauser C. L., Benford G., Matos M., Mueller L. D.2007Hamilton's forces of natural selection after forty years. Evolution 61, 1265–1276 (doi:10.1111/j.1558-5646.2007.00120.x) [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Duvernell D. D., Eanes W. F.2000Adaptive evolution of a candidate gene for aging in Drosophila. Proc. Natl Acad. Sci. USA 97, 10 861–10 865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis R. J., Kang P., Ayyadevara S.2006Quantitative trait loci define genes and pathways underlying genetic variation in longevity. Exp. Gerontol. 41, 1046–1054 (doi:10.1016/j.exger.2006.06.047) [DOI] [PubMed] [Google Scholar]
- Sinclair D. A.2005Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 126, 987–1002 (doi:10.1016/j.mad.2005.03.019) [DOI] [PubMed] [Google Scholar]
- Swindell W. R., Bouzat J. L.2006Inbreeding depression and male survivorship in Drosophila: implications for senescence theory. Genetics 172, 317–327 (doi:10.1534/genetics.105.045740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., Promislow D. E. L., Khazaeli A. A., Curtsinger J. W.1996Age-specific patterns of genetic variance in Drosophila melanogaster. II. Fecundity and its genetic covariance with age-specific mortality. Genetics 143, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W.2006Meeting report for the 4th annual complex trait consortium meeting: from QTLs to systems genetics. Mamm. Genome 17, 2–4 (doi:10.1007/s00335-005-0153-5) [DOI] [PubMed] [Google Scholar]
- Vieira C., Pasyukova E. G., Zeng Z. B., Brant H. J., Lyman R. F., Mackay T. F. C.2000Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154, 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. C.1957Pleitropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 (doi:10.2307/2406060) [Google Scholar]
- Wilson A. J., Charmantier A., Hadfield J. D.2008Evolutionary genetics of ageing in the wild: empirical patterns and future perspectives. Funct. Ecol. 22, 431–442 (doi:10.1111/j.1365-2435.2008.01412.x) [Google Scholar]