Abstract
Recombination between homologous, but non-allelic, stretches of DNA such as gene families, segmental duplications and repeat elements is an important source of mutation. In humans, recent studies have identified short DNA motifs that both determine the location of 40 per cent of meiotic cross-over hotspots and are significantly enriched at the breakpoints of recurrent non-allelic homologous recombination (NAHR) syndromes. Unexpectedly, the most highly penetrant form of the motif occurs on the background of an inactive repeat element family (THE1 elements) and the motif also has strong recombinogenic activity on currently active element families including Alu and LINE2 elements. Analysis of genetic variation among members of these repeat families indicates an important role for NAHR in their evolution. Given the potential for double-strand breaks within repeat DNA to cause pathological rearrangement, the association between repeats and hotspots is surprising. Here we consider possible explanations for why selection acting against NAHR has not eliminated hotspots from repeat DNA including mechanistic constraints, possible benefits to repeat DNA from recruiting hotspots and rapid evolution of the recombination machinery. I suggest that rapid evolution of hotspot motifs may, surprisingly, tend to favour sequences present in repeat DNA and outline the data required to differentiate between hypotheses.
Keywords: recombination hotspot, mutation, repeat element
1. Introduction
It has long been known that illegitimate recombination between paralogous sequences (non-allelic homologous recombination, or NAHR) can be a major source of pathogenic mutation in eukaryotic genomes. In Drosophila, repeat DNA, including transposable elements (TEs) and the short repeats associated with centromeres and telomeres, typically occurs in regions of reduced crossing-over (Charlesworth et al. 1986; Rizzon et al. 2002). There are two possible explanations for the association: reduced selection against NAHR in regions of low recombination or a reduction in the efficacy of selection (Dolgin & Charlesworth 2008). To date, however, there is little consensus as to which force is more important. In contrast, in largely selfing species, such as Arabidopsis thaliana and Caenorhabditis elegans, recombination seems to be less of an important force in shaping TE distribution than gene density (Duret et al. 2000; Wright et al. 2003). In humans, recent efforts to map the fine-scale structure of recombination rate variation (The International HapMap Consortium 2005) have shown a more complex picture with some repeat elements showing increased rates of crossing-over (Myers et al. 2008). These findings suggest a very different model for the coevolution of repeat sequence and recombination in humans. Here, I summarize the evidence in humans that recombination may, in some cases, actually be promoted in certain types of repeat DNA and speculate as to the evolutionary processes that have led to this situation.
2. A sequence motif for human recombination hotspots is highly active in repeat dna
In humans and most other eukaryotes meiotic recombination events are clustered into short 1–2 kb regions known as recombination hotspots (Jeffreys et al. 2001). Such hotspots typically occur every 50–100 kb and lead to a cross-over event approximately once in every 1300 meioses (Myers et al. 2008). However, given that double-strand breaks (DSBs) are resolved as gene conversion events rather than cross-overs in a ratio of 4–15∶1 (Jeffreys & May 2004), DSB formation at hotspots is considerably higher and, for the hottest hotspot in the genome, with a recombination fraction of 1 cM (The International HapMap Consortium 2007; Webb et al. 2008), the rate could be as high as one in two meioses. Although nearly 20 hotspots have been characterized experimentally through studies of sperm and, with lower resolution, through pedigrees (see Coop & Przeworski 2007 for a recent review), much of our understanding of the genomic distribution of hotspots has come from studying patterns of genetic variation (McVean et al. 2004; Myers et al. 2005, 2008). These analyses have demonstrated systematic influences on the location of recombination hotspots, including a tendency to cluster near promoter regions but actively avoid transcribed regions (McVean et al. 2004; The International HapMap Consortium 2007). Comparisons of patterns estimated from sperm and from variation data suggest that male and female hotspots typically coincide (Myers et al. 2005), despite considerable differences at the megabase scale in their genetic maps (Broman et al. 1998; Kong et al. 2002).
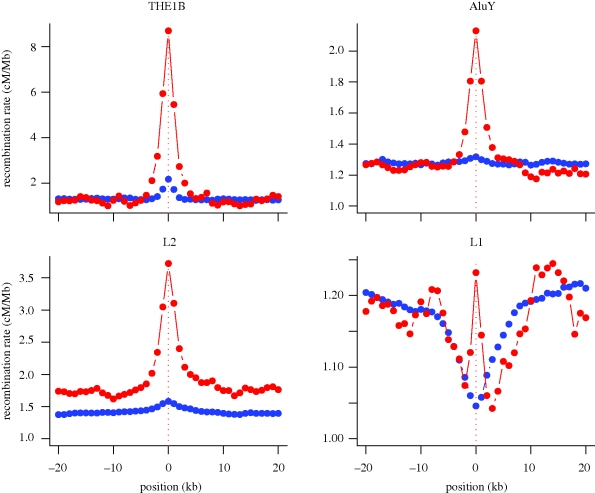
The identification of large numbers of well-mapped recombination hotspots has enabled detailed searches for sequences important in determining their location. Initial analyses identified two sequences, the 7 mer CCTCCCT and the 9 mer CCCCACCCC as being enriched in hotspots (Myers et al. 2005, 2006). Subsequent work, focusing on the 7 mer, extended the motif to a degenerate 13 mer, CCNCCNTNNCCNC (hereafter referred to as the 13 mer motif), with additional, though weaker, context-dependent effects that show a remarkable 3 bp periodicity suggestive of an interaction with zinc-finger DNA-binding proteins (Myers et al. 2008). The activity of the degenerate 13 mer motif is also influenced by extended background and bases present at the degenerate sites. For example, the most penetrant version of the motif, the ‘core 13 mer motif’ CCTCCCTNNCCAC, on the background of a THE1B repeat element leads to a detectable hotspot 70 per cent of the time (Myers et al. 2008). For THE1 elements, the presence of the degenerate motif leads to a 10-fold increase in average recombination rate, compared with a more modest two to threefold increase on L2 elements and three active Alu subfamilies (AluY, AluSg and AlusX; see figure 1). Combined, the presence of the hotspot motif on these highly penetrant backgrounds determines approximately 10 per cent of all hotspots. In contrast, the presence of the core motif in unique DNA leads to a hotspot only 10 per cent of the time and explains only 1.3 per cent of all hotspots (Myers et al. 2008).
Figure 1.
The fine-scale structure of recombination-rate variation around repeat elements in the human genome. For each of the repeat element families shown, the average autosomal recombination rate, measured over 1 kb intervals from the centre of repeats, is shown for all repeats (blue) and those containing the degenerate 13 mer motif CCNCCNTNNCCNC (red). Note the markedly different recombination profiles across repeat families. In each case, however, the presence of the 13 mer motif leads to marked local increase in recombination rate. The fraction of repeats containing the degenerate motif varies from approximately 2 per cent (THE1B, L2, L1) to over 10 per cent (AluY).
Additional support for the role of the 13 mer motif in determining hotspot activity has come from sperm studies. Analysis of experimentally characterized hotspots has identified the 13 mer within 20 bp of the centre of three hotspots and within 300 bp of the centre of a fourth. Of the 17 experimentally characterized hotspots, there is evidence for polymorphism in hotspot activity at four. In two of these (DNA2 and MS32) mutations that co-segregate with hotspot activity disrupt either the degenerate 13 mer motif or a subset of the core 13 mer motif (Jeffreys et al. 1998; Jeffreys & Neumann 2002); in NID1 there is a polymorphism that disrupts the distinct 9 mer described above (Jeffreys & Neumann 2005), and in the fourth there are no apparent sequence differences between the hot and cold alleles (Neumann & Jeffreys 2006). Combined, these results indicate that the 13 mer motif identified is an essential determinant of hotspot activity (for example, through recruiting DSBs) in 40 per cent of hotspots. There is unlikely to be a single explanation for the remaining hotspots. Rather, multiple pathways to hotspot activity must be acting in a manner similar to the situation in yeast (Petes 2001).
In addition to allelic recombination, there is evidence to suggest that the 13 mer motif also plays a role in recurrent NAHR syndromes and other forms of genome instability. In NAHR, cross-over events occurring between paralogous sequences lead to duplication, loss or more complex rearrangements that disrupt gene activity. Analysis of the breakpoints of the six common NAHR disorders with an occurrence rate of 1 in 10 000 to 1 in 2000 where breakpoints have been mapped multiple independent times (Charcot-Marie-Tooth disease: CMT1A; neurofibromatosis: NF1, Sotos syndrome, Smith–Magenis syndrome, Williams–Beuren syndrome, X-linked ichthyosis) revealed the presence of the 13 mer motif in the low-copy repeats in all cases (Myers et al. 2008). Previous work has also established coincidence of hotspots for allelic and non-allelic recombination at the NF1, CMT1A and hereditary neuropathy with pressure palsies NAHR hotspots (Lindsay et al. 2006; Raedt et al. 2006). In addition, the 13 mer motif is found in hypervariable minisatellites where the mutability appears to arise from within the array and is associated with recombination activity (Myers et al. 2008). Unexpectedly, the 13 mer motif also occurs at the breakpoints of the ‘common deletion’ in mitochondrial DNA, a recurrent somatic mutation of 5 kb associated with specific syndromes but which also accumulates in ageing (Myers et al. 2008).
These findings establish a link between the programmed events of DSB formation and recombination (both gene conversion and cross-over) in meiosis and the unprogrammed, often pathogenic processes of NAHR and hypermutability. In the light of such a connection, the presence of highly penetrant forms of the 13 mer motif on repeat elements such as THE1 and LINE2 elements (figure 1) appears to be ‘poor design’. The estimated activities of the different 13 mer motif/background combinations and the rate of gene conversion imply that in every meiosis tens of DSBs will occur in repeat DNA (based on an average of 30 cross-over events per generation, a ratio of gene-conversion to cross-over of 10∶1 and the figure of 10 per cent of hotspots being driven by highly penetrant motif/repeat-DNA combinations). In each case there is the potential for NAHR between paralogous members of the family. Such considerations raise two questions. First, is NAHR occurring among dispersed members of these (typically short) repeats? Second, why do not the deleterious consequences of NAHR select for the avoidance of sequence motifs present in repeat sequence?
3. Nahr among short repeat elements is an important process in genome rearrangement and evolution
Repeat-mediated NAHR deletion and duplication events leading to deleterious phenotypes have been described for many genomic disorders (Deininger & Batzer 1999; Gu et al. 2008). For example, major NAHR hotspots for both Charcot-Marie Tooth Disease and Williams–Beurens Syndrome occur at repeat elements (L2 and AluY, respectively). However, in these and most other cases where NAHR hotspots have been defined, the hotspot is embedded in a much longer low-copy repeat (also referred to as segmental duplications), typically of 10 kb or longer and of extremely high identity; often 99 per cent or higher. Breakpoints of segmental duplications and of rare copy-number variable regions (particularly deletions) are enriched for repeat elements (Cooper et al. 2007; Vissers et al. 2009). However, the presence of microhomology at breakpoints (as opposed to extended homology) suggests a mitotic, as opposed to meiotic, origin for the majority of mutation events and the role of non-homologous end-joining or stalled replication forks (Vissers et al. 2009; Zhang et al. 2009). The implication is that the short-repeat elements that comprise 40 per cent of the human genome are, by themselves, rarely sufficient to generate meiotic NAHR events leading to pathogenic phenotypes.
A different picture arises when considering the potential for NAHR to generate non-pathogenic rearrangements. Comparison of the human and chimpanzee reference genomes identified nearly 500 human-specific deletion events that could be attributed to illegitimate recombination between Alu elements (typically not in extended low-copy repeats) (Sen et al. 2006). This figure suggests a rate of at least one event every 300 meioses or a per-copy rate in the range of 10−9 to 10−8 per generation (assuming a split-time of 4 Myr for humans and chimps, a generation time of 25 years and that the differences in the reference genomes are owing to fixed neutral mutations). Similarly, NAHR between LINE1 elements has been proposed as responsible for 55 human-specific deletions (Han et al. 2008). The extended homology between the breakpoints for these events suggests the role of meiotic recombination and implies that NAHR between repeats in unique DNA occurs at a rate at least comparable with point mutation. In contrast, the rapid structural evolution of subtelomeric regions seems to be driven more by mitotic processes, such as non-homologous end-joining (Linardopoulou et al. 2005).
It is important to note that genome rearrangements (duplication, deletion, inversion) will only result from NAHR if DSBs are resolved as cross-over events. Gene-conversion events, in contrast, will lead to changes within the repeat of little or no functional consequence and may occur at a substantially higher rate. Studies in yeast (Kupiec & Petes 1988a,b) indicate that gene conversion events between dispersed members of Ty TEs, while much reduced in rate compared with allelic conversion events, are much higher in frequency than cross-over events between dispersed repeats. In humans no direct estimates for gene conversion between dispersed repeat elements exist, however, it may be possible to estimate the relative rate of NAHR among members of repeat elements by applying population genetic methods for detecting and estimating the rate of recombination and gene conversion among allelic sequences (McVean et al. 2002). Using such methods indicates that non-allelic conversion events have played an important role in the history of repeat elements (THE1, Alu; table 1). The per-base-pair rate of conversion is estimated to be comparable with or substantially higher than the rate of point mutation (table 1). This analysis is clearly exploratory and requires much additional work to validate the estimates. Nevertheless, the combined evidence suggests that NAHR between dispersed and tandemly arranged members of repeat DNA families could represent a substantial source of mutation in the human genome, perhaps even at a level comparable with point mutation.
Table 1.
Population genetic statistics and inferences about recombination for selected repeat elements on human chromosome 20. Statistics shown are: average pairwise differences (π), number of segregating sites (S), Tajima's D statistic (DTajima), estimate of population-scaled recombination rate (γ), correlation between physical distance and LD as measured by the r2 statistic (Cor(r2, d)), p-value for the likelihood permutation test of McVean et al. (2002) (pLPT), p-value for the permutation test comparing the relationship between physical distance and LD (pr2). L2 and L1 elements excluded because of uncertainty in alignment.
| repeat name | length consensus | number (chr20)a | π | Sb | DTajima | γc | Cor (r2, d) | pLPT | pr2 |
|---|---|---|---|---|---|---|---|---|---|
| THE1B | 364 | 281 | 0.12 | 278 | −0.19 | 0.32 | −0.12 | 0.001 | 0.001 |
| AluY | 282 | 254 | 0.09 | 272 | −1.33 | 1.45 | −0.09 | 0.001 | 0.001 |
| AluSc | 284 | 912 | 0.10 | 206 | −0.02 | 0.27 | −0.16 | 0.001 | 0.001 |
aOnly includes repeat elements with matches to at least 80 per cent of the consensus bases.
bIndels treated as missing data. Only sites with minor allele frequency > 0.05 used for estimating recombination rate.
cEstimate of the population-scaled gene-conversion rate 4Nec, where c is the per-base-pair rate of recombination (here initiation of NAHR).
4. Why does not selection eliminate hotspot mechanisms that target repeat dna?
The evidence discussed in the previous section suggests that NAHR does occur between members of short (approx. 300 bp) repeat element families that show sequence identity typically in the range of 80–95% and that are not necessarily embedded in regions of higher sequence identity. This conclusion raises the question of why the recombination machinery has not evolved to avoid initiating DSBs within repeats.
There are three possible explanations. First, there may be selection against NAHR among repeats but no alternative mechanism is available. Such an explanation seems unlikely given that many, perhaps the majority, of recombination hotspots in humans (and other eukaryotes) are not initiated in repeat DNA. Second, there may be some selective benefit to the repeat DNA for an association with hotspots, for example by enabling their spread through the genome. However, while active elements could benefit (e.g. through generating open chromatin and the potential for transcription or conversion within meiotic cells), this hypothesis seems at odds with the observation that the most recombinogenic motif/repeat background combination is on the inactive THE1 elements, while other elements (e.g. LINE1) show strong local suppression of cross-over hotspots despite being active (figure 1). The third explanation is that the association of hotspots with repeat DNA is driven by other factors and that while it does generate a mutational load, this load is sufficiently small that selection for alternative mechanisms is weak.
But what kind of evolutionary process could ‘drive’ the recombination machinery to one in which a substantial fraction of DSBs occur in repeat DNA? One possible clue is the remarkably rapid evolution seen at primate hotspots. Between humans and chimpanzees there appears to be little or no sharing of cross-over hotspots (Ptak et al. 2004, 2005; Winckler et al. 2005). These findings potentially point to a systematic change in the recombination machinery such that the primary hotspot motif in chimps (if there is one at all) is likely to be different from that in humans. To date, the reason for such a change is unclear, but it is well known that recombination hotspots carry an inherent self-destructive drive that could promote rapid evolution of the system. Specifically, cis-acting polymorphism within a hotspot can influence the tendency of a chromatid to experience DSBs as seen, for example, in males heterozygous for mutations that abolish the short motif at the DNA2 hotspot (Jeffreys & Neumann 2002). In such cases the ‘hotter’ allele will typically be the one that experiences DSB formation and it will be repaired by (and consequently converted to) the ‘colder’ allele (Boulton et al. 1997; Jeffreys & Neumann 2002; Ptak et al. 2005; Coop & Myers 2007) leading to drive against the hot allele at the population level. This phenomenon has led to the ‘hotspot paradox’ that poses the question of why such drive has not eliminated all hotspots (Boulton et al. 1997). One solution to the paradox is that as the number of active hotspot motifs decreases, there is an increasing selective advantage (arising from the failure to achieve appropriate meiotic disjunction) for a change in the machinery to recognize a different primary motif and activate a new set of hotspots.
In what direction might one expect a motif that recruits hotspots to evolve? While the space of possible motifs is huge, there are important constraints that may shape the direction of evolution. Specifically, hotspot motifs must be common, dispersed across the genome, not have some other function (e.g. binding sites for transcription factors) and, at least from the data available in humans, avoid genes. Short repeat sequences, particularly families of extinct TEs (including long terminal repeats), are likely to contain many such motifs. Consequently, as the selection pressure to acquire new hotspots becomes large, the cost associated with any NAHR-mediated mutation arising from using motifs present in repeat DNA is overwhelmed. Over longer evolutionary time scales, the motif and repeat families involved may well change, but it is at least plausible that the rapid evolution of recombination machinery may draw hotspot motifs towards sequences common in repeat DNA.
5. Next steps
The suggestion here is that strong selection pressure on the recombination machinery to change may draw motif-based mechanisms for hotspot formation towards repeat DNA. This is a highly speculative suggestion, but also one that is readily testable with appropriate comparative data on the genome-wide location of recombination hotspots. In primates, efforts are underway to collect genome-wide polymorphism data from a number of species, which can be used to identify and localize recombination hotspots. However, it seems essential to have such information for more closely related groups of species, such as mice. Such data will enable us to ask whether hotspots are general features of eukaryotic genomes, whether there are typically sequence motifs associated with hotspots, whether these evolve rapidly and whether highly penetrant forms of such motifs often occur in repeat-DNA.
However, the obvious alternative idea, that TEs (or the sequences in them) in some way benefit from incorporating such motifs deserves scrutiny. Therefore, we require a better understanding of the mechanistic relationship between the 13 mer motif and hotspot activity so that we can ask what, if anything, binds to the 13 mer, whether it influences transcription in the germ line and whether its presence is associated with TE activity. Irrespective of which hypothesis turns out to be correct, the paradoxical association between recombination hotspots and repeat DNA in humans substantially shifts the way in which we need to think about the general relationship between recombination, repeat DNA and genome evolution.
Acknowledgements
I would like to thank Simon Myers, Adam Auton, Fyodor Kondrashov, Bill Hill, Laurence Loewe and an anonymous reviewer for discussion and comments on the manuscript. I would also like to thank Brian Charlesworth who inspired me and all around him with his intellect, rigour and fondness for comic verse.
Footnotes
One contribution of 16 to a Theme Issue ‘The population genetics of mutations: good, bad and indifferent’ dedicated to Brian Charlesworth on his 65th birthday.
References
- Boulton A., Myers R. S., Redfield R. J.1997The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl Acad. Sci. USA 94, 8058–8063 (doi:10.1073/pnas.94.15.8058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Murray J. C., Sheffield V. C., White R. L., Weber J. L.1998Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63, 861–869 (doi:10.1086/302011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., Stephan W.1986The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112, 947–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Myers S. R.2007Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 3, e35 (doi:10.1371/journal.pgen.0030035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Przeworski M.2007An evolutionary view of human recombination. Nat. Rev. Genet. 8, 23–34 (doi:10.1038/nrg1947) [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Nickerson D. A., Eichler E. E.2007Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 39, S22–S29 (doi:10.1038/ng2054) [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A.1999Alu repeats and human disease. Mol. Genet. Metab. 67, 183–193 (doi:10.1006/mgme.1999.2864) [DOI] [PubMed] [Google Scholar]
- Dolgin E. S., Charlesworth B.2008The effects of recombination rate on the distribution and abundance of transposable elements. Genetics 178, 2169–2177 (doi:10.1534/genetics.107.082743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Marais G., Biemont C.2000Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 156, 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Zhang F., Lupski J. R.2008Mechanisms for human genomic rearrangements. Pathogenetics 1, 4 (doi:10.1186/1755-8417-1-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Lee J., Meyer T. J., Remedios P., Goodwin L., Batzer M. A.2008L1 recombination-associated deletions generate human genomic variation. Proc. Natl Acad. Sci. USA 105, 19 366–19 371 (doi:10.1073/pnas.0807866105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., May C. A.2004Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36, 151–156 (doi:10.1038/ng1287) [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Neumann R.2002Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 31, 267–271 (doi:10.1038/ng910) [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Neumann R.2005Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum. Mol. Genet. 14, 2277–2287 (doi:10.1093/hmg/ddi232) [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Murray J., Neumann R.1998High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell. 2, 267–273 (doi:10.1016/S1097-2765(00)80138-0) [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Kauppi L., Neumann R.2001Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29, 217–222 (doi:10.1038/ng1001-217) [DOI] [PubMed] [Google Scholar]
- Kong A., et al. 2002A high-resolution recombination map of the human genome. Nat. Genet. 31, 241–247 [DOI] [PubMed] [Google Scholar]
- Kupiec M., Petes T. D.1988aAllelic and ectopic recombination between Ty elements in yeast. Genetics 119, 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M., Petes T. D.1988bMeiotic recombination between repeated transposable elements in Saccharomyces cerevisiae. Mol. Cell Biol. 8, 2942–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E. V., Williams E. M., Fan Y., Friedman C., Young J. M., Trask B. J.2005Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437, 94–100 (doi:10.1038/nature04029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S. J., Khajavi M., Lupski J. R., Hurles M. E.2006A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 79, 890–902 (doi:10.1086/508709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G., Awadalla P., Fearnhead P.2002A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160, 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A., Myers S. R., Hunt S., Deloukas P., Bentley D. R., Donnelly P.2004The fine-scale structure of recombination rate variation in the human genome. Science 304, 581–584 (doi:10.1126/science.1092500) [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P.2005A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (doi:10.1126/science.1117196) [DOI] [PubMed] [Google Scholar]
- Myers S., Spencer C. C., Auton A., Bottolo L., Freeman C., Donnelly P., McVean G.2006The distribution and causes of meiotic recombination in the human genome. Biochem. Soc. Trans. 34, 526–530 [DOI] [PubMed] [Google Scholar]
- Myers S., Freeman C., Auton A., Donnelly P., McVean G.2008A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 40, 1124–1129 (doi:10.1038/ng.213) [DOI] [PubMed] [Google Scholar]
- Neumann R., Jeffreys A. J.2006Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum. Mol. Genet. 15, 1401–1411 (doi:10.1093/hmg/ddl063) [DOI] [PubMed] [Google Scholar]
- Petes T. D.2001Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369 (doi:10.1038/35072078) [DOI] [PubMed] [Google Scholar]
- Ptak S. E., Roeder A. D., Stephens M., Gilad Y., Paabo S., Przeworski M.2004Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2, e155 (doi:10.1371/journal.pbio.0020155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak S. E., Hinds D. A., Koehler K., Nickel B., Patil N., Ballinger D. G., Przeworski M., Frazer K. A., Paabo S.2005Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37, 429–434 (doi:10.1038/ng1529) [DOI] [PubMed] [Google Scholar]
- Raedt T. D., et al. 2006Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat. Genet. 38, 1419–1423 (doi:10.1038/ng1920) [DOI] [PubMed] [Google Scholar]
- Rizzon C., Marais G., Gouy M., Biemont C.2002Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S. K., et al. 2006Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 79, 41–53 (doi:10.1086/504600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Hapmap Consortium. 2005A haplotype map of the human genome. Nature 437, 1299–1320 (doi:10.1038/nature04226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Hapmap Consortium 2007A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (doi:10.1038/nature06258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers L. E., et al. 2009Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum. Mol. Genet. 18, 3579–3593 (doi:10.1093/hmg/ddp306) [DOI] [PubMed] [Google Scholar]
- Webb A. J., Berg I. L., Jeffreys A.2008Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proc. Natl Acad. Sci. USA 105, 10 471–10 476 (doi:10.1073/pnas.0804933105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler W., et al. 2005Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308, 107–111 (doi:10.1126/science.1105322) [DOI] [PubMed] [Google Scholar]
- Wright S. I., Agrawal N., Bureau T. E.2003Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13, 1897–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Khajavi M., Connolly A. M., Towne C. F., Batish S. D., Lupski J. R.2009The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 41, 849–853 (doi:10.1038/ng.399) [DOI] [PMC free article] [PubMed] [Google Scholar]