Abstract
We discuss the dynamics of adaptive evolution in asexual (clonal) populations. The classical ‘periodic selection’ model of clonal evolution assumed that beneficial mutations are very rare and therefore substitute unfettered into populations as occasional, isolated events. Newer models allow for the possibility that beneficial mutations are sufficiently common to coexist and compete for fixation within populations. Experimental evolution studies in microbes provide empirical support for stochastic models in which both selection and mutation are strong effects and clones compete for fixation; however, the relative importance of competition among clones bearing mutations of different selective effects versus competition among clones bearing multiple mutations remains unresolved. We provide some new theoretical results, moreover, suggesting that population dynamics consistent with the periodic selection model can arise even in a deterministic model that can accommodate a very high beneficial mutation rate.
Keywords: beneficial mutations, periodic selection, clonal interference, multiple mutations, asexual populations
1. Introduction
Beneficial mutations are the very stuff of adaptive evolution, yet as Orr (2010) has noted, population genetics has focused most of its attention to date on deleterious and neutral mutations. The historical neglect of beneficial mutations is partly attributable to their rarity compared with deleterious and neutral mutations, but it is also true that we have lacked ready means to detect beneficial mutations. Recently, experimental evolution approaches have provided evidence that beneficial mutations are more abundant than previously suspected and have stimulated theoretical and experimental interest in the consequences of abundant beneficial mutations. Interest has been largely focused on asexual (clonal) populations, for perhaps two reasons. The first reason is practical: experimental evolution studies are most readily carried out with microbes, which are more easily propagated asexually. The second reason is theoretical and is more significant: clonality is expected to have a profound effect on the substitution of beneficial mutations into a population (Fisher 1930; Muller 1932).
In a classic paper, Muller (1932) contrasted the genetics of adaptation in sexual and asexual populations: when beneficial mutations are extremely rare, the rates of adaptation of sexual and asexual populations, all else being equal, will be the same and will depend on the waiting time between isolated mutations; however, when beneficial mutations are common, sexual populations can evolve by combining coexisting mutations from different genetic backgrounds, but asexual populations must wait for different mutations to arise on the same genetic background. An adaptation of Muller's (1932) figure comparing the fate of beneficial mutations in sexual and asexual populations (Crow & Kimura 1965, 1970) has appeared in numerous textbooks and is familiar to many biologists.
Despite the fundamental insight of Fisher and Muller (and, perhaps, for lack of compelling evidence that beneficial mutations are sufficiently common to coexist in asexual populations), early work on the adaptive evolution of clonal microbial populations emphasized the rarity of beneficial mutations. In particular, the notion that the evolution of such populations is characterized by ‘periodic selection’—replacement of a population by the clonal descendants of a rare beneficial mutation, followed by a wait until the next such replacement and so forth—took hold in the 1950s, seemingly supported by some of the earliest experimental evolution studies in bacteria (Novick & Szilard 1950; Atwood et al. 1951a,b) and later results in yeast (Paquin & Adams 1983). Indeed, periodic fixation of rare beneficial mutations was still a sufficiently influential concept in the 1990s to be assumed as the likely explanation for a ‘punctuated’ pattern of stepwise evolution of fitness and cell size increases observed in an experimental Escherichia coli population (Elena et al. 1996). But emerging observations were suggesting that multiple beneficial mutations might actually be contending for fixation in such populations. Molecular analyses, for example, were uncovering phenomena that seemed to indicate evolutionary ‘leapfrogging’ of clones: events in which a numerically dominant genotype was rapidly supplanted by a less-frequent genotype where both appeared to have derived independently from a common ancestor (Holmes et al. 1992; Papadopoulos et al. 1999), as if the less-frequent genotype harboured a beneficial mutation stronger in its effect than the mutation that had propelled the dominant genotype to high frequency in the first place. Influenced by such observations, Gerrish & Lenski (1998) developed a theory of adaptation in asexual populations in which several single beneficial mutations of differing effect contend for fixation, a process that they termed ‘clonal interference’. Earlier theoretical work, in fact, had explored the implications of interference between linked beneficial mutations in sexual populations (Hill & Robertson 1966; Felsenstein 1974; Barton 1995), which became known in that context as the ‘Hill–Robertson effect’.
In what follows, we briefly review some of the more recent developments in the population genetics of adaptation in asexuals. We assume that population sizes and mutation rates are such that rapid fitness degradation owing to the accumulation of deleterious mutations (see Loewe & Hill 2010) is not a concern; we focus, instead, on the dynamics of beneficial mutations. We emphasize empirical results from experimental populations of microbes, as this is where most of the important work in this area has taken place recently. However, we note at the outset that the extent to which beneficial mutation rates and distributions of beneficial mutational effects observed in experimental populations of microbes resemble those in natural populations, microbial or otherwise, remains essentially unknown.
As a framework for discussion, we consider three regimes of beneficial mutation and selection (following terminology introduced by Gillespie 1983, 1984, 1991) under which an asexual population might be evolving: (i) Strong selection, weak mutation (SSWM)—in this regime, beneficial mutations are rare and their average selective effect is substantial; (ii) Weak selection, strong mutation (WSSM)—in this regime, beneficial mutations are extremely common but their average selective effect is small; and (iii) Strong selection, strong mutation (SSSM)—in this regime, beneficial mutations are common enough to co-occur in a population and their selective effects can be substantial. We note that these categories reflect a combination of: (i) assumptions about the mutation rate necessary for the construction of the corresponding models and (ii) constraints on selection that make the models fit observed data.
The most significant recent theoretical and experimental work has focused on the SSSM regime, and this is where we concentrate our attention in the review section of this paper. At the end of the paper, however, we present some new theoretical results suggesting that evolutionary dynamics in the WSSM regime can mimic behaviour previously supposed to support the periodic selection model.
2. Adaptation in asexual populations: the dynamical consequences of beneficial mutations
The substitution of a beneficial mutation in a (real) population involves both genetic drift and selection. In a finite population, a newly arisen beneficial mutation has only a small probability of reaching a frequency at which its fate can be governed deterministically by selection. Haldane (1927) first showed that this probability is approximately 2s, if a mutation is of small beneficial effect s and offspring number is Poisson distributed. A beneficial mutation that has survived drift remains polymorphic for some period of time as it rises in frequency under the influence of selection. In a sexual population with free recombination, the ultimate fixation of such a beneficial mutation is certain, and the time to fixation is governed by s. In an asexual population, however, fixation of a beneficial mutation that has survived drift is not certain, and the time to fixation (assuming it occurs) is not governed solely by s: both depend, instead, on whether competing beneficial mutations are present in the population. Whether competing beneficial mutations are present depends, in turn, on the relationship between the typical selection coefficient associated with a beneficial mutation, the population size (N), and the rate at which beneficial mutations arise in individual genomes (Ub). Stronger beneficial mutations have a higher probability of surviving drift; they also sweep towards fixation more rapidly, allowing a smaller window of generations in which competing beneficial mutations might arise and survive drift. The total supply rate of beneficial mutations that survive drift in the population is the product 2sNUb, and the total number of replications that could potentially give rise to a competing mutation before a particular beneficial mutation of interest reaches majority frequency in the population is ln (Ns/2)/s. The expected number of competing beneficial mutations is thus given by the product of these two quantities, which is 2NUb ln (Ns/2), and the probability that a given beneficial mutation meets no superior competitors on its way to fixation is inversely related to this expected number.
(a). The strong selection, weak mutation regime
Assuming that selection and drift operate independently of one another, beneficial mutations will tend to survive drift and substitute singly in an asexual population when 2NUb ≪ 1/ln (Ns/2) (Gerrish & Lenski 1998; Desai & Fisher 2007; Desai et al. 2007); this is the SSWM regime. Under this regime, the population will evolve via a train of isolated adaptive substitutions, consistent with the periodic selection model; the rate at which such substitutions occur, and hence the rate of adaptation, will be limited by the rate of mutation. Each adaptive substitution is predicted to decrease variation in the population sharply (Koch 1974; Levin 1981). As mentioned previously, the periodic selection model was supported by pioneering evolution experiments in microbes in the 1950s through the 1990s; interestingly, however, the same experimental approaches have more recently been used to infer that beneficial mutations are too common for simple periodic selection to occur.
(b). The weak selection, strong mutation regime
At the other extreme from the SSWM regime lies the condition in which 2NUb ≫ 1/ln (Ns/2). Here, beneficial mutations are so common, relative to their selective impact, that it is to be expected that very many beneficial mutations are present in a population at all times and the rate of adaptation is wholly limited by the rate at which selection can incorporate these beneficial mutations into the population; this is the WSSM regime. Models of the WSSM regime are highly simplified theoretical caricatures of evolution at high NUb whose justification is nevertheless gaining some credibility as evidence for high beneficial mutation rates seems to be accumulating (see below). In these models, evolution of an asexual population is approximated by a deterministic model that represents the population fitness distribution through time as a travelling wave (Tsimring et al. 1996; Rouzine et al. 2003, 2008; Gerrish et al. 2007). We present some new theoretical results based on this approach below, and so we postpone further description of modelling approaches in this regime until then.
(c). The strong selection, strong mutation regime
Along the continuum from mutation limitation to selection limitation, the complex clonal dynamics originally envisioned by Fisher and Muller begin to arise when beneficial mutations become common enough to interfere with one another (2NUb > 1/ln (Ns/2)). This realm, the SSSM regime, is characterized by chance co-occurrence and competition among beneficial mutations; here, stochastic modelling approaches would seem most appropriate. Two such approaches were recently taken to model the adaptive evolution of an asexual population in this regime. Gerrish & Lenski (1998) and Wilke (2004) considered competition among single beneficial mutations of differing, continuously distributed effects, under the simplifying assumption that mutations ultimately fix one at a time in a population. In contrast, Desai and co-workers (Desai & Fisher 2007; Desai et al. 2007) more recently modelled competition among multiple-mutant clones, under the simplifying assumption that all beneficial mutations are of the same effect. Under both models, some beneficial mutations are inevitably ‘wasted’ in an asexual population as a consequence of clonal competition; the models do, however, make differing predictions as to the dynamics of adaptation and the maintenance of variation in asexual populations. In discussing these models further, we will refer to them separately as the ‘original clonal interference’ and ‘original multiple mutations’ models. In doing so, we do not mean to imply that these models are mutually exclusive representations of evolution in asexual populations. As we describe below, recent simulation studies indicate that the processes described by both models—competition among single mutants of different effect versus competition among clones bearing different numbers of mutations of similar effect—can both occur in asexual populations, with their relative importance depending on the distribution of beneficial mutational effects.
The central concept of the original clonal interference model of Gerrish & Lenski (1998) is that an asexual population may harbour several separate ‘contending mutations’ that interfere with each others' fixation. As a simplified two-locus illustration of this, consider an ancestral haploid asexual genotype ab: at some time, a beneficial mutation arises in this ancestral genotype to give rise to the more-fit genotype Ab. If the A mutation survives drift, it can then spread deterministically through the population. While the population is still polymorphic for the ab and Ab backgrounds, however, a yet fitter mutation might arise on the ancestral ab background and survive genetic drift, yielding genotype aB that now interferes with the fixation of genotype Ab, ultimately supplanting it. Thus, in this scheme, a beneficial mutation is fixed in a population only if it survives drift and no superior, interfering mutation appears and survives drift in the time interval required for its fixation. Using this basic framework, and assuming that the fitness effects of beneficial mutations are exponentially distributed, Gerrish and Lenski derived several results concerning the dynamics of adaptation in asexual populations. Perhaps the most significant of these, in their implications for the dynamics of asexual evolution, were as follows: (i) the fixation probability of a given beneficial mutation is a decreasing function of both the population size N and beneficial mutation rate Ub. (ii) Substitutions will occur as discrete, rare events, no matter how frequently beneficial mutations arise. This implies that periodic-selection-like dynamics (e.g. periodic declines in variation) are to be expected even when beneficial mutations are common enough to coexist in a population. (iii) The rate of fitness increase in a population will be an increasing function of population size and mutation rate but will depend only weakly on these parameters when their product NUb, the beneficial mutation supply rate, is already large.
Numerous predictions for the dynamics of adaptation in asexuals emerged from the original clonal interference model of Gerrish & Lenski (1998), and the literature devoted to tests of these predictions is substantial. Predictions and supporting studies include the following: the rate of adaptation of an asexual population should increase in a less-than-linear manner with increasing NUb, as implied above (Miralles et al. 1999, 2000; de Visser et al. 1999; Colegrave 2002; de Visser & Rozen 2005). Multiple clonal backgrounds within an evolving asexual population should increase in fitness simultaneously as they acquire beneficial mutations in parallel (Notley-Mcrobb & Ferenci 1999a,b, 2000; Shaver et al. 2002; de Visser & Rozen 2005; Hegreness et al. 2006; Kao & Sherlock 2008; Pepin & Wichman 2008). Selectively favoured clones may be transiently common in populations, only to be displaced by later-arising, fitter clones (Papadopoulos et al. 1999; Wichman et al. 1999; Holder & Bull 2001; Imhof & Schlötterer 2001; Hegreness et al. 2006; Bollback & Huelsenbeck 2007; Kao & Sherlock 2008; Pepin & Wichman 2008). Following from the previous prediction, those clones that do sweep to fixation in a population should take a longer time to fix than predicted by the net fitness gain accompanying their fixation (Shaver et al. 2002; de Visser & Rozen 2006). Finally, the frequency trajectory of a clone that does rise to selective fixation may exhibit erratic dynamics as its fitness advantage fluctuates relative to that of competing clones (Shaver et al. 2002; de Visser & Rozen 2006; Kao & Sherlock 2008).
An assumption of the original clonal interference model is that contending mutations always arise from the ancestral genotype—the ab background in the simplified example given above—rather than on the background of a currently spreading beneficial mutation (e.g. the Ab or aB backgrounds) in the population. Under this assumption, competition in the population is constrained to occur only among clones bearing single beneficial mutations, which greatly simplifies analysis of the dynamics. The truth is more complicated; however, as noted by several workers (Kim & Orr 2005; Barrett et al. 2006; Desai & Fisher 2007; Desai et al. 2007; Fogle et al. 2008), genotypes containing multiple newly arisen beneficial mutations are very likely to be present in populations whenever NUb is sufficiently high for contending mutations to be present. Kim & Orr (2005), for example, analysed a two-locus model in which, at very high NUb, recurrent mutation creates multiple-mutation genotypes at a sufficiently high rate to alleviate the effect of clonal interference; their predictions found some support in a study of adaptation in very large populations of an RNA virus with an extremely high mutation rate (Bollback & Huelsenbeck 2007).
Still more recently, Desai and co-workers (Desai & Fisher 2007; Desai et al. 2007) analysed a general model of evolution in an asexual population when multiple-mutant genotypes are allowed to arise and coexist. To render their model analytically tractable, Desai et al. (2007) made the simplifying assumption that all beneficial mutations can be considered as having a single characteristic selective value (see also Haigh 1978). This characteristic value is determined, in part, by the biasing effect of clonal interference acting on the distribution of effects of newly arising beneficial mutations. Mutations of smaller selective value are unlikely to survive clonal interference, whereas those of larger selective value are unlikely to arise in the first place; the distribution of mutations that contribute to adaptation is thus argued to be unimodal and centred strongly on a characteristic value. The validity of this simplification turns out to depend on the distribution of effects of beneficial mutations—a subject to which we return below.
The original multiple-mutations model of Desai et al. (2007) makes some quantitative predictions for the dynamics of evolution that differ from those of the original clonal interference model of Gerrish & Lenski (1998): (i) in contrast to the original clonal interference model, at high NUb substitutions will not occur as isolated events. Instead, a ‘nose’ of higher fitness, multiple-mutation genotypes will be continually generated by mutation and will lead the fitness distribution of the population smoothly towards higher values. (ii) Following from (i), variance for fitness in the population as a whole will not fluctuate with isolated substitutions, but will instead be maintained at a relatively constant value that depends on population size, mutation rate and average selection coefficient. (iii) The speed of adaptation will increase logarithmically with N and with Ub in marked contrast with the speed of adaptation under simple periodic selection, which is predicted to increase as NUb. When contrasted with the original clonal interference model, however, the difference is less marked; there, the speed of adaptation is predicted to increase logarithmically with N lnN and with Ub (Gerrish & Lenski 1998; Wilke 2004).
To test the original multiple-mutations model, Desai et al. (2007) evolved diploid populations of yeast (Saccharomyces cerevisiae) for 500 generations in the laboratory with six different combinations of population size and genomic mutation rate, measuring average fitnesses at regular intervals and the distribution of fitnesses at the endpoint. In contrast to the predictions of the original clonal interference model, they observed a smooth fitness increase in all populations. Moreover, they observed fitness increases of 4–7 per cent in large populations with high mutation rates, and they argued that successive single substitutions of beneficial mutations—as required under either the periodic selection model or the original clonal interference model—would have required far more than 500 generations to affect such a fitness increase. Analysis of fitness distributions at the end of the experiment showed that large populations had higher fitness variance than smaller populations as predicted by the theory, and that no population showed evidence of being dominated by a single clone as expected from periodic selection or the original clonal interference model.
Clearly, competition among clones bearing multiple mutations is important in the evolution of asexual populations at high NUb; indeed, as the resolution of methods for characterizing experimental evolution at the genomic level improves, studies documenting dynamics consistent with both the original clonal interference model and the original multiple-mutations model are emerging (see below). As noted by Desai et al. (2007), these models are incomplete representations of asexual evolution, each ignoring the effect that dominates the other: the original clonal interference model ignores competition among multiple-mutant clones; the original multiple-mutations model ignores competition among mutations of different effect. No analytical treatment that encompasses both effects has yet emerged, although a forthcoming modification of the original Gerrish & Lenski model based on a finite set of linked sites under time-dependent selection may provide a first step in that direction (Michael Lassig, personal communication; S. Schiffels, G. Szollosi, V. Mustonen & M. Lassig, in preparation.). Recent simulation studies (Park & Krug 2007; Fogle et al. 2008) have indicated that the relative importance of differing mutational effects versus multiple mutations depends critically on the incoming distribution of effects of beneficial mutations. If large-effect mutations are relatively common—if the distribution is less steep than exponential—then competition among clones bearing single mutations of differing effect can dominate asexual evolution in large populations; if large-effect beneficial mutations are quite rare—if the distribution is steeper than exponential—then competition among clones bearing multiple mutations of similar effect can be more important. Unfortunately, as noted by Orr (2010), we still lack the data to draw general empirical conclusions about the distribution of beneficial mutational effects, despite some promising recent approaches (e.g. Beisel et al. 2007; Rokyta et al. 2008); indeed, this distribution may well depend on the state of adaptation to the environment in which it is estimated (Martin & Lenormand 2008).
3. How common are beneficial mutations?
The development of the complex theories of asexual evolution reviewed above occurred in parallel with work in experimental populations suggesting that beneficial mutation rates are higher than previously suspected. This is not so surprising, as clonal competition in evolving populations would never have been detected and studied experimentally unless NUb in real populations were sufficiently high to provide for coexisting beneficial mutations. But consideration of the effect of clonal competition on adaptive substitutions has itself led to upward revision of estimated Ub values. Estimates from experimental populations of RNA virus (Miralles et al. 1999) and E. coli and Pseudomonas fluorescens bacteria (Gerrish & Lenski 1998; Imhof & Schlötterer 2001; Rozen et al. 2002; Kassen & Bataillon 2006) previously suggested Ub values of 10−9 or 10−8. These studies, however, were carried out with large (107 or more) effective population sizes and in strains that had not previously undergone genetic adaptation to the growth environment used. Although NUb is expected to be quite high under such circumstances, clonal interference, with its attendant wastage of small-effect beneficial mutations, is predicted to be very strong. If drift and clonal interference were not accounted for in making these estimates, this would arguably lead to an underestimate of Ub and an overestimate of the average effect of beneficial mutations. In fact, drift and clonal interference were accounted for, but a downward bias in the estimates still exists because of assumptions of the models used (see below). More recent experimental work in E. coli employing smaller effective populations (approx. 104) and strains that were previously adapted to the growth environment has suggested Ub ≈ 2 × 10−5 (Perfeito et al. 2007)—some 1000-fold higher than previous estimates, presumably owing to the detection of abundant mutations of small effect that were being outcompeted in larger populations. A similarly high rate of beneficial mutation in S. cerevisiae is suggested by the analysis of experimental populations by Desai et al. (2007), who find Ub ≈ 2.4 × 10−5. Assuming that total genomic mutation rates in E. coli and yeast are equal and approximately 3 × 10−3 as calculated by Drake (Drake et al. 1998; Drake 2009), these estimates imply that almost 1 per cent of all mutations in experimental populations of these organisms are beneficial. Some mutation-accumulation studies in yeast have suggested that beneficial mutations can be even more common: Wloch et al. (2001) found that almost 2 per cent of mutations in haploid yeast lines were beneficial, and Joseph & Hall (2004) concluded that 5.75 per cent of mutations in diploid yeast lines were beneficial. Even leaving room for the possibility that these latter estimates are suspiciously high, there seems to be growing evidence that beneficial mutation rates are much higher than previously thought, at least in experimental populations.
The advent of rapid and relatively inexpensive methods of genome sequencing and characterization is making it increasingly feasible to identify mutations, beneficial and otherwise, in genomes from evolved experimental populations. Much work in this area has focused on viruses with genomes in the order of a few kilobases in length (e.g. Bull et al. 2003; Bollback & Huelsenbeck 2007; Pepin & Wichman 2008; Rokyta et al. 2008, 2009; Betancourt 2009), but very recent work has begun characterizing beneficial mutations in experimental populations of bacteria and yeast (e.g. Fiegna et al. 2006; Hegreness et al. 2006; Herring et al. 2006; Gresham et al. 2008; Kao & Sherlock 2008; Barrick & Lenski 2009; Barrick et al. 2009). While it seems too early to draw firm general conclusions about the dynamics of beneficial mutations in asexual populations from these studies, some interesting results have recently emerged in support of both clonal interference and multiple-mutation dynamics. For example, Betancourt (2009) sequenced evolved populations of an RNA bacteriophage and found evidence that beneficial mutations are sufficiently abundant for selective substitutions to overlap in time within a population, consistent with the multiple-mutations model. Very recently, Barrick & Lenski (2009) used whole-genome sequencing to identify 45 mutations in an experimental E. coli population that had evolved for 20 000 generations in a constant medium. They show that substitution of these mutations occurred at a roughly constant rate. Such constancy of substitution is predicted by genetic drift, but other evidence indicates that almost all of the mutations were beneficial. Indeed, a re-analysis of Barrick and Lenski's data (Philip J. Gerrish 2009, unpublished data) suggests that the substitution of beneficial mutations in this population was significantly underdispersed temporally, as predicted by clonal interference analysis (Gerrish 2001). As more data of this kind become available, it should be increasingly possible to test predictions concerning the dynamics of asexual adaptation at the genome level.
4. Reconciling discrepancies in estimates of ub: some results from an encompassing deterministic treatment
The estimates of Ub from experimental evolution studies presented in the foregoing section depend on a model that describes the mapping between beneficial mutations that arise in the population and those that can be observed. The idea behind the approach of Perfeito et al. (2007) was to eliminate as much bias between existing and observed beneficial mutations as possible by reducing the population size and hence reducing the effect of clonal interference. In this way, Perfeito et al. obtained estimates of Ub that were orders of magnitude higher than previous estimates. At least two of these previous estimates (Gerrish & Lenski 1998; Rozen et al. 2002) were obtained by taking data from large populations and transforming these data into estimates of Ub using a likelihood model that accounted for both drift and clonal interference. This model, however, made an a priori assumption that Ub was roughly of the same order of magnitude as 1/2N ln (Ns/2); it is no surprise, therefore, that the highest likelihood peak obtained was in this vicinity. (A second peak was in fact observed at much higher Ub and lower average selective advantage, but this peak was not reported because it was significantly lower than the other peak—again, most likely due to the model's a priori assumptions about Ub.)
Here, we provide some new theoretical analysis which shows that the deterministic ‘solitary wave’ approach mentioned above may be adapted so that it applies equally well to the territory of intermediate NUb. Our adaptation of the solitary wave model differs from previous adaptations (Desai & Fisher 2007; Desai et al. 2007; Rouzine et al. 2008) in that our model has no stochastic component; as a result, our model has the advantage of being much simpler to write down but the disadvantage of being less satisfying from a rigorous theoretical standpoint. This new model is not technically restricted to a limited range of NUb and therefore in principle does not require any a priori assumptions about the range of values in which these estimates are likely to reside. Indeed, we have found that data used previously to make a very low estimate of Ub (Elena et al. 1996; Gerrish & Lenski 1998) can be re-analysed using this model to obtain a much higher estimate of Ub consistent with other, more recent estimates (Wloch et al. 2001; Joseph & Hall 2004; Desai et al. 2007; Perfeito et al. 2007). The fundamental assumption of this new model is that the particulate acquisition of beneficial mutations may be approximated as a continuous ‘flux’. This assumption works best for high Ub because the particulate acquisition of beneficial mutations will be better approximated as a flux when the rate of acquisition is high. But still, this limitation is much less restrictive than the a priori constraints imposed on estimates by other models, suggesting that this model might offer a way of obtaining rough estimates before more detailed models are used to refine them. Interestingly, the dynamics of fitness evolution under this new model look remarkably like those predicted by the periodic selection model, despite the model's deterministic formulation and the attendant assumptions that are implicit in such formulations.
Under the assumptions of infinite population size and infinite genome size, adaptation can be modelled compactly with the partial integro-differential equation
| 4.1 |
where u(x, t) is the probability density of individuals in the population with fitness x at time t, and  ; M is the mutation operator, defined as
; M is the mutation operator, defined as
 |
where μ is genomic mutation rate, fB fraction of mutations that increase fitness (beneficial), fD fraction of mutations that decrease fitness (deleterious), gB distribution of effects of beneficial mutations (probability density), and gD distribution of effects of deleterious mutations (probability density).
Equation (4.1) gives rise to a ‘solitary wave’ (Tsimring et al. 1996; Rouzine et al. 2003, 2008; Gerrish et al. 2007)—a single moving peak—that progresses in the direction of increasing fitness, x, and converges to a fixed shape, i.e.  , where c is the wave velocity. In this model, the dynamics of mean fitness converge to a linear dependence on time:
, where c is the wave velocity. In this model, the dynamics of mean fitness converge to a linear dependence on time:  , indicating that the ‘solitary wave’ moves with constant velocity in the direction of increasing fitness (dashed line in figure 1).
, indicating that the ‘solitary wave’ moves with constant velocity in the direction of increasing fitness (dashed line in figure 1).
Figure 1.
A representative numerical solution of equation (4.2), obtained by standard trapezoidal integration in x and by Runge–Kutta integration in t. The upper solid line is mean fitness; the lower solid line is fitness variance. The dotted line is the frequency of a neutral variant in the population; this is qualitatively similar to observations in experimental yeast populations (Paquin & Adams 1983). Parameter values are: N = 106, Ub = μfB = 10−7, UD = μfD = 0.01, means and variances of distributions gB and gD were 0.03, and these distributions were truncated at 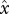 such that
such that  . Mutation effects were assumed to have exponential distributions. The upper dashed line plots mean fitness under the assumption N = ∞; these dynamics are consistent with the multiple-mutations model of Desai & Fisher (2007) and Desai et al. (2007).
. Mutation effects were assumed to have exponential distributions. The upper dashed line plots mean fitness under the assumption N = ∞; these dynamics are consistent with the multiple-mutations model of Desai & Fisher (2007) and Desai et al. (2007).
The above deterministic model applies most naturally to situations in which NUb is very high; in fact, the model assumes NUb to be infinite (because N is infinite). Nevertheless, the model can be adapted to situations of intermediate NUb by introducing a simple rule. If we assume that the population is now finite and consists of N individuals, then any fitness class whose frequency is less than 1/N contains less than one individual, and we make the reasonable assumption that less than one individual cannot replicate (Kepler & Perelson 1995). Under this new rule, the governing equation now reads
| 4.2 |
where
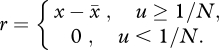 |
This new model is plotted in figure 1. Two easily measured quantities—fitness and the frequency of a neutral variant—display dynamics that are suggestive of simple periodic selection described above as characterizing the SSWM regime. The underlying population dynamics that produce these patterns, however, are quite different from those that generate simple periodic selection patterns. In simple periodic selection, single homogeneous mutant lineages sweep to fixation as rare, isolated events. In the deterministic model presented above, on the other hand, there is considerable demographic turmoil of populations during a ‘step-up’ in fitness, and many fitness classes decrease while new ones increase. Yet, despite this turnover, the mean covariance between the frequency gradients of the fastest-increasing fitness class and the rest of the population is never negative (not shown)—suggesting that what is substituting during a ‘step’ is not a single homogeneous lineage but a ‘cloud’ of mutants.
The ‘fitness steps’ in E. coli populations as reported in Elena et al. (1996) were previously used to obtain estimates of beneficial mutation rate, 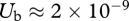 , and mean fitness effect of beneficial mutations,
, and mean fitness effect of beneficial mutations, 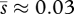 , by fitting the data to a stochastic clonal interference model (Gerrish & Lenski 1998). In contrast, a least squares fit of the same data to the above deterministic model gives the single combination of parameter estimates
, by fitting the data to a stochastic clonal interference model (Gerrish & Lenski 1998). In contrast, a least squares fit of the same data to the above deterministic model gives the single combination of parameter estimates  and
and  . Overall, this is more in line with estimates of Perfeito et al. (
. Overall, this is more in line with estimates of Perfeito et al. ( and
and  ) for a similar E. coli population and with other recent high estimates of Ub mentioned above.
) for a similar E. coli population and with other recent high estimates of Ub mentioned above.
5. Conclusion
Our understanding of the dynamics of adaptation in asexual populations has moved from a model that emphasized the substitution of single beneficial mutations (periodic selection), to a model that emphasized competition among single beneficial mutations (the original clonal interference model), to models that emphasize an abundance of beneficial mutations both among and within genotypes segregating in a population (the original multiple-mutations model and subsequent simulations; results in this paper). Pioneering microbial evolution experiments produced dynamics that were consistent with the periodic selection model, but very similar subsequent experiments were instead interpreted as providing support for abundant beneficial mutations. The original clonal interference theory provides one explanation for how periodic-selection-like dynamics can emerge despite abundant beneficial mutations: among the contending mutations that arise and compete by chance in a population in which both selection and mutation are strong effects, rare superior competitors can prevail quite swiftly. However, as beneficial mutations become more common in a population, it becomes increasingly likely that genotypes bearing multiple beneficial mutations prevail rather than genotypes bearing individual mutations. The original multiple-mutations theory assumes that all mutations are of the same beneficial effect and that clonal competition occurs among genotypes bearing different numbers of beneficial mutations. Under these circumstances, a ‘nose’ of high-fitness, multiple-mutation genotypes is predicted to lead the fitness distribution of an asexual population smoothly towards higher values. But the original multiple-mutations theory, while it corrects a shortcoming of the original clonal interference model, introduces a shortcoming of its own: without the possibility for beneficial mutations of different effect, the potential for the emergence of strongly superior clonal competitors is greatly reduced. Development of a stochastic analytical theory that encompasses both clonal interference and multiple-mutation dynamics is a daunting prospect, but approaches using computer simulation indicate that both competition among mutations of differing effect and competition among clones bearing multiple beneficial mutations can be expected to take place in asexual populations; the relative importance of these processes depends on the shape of the distribution of beneficial mutational effects, which remains unresolved in real populations. Finally, we show that periodic-selection-like dynamics can emerge from a deterministic approximation of asexual evolution in the limit when beneficial mutations are extremely common relative to their selective effects, and we note that such a model provides an estimate of Ub for an evolving E. coli population that is more in line with recent high estimates of Ub in E. coli and yeast.
Acknowledgements
We thank two anonymous reviewers, Bill Hill and Laurence Loewe for their helpful critical comments. P.D.S. thanks Brian Charlesworth for his mentorship and his friendship over the years. P.J.G. and P.D.S. are supported by National Institutes of Health Grant GM079483-01A2.
Footnotes
One contribution of 16 to a Theme Issue ‘The population genetics of mutations: good, bad and indifferent’ dedicated to Brian Charlesworth on his 65th birthday.
References
- Atwood K. C., Schneider L. K., Ryan F. J.1951aPeriodic selection in Escherichia coli. Proc. Natl Acad. Sci. USA 37, 146–155 (doi:10.1073/pnas.37.3.146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood K. C., Schneider L. K., Ryan F. J.1951bSelective mechanisms in bacteria. Cold Spring Harb. Symp. Quant. Biol. 16, 345–355 (doi:10.1101/SQB.1951.016.01.026) [DOI] [PubMed] [Google Scholar]
- Barrett R. D. H., M'gonigle L. K., Otto S. P.2006The distribution of mutational effects under strong selection. Genetics 174, 2071–2079 (doi:10.1534/genetics.106.062406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Lenski R. E.2009Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. (doi:10.1101/sqb.2009.74.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Yu D.-S., Yoon S. H., Jeong H., Tae Kwang T. K. O., Schneider D., Lenski R. E., Kim J. F.2009Genome evolution and adaptation in a long-term experiment with E. coli. Nature 461, 1243–1247 (doi:10.1038/nature08480) [DOI] [PubMed] [Google Scholar]
- Barton N. H.1995Linkage and the limits to natural selection. Genetics 140, 821–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C. J., Rokyta D. R., Wichman H. A., Joyce P.2007Testing the extreme value domain of attraction for distributions of beneficial fitness effects. Genetics 176, 2441–2449 (doi:10.1534/genetics.106.068585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J.2009Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage MS2. Genetics 181, 1535–1544 (doi:10.1534/genetics.107.085837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback J. P., Huelsenbeck J. P.2007Clonal interference is alleviated by high mutation rates in large populations. Mol. Biol. Evol. 24, 1397–1406 (doi:10.1093/molbev/msm056) [DOI] [PubMed] [Google Scholar]
- Bull J. J., Badgett M. R., Rokyta D., Molineux I. J.2003Experimental evolution yields hundreds of mutations in a functional viral genome. J. Mol. Evol. 57, 241–248 (doi:10.1007/s00239-003-2470-1) [DOI] [PubMed] [Google Scholar]
- Colegrave N.2002Sex releases the speed limit on evolution. Nature 420, 664–666 (doi:10.1038/nature01191) [DOI] [PubMed] [Google Scholar]
- Crow J. F., Kimura K.1965Evolution in sexual and asexual populations. Am. Nat. 99, 439–450 (doi:10.1086/282389) [Google Scholar]
- Crow J. F., Kimura M.1970An introduction to population genetics theory. New York, NY: Harper and Row [Google Scholar]
- de Visser J. A. G. M., Rozen D. E.2005Limits to adaptation in asexual populations. J. Evol. Biol. 18, 779–788 (doi:10.1111/j.1420-9101.2005.00879.x) [DOI] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Rozen D. E.2006Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172, 2093–2100 (doi:10.1534/genetics.105.052373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E.1999Diminishing returns from mutation supply rate in asexual populations. Science 283, 404–406 (doi:10.1126/science.283.5400.404) [DOI] [PubMed] [Google Scholar]
- Desai M., Fisher D. S.2007Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176, 1759–1798 (doi:10.1534/genetics.106.067678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Fisher D. S., Murray A. W.2007The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17, 385–394 (doi:10.1016/j.cub.2007.01.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W.2009Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet. 5, e1000520 (doi:10.1371/journal.pgen.1000520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D., Crow J. F.1998Rates of spontaneous mutation. Genetics 148, 1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S. F., Cooper V. S., Lenski R. E.1996Punctuated evolution caused by selection of rare beneficial mutations. Science 272, 1802–1804 (doi:10.1126/science.272.5269.1802) [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1974The evolutionary advantage of recombination. Genetics 78, 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegna F., Yu Y. T., Kadam S. V., Velicer G. V.2006Evolution of an obligate social cheater to a superior competitor. Nature 441, 310–314 (doi:10.1038/nature04677) [DOI] [PubMed] [Google Scholar]
- Fisher R. A.1930The genetical theory of natural selection. Oxford, UK: Clarendon Press [Google Scholar]
- Fogle C. A., Nagle J. L., Desai M.2008Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics 180, 2163–2173 (doi:10.1534/genetics.108.090019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish P. J.2001The rhythm of microbial adaptation. Nature 413, 299–302 (doi:10.1038/35095046) [DOI] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E.1998The fate of competing beneficial mutations in an asexual population. Genetica 102/103, 127–144 (doi:10.1023/A:1017067816551) [PubMed] [Google Scholar]
- Gerrish P. J., Colato A., Perelson A. S., Sniegowski P. D.2007Complete genetic linkage can subvert natural selection. Proc. Natl Acad. Sci. USA 104, 6266–6271 (doi:10.1073/pnas.0607280104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H.1983A simple stochastic gene substitution model. Theor. Popul. Biol. 23, 202–215 (doi:10.1016/0040-5809(83)90014-X) [DOI] [PubMed] [Google Scholar]
- Gillespie J. H.1984Molecular evolution over the mutational landscape. Evolution 38, 1116–1129 (doi:10.2307/2408444) [DOI] [PubMed] [Google Scholar]
- Gillespie J. H.1991The causes of molecular evolution. New York, NY: Oxford University Press [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., Ward A., Desevo C. G., Botstein D., Dunham M. J.2008The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4, e1000303 (doi:10.1371/journal.pgen.10003031116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J.1978The accumulation of deleterious genes in a population—Muller's Ratchet. Theor. Popul. Biol. 14, 251–267 (doi:10.1016/0040-5809(78)90027-8) [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S.1927A mathematical theory of natural and artificial selection. Part V. Selection and mutation. Proc. Camb. Phil. Soc. 23, 838–844 (doi:10.1017/S0305004100015644) [Google Scholar]
- Hegreness M., Shoresh N., Hartl D., Kishony R.2006An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311, 1615–1617 (doi:10.1126/science.1122469) [DOI] [PubMed] [Google Scholar]
- Herring C. D., et al. 2006Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 38, 1406–1412 (doi:10.1038/ng1906) [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A.1966The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (doi:10.1017/S0016672300010156) [PubMed] [Google Scholar]
- Holder K. K., Bull J. J.2001Profiles of adaptation in two similar viruses. Genetics 159, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C., Zhang L. Q., Simmonds P., Ludlam C. A., Brown A. J. L.1992Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type I within a single infected patient. Proc. Natl Acad. Sci. USA 89, 4835–4839 (doi:10.1073/pnas.89.11.4835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof M., Schlötterer C.2001Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl Acad. Sci. USA 98, 1113–1117 (doi:10.1073/pnas.98.3.1113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. B., Hall D. W.2004Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics 168, 1817–1825 (doi:10.1534/genetics.104.033761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K. C., Sherlock G.2008Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40, 1499–1504 (doi:10.1038/ng.280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., Bataillon T.2006Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38, 484–488 (doi:10.1038/ng1751) [DOI] [PubMed] [Google Scholar]
- Kepler T. B., Perelson A. S.1995Modeling and optimization of populations subject to time-dependent mutation. Proc. Natl Acad. Sci. USA 92, 8219–8223 (doi:10.1073/pnas.92.18.8219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Orr H. A.2005Adaptation in sexuals vs. asexuals: clonal interference and the Fisher–Muller model. Genetics 171, 1377–1386 (doi:10.1534/genetics.105.045252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A.1974The pertinence of the periodic selection phenomenon to prokaryotic evolution. Genetics 77, 127–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R.1981Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Hill W. G.2010The population genetics of mutations: good, bad and indifferent. Phil. Trans. R. Soc. B 365, 1153–1167 (doi:10.1098/rstb.2009.0317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T.2008The distribution of beneficial and fixed mutation fitness effects close to an optimum. Genetics 179, 907–916 (doi:10.1534/genetics.108.087122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles R., Gerrish P. J., Moya A., Elena S. F.1999Clonal interference and the evolution of RNA viruses. Science 285, 1745–1747 (doi:10.1126/science.285.5434.1745) [DOI] [PubMed] [Google Scholar]
- Miralles R., Moya A., Elena S. F.2000Diminishing returns of population size in the rate of RNA virus adaptation. J. Virol. 74, 3566–3571 (doi:10.1128/JVI.74.8.3566-3571.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J.1932Some genetic aspects of sex. Am. Nat. 66, 118–138 (doi:10.1086/280418) [Google Scholar]
- Notley-Mcrobb L., Ferenci T.1999aAdaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ. Microbiol. 1, 33–43 (doi:10.1046/j.1462-2920.1999.00002.x) [DOI] [PubMed] [Google Scholar]
- Notley-Mcrobb L., Ferenci T.1999bThe generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1, 45–52 (doi:10.1046/j.1462-2920.1999.00003.x) [DOI] [PubMed] [Google Scholar]
- Notley-Mcrobb L., Ferenci T.2000Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics 156, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A., Szilard L.1950Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl Acad. Sci. USA 36, 708–719 (doi:10.1073/pnas.36.12.708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A.2010The population genetics of beneficial mutations. Phil. Trans. R. Soc. B 365, 1195–1201 (doi:10.1098/rstb.2009.0282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos D., Schneider D., Meier-Eiss J., Arber W., Lenski R. E., Blot M.1999Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl Acad. Sci. USA 96, 3807–3812 (doi:10.1073/pnas.96.7.3807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C., Adams J.1983Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature 302, 495–500 (doi:10.1038/302495a0) [DOI] [PubMed] [Google Scholar]
- Park S.-C., Krug J.2007Clonal interference in large populations. Proc. Natl Acad. Sci. USA 104, 18 135–18 140 (doi:10.1073/pnas.0705778104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin K. M., Wichman H. A.2008Experimental evolution and genome sequencing reveal variation in levels of clonal interference in large populations of bacteriophage φX174. BMC Evol. Biol. 8, 85 (doi:10.1186/1471-2148-8-85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L., Fernandes L., Mota C., Gordo I.2007Adaptive mutations in bacteria: high rate and small effects. Science 317, 813–815 (doi:10.1126/science.1142284) [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Beisel C. J., Joyce P., Ferris M. T., Burch C. L., Wichman H. A.2008Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67, 368–376 (doi:10.1007/s00239-008-9153-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D. R., Abdo Z., Wichman H. A.2009The genetics of adaptation for eight microvirid bacteriophages. J. Mol. Evol. 69, 229–239 (doi:10.1007/s00239-009-9267-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I. M., Wakeley J., Coffin J. M.2003The solitary wave of asexual evolution. Proc. Natl Acad. Sci. USA 100, 587–592 (doi:10.1073/pnas.242719299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I. M., Brunet É., Wilke C. O.2008The traveling wave approach to asexual evolution: Muller's ratchet and speed of adaptation. Theor. Popul. Biol. 73, 24–46 (doi:10.1016/j.tpb.2007.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A. G. M., Gerrish P. J.2002Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12, 1040–1045 (doi:10.1016/S0960-9822(02)00896-5) [DOI] [PubMed] [Google Scholar]
- Shaver A. C., Dombrowski P. G., Sweeney J. Y., Treis T., Zappala R. M., Sniegowski P. D.2002Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimring L. S., Levine H., Kessler D. A.1996RNA virus evolution via a fitness-space model. Phys. Rev. Lett. 76, 4440–4443 (doi:10.1103/PhysRevLett.76.4440) [DOI] [PubMed] [Google Scholar]
- Wichman H. A., Badgett M. R., Scott L. A., Boulianne C. M., Bull J. J.1999Different trajectories of parallel evolution during viral adaptation. Science 285, 422–424 (doi:10.1126/science.285.5426.422) [DOI] [PubMed] [Google Scholar]
- Wilke C. O.2004The speed of adaptation in large asexual populations. Genetics 167, 2045–2053 (doi:10.1534/genetics.104.027136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloch D. M., Szafraniec K., Borts R. H., Korona R.2001Direct estimate of the mutation rate and the distribution of fitness effects in the yeast Saccharomyces cerevisiae. Genetics 159, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]



