Abstract
The frequencies of alternative synonymous codons vary both among species and among genes from the same genome. These patterns have been inferred to reflect the action of natural selection. Here we evaluate this in bacteria. While intragenomic variation in many species is consistent with selection favouring translationally optimal codons, much of the variation among species appears to be due to biased patterns of mutation. The strength of selection on codon usage can be estimated by two different approaches. First, the extent of bias in favour of translationally optimal codons in highly expressed genes, compared to that in genes where selection is weak, reveals the long-term effectiveness of selection. Here we show that the strength of selected codon usage bias is highly correlated with bacterial growth rate, suggesting that selection has favoured translational efficiency. Second, the pattern of bias towards optimal codons at polymorphic sites reveals the ongoing action of selection. Using this approach we obtained results that were completely consistent with the first method; importantly, the frequency spectra of optimal codons at polymorphic sites were similar to those predicted under an equilibrium model. Highly expressed genes in Escherichia coli appear to be under continuing strong selection, whereas selection is very weak in genes expressed at low levels.
Keywords: codon usage, bacteria, population genetics, selection, mutation bias
1. Introduction
When the genetic code was decrypted in the 1960s, it became apparent that most amino acids are encoded by multiple (two to six) codons, which typically differ only at the third nucleotide of the codon. With the introduction of DNA sequencing in the late 1970s, it emerged that these alternative synonymous codons are not used with equal frequencies. Two phenomena were soon apparent: patterns of codon usage vary among species (Grantham et al. 1980), and in the model bacterium Escherichia coli (for which most data were available), codon usage is more biased in genes expressed at higher levels (Post & Nomura 1980; Gouy & Gautier 1982; see table 1). Both phenomena were interpreted as reflecting the action of natural selection.
Table 1.
Codon usage in E. coli. Codon usage is compared between a set of 40 highly expressed genes (high; see Sharp et al. 2005) and the genome as a whole (all); the data are relative synonymous codon usage values (the ratio of the observed number to that expected if all codons for an amino acid were used equally). Nineteen codons occurring at significantly higher frequencies (see Henry & Sharp 2007) in the high dataset are shown in bold. The data are for E. coli strain K-12 MG1655 (accession number U00096).
| high | all | high | all | high | all | high | all | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 0.45 | 1.15 | Ser | UCU | 2.54 | 0.87 | Tyr | UAU | 0.48 | 1.14 | Cys | UGU | 0.77 | 0.89 |
| Phe | UUC | 1.55 | 0.85 | Ser | UCC | 1.52 | 0.89 | Tyr | UAC | 1.52 | 0.86 | Cys | UGC | 1.23 | 1.11 |
| Leu | UUA | 0.14 | 0.79 | Ser | UCA | 0.19 | 0.74 | Ter | UAA | 2.85 | 1.89 | Ter | UGA | 0.15 | 0.88 |
| Leu | UUG | 0.25 | 0.77 | Ser | UCG | 0.06 | 0.92 | Ter | UAG | 0 | 0.23 | Trp | UGG | 1.00 | 1.00 |
| Leu | CUU | 0.30 | 0.62 | Pro | CCU | 0.59 | 0.64 | His | CAU | 0.61 | 1.14 | Arg | CGU | 4.13 | 2.27 |
| Leu | CUC | 0.23 | 0.63 | Pro | CCC | 0.05 | 0.50 | His | CAC | 1.39 | 0.86 | Arg | CGC | 1.80 | 2.39 |
| Leu | CUA | 0.01 | 0.22 | Pro | CCA | 0.52 | 0.76 | Gln | CAA | 0.37 | 0.69 | Arg | CGA | 0 | 0.39 |
| Leu | CUG | 5.07 | 2.97 | Pro | CCG | 2.85 | 2.10 | Gln | CAG | 1.63 | 1.31 | Arg | CGG | 0.03 | 0.59 |
| Ile | AUU | 0.72 | 1.52 | Thr | ACU | 1.89 | 0.67 | Asn | AAU | 0.25 | 0.90 | Ser | AGU | 0.29 | 0.91 |
| Ile | AUC | 2.27 | 1.26 | Thr | ACC | 1.75 | 1.74 | Asn | AAC | 1.75 | 1.10 | Ser | AGC | 1.40 | 1.66 |
| Ile | AUA | 0.01 | 0.22 | Thr | ACA | 0.19 | 0.53 | Lys | AAA | 1.45 | 1.53 | Arg | AGA | 0.03 | 0.23 |
| Met | AUG | 1.00 | 1.00 | Thr | ACG | 0.17 | 1.07 | Lys | AAG | 0.55 | 0.47 | Arg | AGG | 0 | 0.14 |
| Val | GUU | 2.07 | 1.03 | Ala | GCU | 1.83 | 0.65 | Asp | GAU | 0.69 | 1.26 | Gly | GGU | 2.53 | 1.35 |
| Val | GUC | 0.30 | 0.86 | Ala | GCC | 0.32 | 1.08 | Asp | GAC | 1.31 | 0.74 | Gly | GGC | 1.39 | 1.61 |
| Val | GUA | 1.14 | 0.62 | Ala | GCA | 1.09 | 0.85 | Glu | GAA | 1.51 | 1.38 | Gly | GGA | 0.03 | 0.44 |
| Val | GUG | 0.48 | 1.49 | Ala | GCG | 0.76 | 1.42 | Glu | GAG | 0.49 | 0.62 | Gly | GGG | 0.06 | 0.60 |
The selective differences among synonymous codons reflect two aspects of the transfer RNA (tRNA) population present in the cell (Ikemura 1985). First, for some amino acids there are multiple species of tRNAs with different anticodons, and it is those codons translated by the most abundant tRNA species which are preferred in highly expressed genes. For example, there are five different Leu tRNAs in E. coli, but that with anticodon CAG is much more abundant than the others. This anticodon is complementary to the codon CUG, which is used nearly 20 times more often than any of the other five Leu codons in highly expressed genes (table 1). Second, many tRNAs can translate more than one codon, but with variable ability; the codon best recognized by the anticodon is preferred in highly expressed genes. For example, there is a single Phe tRNA in E. coli, with anticodon GAA, which translates both UUU and UUC; however, UUC is perfectly complementary to the anticodon, and is used about three times more often than UUU in highly expressed genes (table 1). Thus, from knowledge of the tRNA population it is possible to predict which codons are translationally optimal; namely, those that are best recognized by the most abundant tRNA species.
There has been much debate about exactly why translationally optimal codons are selected. The traditional view is that use of optimal codons increases the efficiency of translation (Ehrenberg & Kurland 1984; Andersson & Kurland 1990). Ribosomes constitute about two-thirds of the protein content of an E. coli cell when growing rapidly (Pedersen et al. 1978), and the abundance of ribosomes may be the main factor limiting growth rate. Optimal codons may be translated faster than non-optimal codons (Sørensen & Pedersen 1991), such that ribosomes move faster along an mRNA containing more optimal codons, and the ribosomes are more quickly released to be available to translate other mRNAs. Thus, use of optimal codons, especially in genes expressed at high levels encoding mRNAs that must be translated more often, allows more efficient use of ribosomes and leads to faster growth rate (Kudla et al. 2009), conferring an obvious selective advantage, at least in bacteria occupying certain niches.
An alternative view is that the use of optimal codons increases the accuracy of translation. Sites where the identity of the amino acid is more critical for protein function are expected to be more conserved across species, and also expected to be the sites where accuracy of translation is more important. The fruitfly, Drosophila melanogaster, exhibits stronger codon usage bias in more highly expressed genes, analogous to the situation in E. coli (Shields et al. 1988; Duret & Mouchiroud 1999), and it was found that codons for conserved amino acids have stronger codon bias in D. melanogaster (Akashi 1994). This accuracy hypothesis has the potential to explain the observation, otherwise surprising, that rates of non-synonymous and synonymous nucleotide substitution are correlated across genes in comparisons between E. coli and its close relative Salmonella enterica (Sharp 1991). Based on a variety of observations, some authors have concluded that translational accuracy is the primary object of codon selection in E. coli (Stoletzki & Eyre-Walker 2006), and indeed the dominant constraint on gene sequence evolution across both bacteria and eukaryotes (Drummond & Wilke 2008).
In this article we will focus on bacteria, examining the extent to which natural selection is responsible for the variations in codon usage seen among species and within genomes. In particular, we will contrast the results of two different approaches to estimating the strength of selection on codon usage bias. Finally, we will discuss the implications of the results, including their relevance to the efficiency versus accuracy debate introduced above.
2. Variation in codon usage bias among bacteria
Analyses of bacteria other than E. coli have revealed that codon usage patterns vary among species in a number of ways. Most of the differences appear to be due, ultimately, to variations in mutation biases. First, it had been known for half a century that base composition, summarized by G + C content in double-stranded DNA, varies greatly among bacteria (Belozersky & Spirin 1958). Among published bacterial genome sequences, values of G + C content range from 17 per cent in Carsonella ruddii (Nakabachi et al. 2006) to 73 per cent in Frankia alni (Normand et al. 2007). This variation has long been viewed as the primary influence on codon usage differences between species of bacteria (Bibb et al. 1984; Muto & Osawa 1987). This has been confirmed by multivariate analyses comparing total genomic codon usage among bacteria, which showed that the single most important source of variation is G + C content (Lynn et al. 2002; Chen et al. 2004). It has often been speculated that this variation reflects the action of selection. In particular, it has been suggested that there would be pressure on thermophilic bacteria to have more G + C-rich genomes, because they are more thermostable (Bernardi & Bernardi 1986; Musto et al. 2004). However, most analyses have failed to find any correlation between growth temperature and genomic G + C content (e.g. Galtier & Lobry 1997; Lynn et al. 2002). Overall, the variation in G + C content is most simply explained by subtle but persistent mutation biases (Sueoka 1962).
Second, genome sequencing has revealed that in many bacteria base composition varies systematically between the leading and lagging strands of replication, with the leading strand being more G + T-rich (Lobry 1996; McLean et al. 1998). This strand-specific bias impacts codon usage, but the strength of the effect varies considerably among species. In the spirochaetes Borrelia burgdorferi (the cause of Lyme disease) and Treponema denticola (the cause of syphilis), strand-specific bias dominates codon usage variation among genes (Lafay et al. 1999); in other species the effect is much weaker, or undetectable (Kloster & Tang 2008). The source of this strand-specific bias has been debated, but the predominant ideas concern mutation biases. The leading and lagging strands are replicated by different mechanisms with different mutation rates (Fijalkowska et al. 1998), which could lead to the observed differences in base composition. Alternatively, since there is an excess of genes located on the leading strand in many bacteria (Brewer 1988; Tillier & Collins 2000), biases in transcription-coupled repair could lead to a skew between the strands in nucleotide composition (Francino et al. 1996).
Third, for some amino acids, the identity of the translationally optimal codon varies among species. For example, in Clostridium perfringens, the codons heavily used in highly expressed genes (Musto et al. 2003) differ from those in E. coli (table 1) for six amino acids. These differences are correlated with changes in tRNA populations. While tRNA abundances have been measured for very few species, it is known that tRNA abundance is correlated with tRNA gene copy number (Kanaya et al. 1999), and so the latter may be used to predict the most abundant tRNAs. In the E. coli genome, where there are eight genes encoding five different Leu tRNAs, four genes encode the tRNA with the CAG anticodon (mentioned above as being the most abundant Leu tRNA species in E. coli). The C. perfringens genome also contains eight Leu tRNA genes (for four different tRNAs), but four encode the tRNA with anticodon UAA; the heavily used Leu codon is UUA, perfectly complementary to this predicted most abundant tRNA. Thus, there is co-adaptation between the codon usage of highly expressed genes and the tRNA population in both species, but the identity of the co-adapted state differs. Exactly how this divergence can occur is unclear, but it has been hypothesized that it could be driven by pressure from biased mutation patterns (Shields 1990).
Fourth, not all bacterial species exhibit the same clear trend in codon usage patterns associated with gene expression level. For example, in Helicobacter pylori (a bacterium that causes stomach ulcers), there is at most a very minor difference in codon usage between highly expressed and other genes (Lafay et al. 2000; Kloster & Tang 2008), while in B. burgdorferi most of the highly expressed genes are located on the leading strand of replication, and have G + T-rich codon usage that does not differ from other genes on that strand (Lafay et al. 1999). This difference among species most likely reflects variation in the extent to which natural selection is effective in shaping codon usage; this is the subject of the next two sections.
3. Variation in the strength of selected codon usage bias among bacteria
We have previously examined the strength of selected codon usage bias in 80 distinct bacterial species with genome sequences available (Sharp et al. 2005). To quantify the strength of selected codon usage bias, we modified a population genetic model (Bulmer 1991). The strength of past selection on codon usage can be estimated from the frequency of optimal codons in a gene, if the expected frequency of those codons in the absence of selection is known. Since, for some amino acids, the identity of the optimal codon varies among species, we focused on four amino acids where it is expected that the same codon would always be favoured by selection. For example, the only Phe tRNA genes known across bacteria have GAA at the anticodon site, and so UUC is always expected to be favoured over UUU, when selection is effective. Similarly, for Tyr, Asn and Ile, G at the critical position of the anticodon should always lead to preference for the C-ending rather than the U-ending codon. To determine the frequency of optimal codons in genes potentially under strong selection, we examined a standard set of 40 highly expressed genes (encoding translation elongation factors and ribosomal proteins) found in all bacterial species; these genes encode proteins with around 104–105 copies in the E. coli cell (Ishihama et al. 2008). To define an analogous set of genes present, and expressed at low levels, in all bacteria is more difficult. So we used the codon usage of the genome as a whole as an estimate of the pattern of codon usage when selection is weak; this can be justified because only a minority of genes within a genome are highly expressed. Comparison of codon usage between these two datasets yields an estimate of the compound parameter S = 2Nes, where Ne is the effective population size and s is the selective difference between optimal and non-optimal codons. Thus, S might vary among species because there have been differences in either their population sizes or the strength of selection.
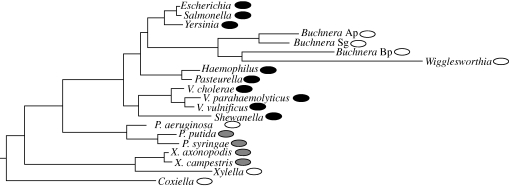
Application of this approach to 80 bacterial genomes revealed considerable variation among species (Sharp et al. 2005). The S value for E. coli was 1.49. In 24 species (30% of the total), including H. pylori, the S value was not significantly greater than zero, providing no evidence for selected codon usage bias. Thirty species (37.5%) had S values greater than 1, with the highest value (2.65) seen in Clostridium perfingens, a widespread bacterium that causes a variety of diseases but is most famous as a ‘flesh-eating bug’. The 80 species examined included variable numbers of representatives from 14 different major lineages (phyla) of bacteria. There was clear phylogenetic clustering of species with high or low S values, but species with strongly selected codon usage bias occurred in several different phyla. Of the 20 species from the gamma proteobacteria (which includes E. coli), nine had S values greater than 1. These nine species form a clade together with a lineage comprised of four species with low S values (figure 1). Thus, it appears that strongly selected codon usage bias evolved on the branch leading to this clade (which includes the orders Enterobacteriales, Pasteurellales, Vibrionales and Alteromonadales) and was subsequently lost on the lineage including Buchnera species and Wigglesworthia. Buchnera species and Wigglesworthia are endosymbionts of insects, which have undergone genome reduction and apparently a general relaxation of genomic selection pressures owing to reduced effective population sizes (Moran & Wernegreen 2000; Wernegreen & Funk 2004); one symptom of this is their long branch lengths in the evolutionary tree (figure 1), reflecting an increased rate of molecular evolution.
Figure 1.
Variation in the strength of selected codon usage bias (S) in gamma proteobacteria. Species are denoted by their genus names, except where there are multiple species from a genus; the abbreviated genus names are Vibrio, Pseudomonas and Xanthomonas. The three Buchnera strains are species infecting different aphid hosts. Shaded ovals next to species names indicate the magnitude of S: white (S < 0.2), grey (0.2 < S < 1.0), black (S > 1.0). Phylogenetic relationships and S values were taken from Sharp et al. (2005). Note that the clustering of Wigglesworthia with Buchnera species may be a phylogenetic artefact (Herbeck et al. 2005); if so, reduced S values evolved independently in the two lineages.
Across the 80 species, values of S were found to be strongly positively correlated with both the number of rRNA operons and the number of tRNA genes in the genome, even after correction for the underlying phylogenetic relationships among species. Many of the species with low S values had only one rRNA operon and a minimal complement of (around 30–40) tRNA genes. In contrast, the E. coli genome has seven rRNA operons and 86 tRNA genes. These results were interpreted as reflecting selection for a co-adapted suite of genomic characteristics required for rapid growth (Sharp et al. 2005). For example, C. perfringens has 10 rRNA operons and 96 tRNA genes and can replicate in only 7 min under ideal conditions (Labbe & Huang 1995).
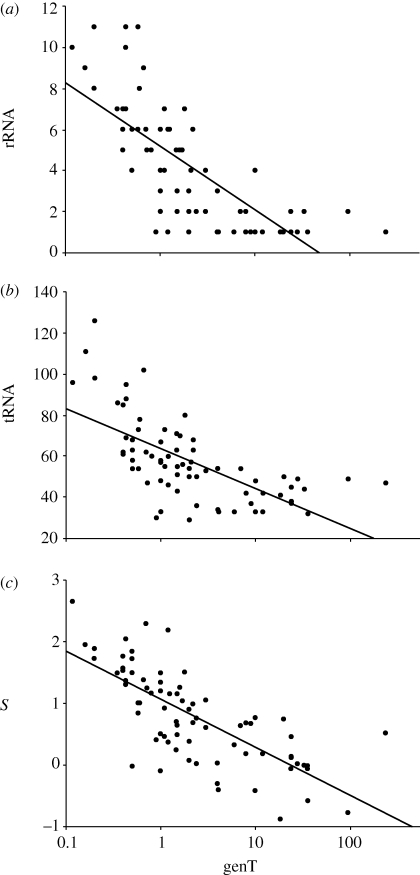
To test this association with growth rate, we have used minimum generation time data for 76 of these 80 species, drawn from the compilations made by E.P.C. Rocha (Rocha 2004; Coutourier & Rocha 2006). rRNA operon number, tRNA gene number and S values are all strongly negatively correlated with generation time (figure 2). Using independent contrasts to overcome the fact that the data points are linked by an underlying phylogeny (Felsenstein 1985), the correlation coefficients for rRNA, tRNA and S are 0.35, 0.27 and 0.49, respectively, and all are highly significant (p < 0.01). Thus, selection for rapid growth appears to have selected for an increase in the number of rRNA operons and tRNA genes, and for codon usage more strongly biased towards translationally optimal codons.
Figure 2.
Correlations of (a) rRNA operon copy number, (b) tRNA gene copy number and (c) the strength of selected codon usage bias (S), with generation time in bacteria. The minimum generation time (in hours) is plotted on a logarithmic scale.
The observation that closely related species tend to have similar S values (as in figure 1) may reflect similarity of lifestyles, such that closely related bacteria are subject to similar strengths of selection for rapid growth. However, it is also likely that codon usage patterns change relatively slowly. Some of the outlier species in figure 2 could be on lineages that have recently entered a new niche. If a species changed from a lifestyle where rapid growth was advantageous, to one where it was not, it would take some time for strongly selected codon usage bias to decay. That is, the values of S reflect selection on codon usage over a long evolutionary period, but not necessarily the current strength of selection.
4. Variation in the strength of selection on codon usage bias among bacteria
An alternative approach, which aims to estimate the strength of current selection on codon usage bias, is to examine the frequency spectrum of optimal codons across polymorphic sites. In an equilibrium population, assuming an infinite sites model and free recombination among sites, the effect of selection on the frequency spectrum can be predicted (McVean & Charlesworth 1999). In the absence of selection, the distribution is expected to be U-shaped with a mean of 0.5, but as the strength of selection is increased, the distribution becomes skewed towards higher frequencies of optimal codons. Importantly, this distribution is not expected to be influenced by mutation biases (McVean & Charlesworth 1999). The observed distribution of allele frequencies can be compared to those predicted for different values of 2Nes, to obtain the maximum likelihood estimate of this compound parameter, termed gamma (Cutter & Charlesworth 2006). Note that both gamma and S (from the previous section) are estimates of 2Nes, but gamma differs from S in reflecting current, ongoing selection. Cutter & Charlesworth (2006) applied this approach to gene sequences from a eukaryote (Caenorhabditis remanei), and found a strong correlation between estimates of gamma and the strength of codon usage bias reflecting long-term evolution (as summarized here by S). Here, we use a similar approach to analyse bacterial codon usage.
We applied the method to 25 genome sequences of E. coli (including strains of Shigella ‘species’, which lie within the radiation of E. coli). First, we analysed polymorphic codon sites in the same 40 highly expressed genes used to estimate S above. All sites with non-synonymous variation, or more than two alleles, were excluded from the analysis, as were sites where the two alleles were both optimal or both non-optimal codons; the latter included sites encoding Cys and Lys, where no optimal codon was designated (table 1). Among nearly 6000 potentially synonymously variable codons, 194 were segregating for one optimal and one non-optimal codon. The average frequency of optimal codons across the polymorphic sites (qopt) was well in excess of 0.5 and the gamma value was estimated as 1.70 (table 2); this value is not substantially (and certainly not significantly) different from the S value of 1.49 estimated by comparing the codon usage of these 40 genes to that in the genome as a whole.
Table 2.
Estimates of the strength of selection for optimal codons from polymorphism data from E. coli and H. pylori.
| speciesa | Sb | genesc | CAId | sitese | polyf | qoptg | gammah (95% CI) |
|---|---|---|---|---|---|---|---|
| E. coli (n = 25) | 1.49 | 40 high | 0.67 | 5963 | 194 | 0.69 | 1.70 (1.23 to 2.25) |
| 10 low | 0.35 | 7211 | 1255 | 0.50 | −0.04 (−0.20 to 0.12) | ||
| H. pylori (n = 7) | 0.02 | 40 high | n.a. | 1131 | 172 | 0.52 | 0.28 (−0.27 to 0.84) |
aAll complete genome sequences were obtained from the GenBank database (July 2009). For E. coli, where multiple substrains of one strain were present, only one was retained.
bS is an estimate of the strength of selection on optimal codons (=2Nes) from comparisons of codon usage bias.
cThe 40 high genes are a standard set of 40 genes expressed at high levels in all species (Sharp et al. 2005). The 10 low genes are 10 genes expressed at low levels in E. coli (dld, glnE, helD, metL, mutS, rmuC, spoT, uvrD, yebT, yhdP).
dThe CAI is a species-specific measure of selected codon usage bias (Sharp & Li 1987), here calculated for a concatenation of the genes.
eThe number of potentially synonymously variable sites examined.
fThe number of sites polymorphic for optimal and non-optimal codons.
gThe average frequency of optimal codons at sites polymorphic for optimal and non-optimal codons.
hGamma is an estimate (with 95% CI) of the strength of selection on optimal codons (=2Nes) from polymorphism data.
We then examined 10 genes with low codon usage bias and expressed at low levels. From the 20 chromosomal genes encoding proteins with the lowest recorded copy numbers (between 50 and 100 per cell) in Ishihama et al. (2008), we selected those having codon adaptation index (CAI) values between 0.3 and 0.4. The CAI (Sharp & Li 1987) is a widely used species-specific measure of selected codon usage bias, which would take a maximum value of 1 for a gene using only optimal codons. In E. coli K-12, the range (99th percentile) of CAI values is from 0.15 to 0.74, with a median of 0.31. Thus, the 10 genes selected here do not have the lowest CAI values, but a substantial fraction of genes with lower values are either hypothetical or of likely foreign origin (i.e. owing to horizontal gene transfer). These 10 ‘low’ genes exhibited a much higher level of polymorphism for optimal versus non-optimal codons, consistent with much lower levels of constraint on codon usage in these genes; genes with lower codon usage bias also exhibit higher levels of interspecific divergence at synonymous sites (Sharp et al. 1989; but see also Berg & Martelius 1995; Eyre-Walker & Bulmer 1995). The average frequency of optimal codons among polymorphic sites across the 25 strains was very close to the value of 0.5 expected in the absence of selection and consequently the estimated gamma value was very close to zero (table 2).
In contrast, we analysed seven genome sequences of H. pylori, where codon selection has previously been estimated to be very weak (S = 0.02; Sharp et al. 2005). We focused on the same 40 highly expressed genes as above, where selection (if present) should be strongest. In this species there is a difficulty identifying which codons are optimal, because there is little difference between the codon usage of highly expressed and other genes (Lafay et al. 2000). Therefore, we focused on the four amino acids used to derive S values, where the C-ending codons are expected to be optimal for biochemical reasons, even if they are not preferred because selection is ineffective. In contrast to the analysis of 40 highly expressed genes in E. coli, the average frequency of optimal codons across polymorphic sites was only just greater than 0.5, and the estimate of gamma was not significantly greater than zero (table 2).
Finally, we examined the sequences of five genes determined for 247 strains of C. perfringens (Rooney et al. 2006). Sixteen optimal codons for C. perfringens were defined by the same approach as applied to E. coli in table 1. The five genes vary in their strength of codon usage bias (measured by Fop in table 3), apparently reflecting differing levels of expression. These data are limited in terms of the number of polymorphic sites. Nevertheless, values of Fop and of the average frequency of optimal codons across polymorphic sites showed the same rank order across the five genes (table 3). Similar to E. coli, polymorphic sites in genes with low codon usage bias had average frequencies of optimal codons close to 0.5, yielding gamma values close to zero. The most highly expressed gene in the dataset, rplL, is one of the 40 genes in the highly expressed datasets used above, and seems representative of that dataset because its Fop value is very close to that obtained from the 40 genes as a whole (Fop = 0.647). The estimated gamma value for rplL was 3.28; the value has very wide confidence intervals reflecting the small number of polymorphic sites, but again it is quite close to the S value of 2.65 estimated for this species.
Table 3.
Estimates of the strength of selection (gamma) for optimal codons from polymorphism among 247 strains of C. perfringens.
| gene | Fopa | sitesb | qoptc | gammad (95% CI) |
|---|---|---|---|---|
| rplL | 0.65 | 6 | 0.88 | 3.28 (0.48 to 8.77) |
| gyrA | 0.49 | 25 | 0.76 | 1.74 (0.62 to 3.06) |
| colA | 0.42 | 33 | 0.58 | 0.48 (−0.37 to 1.37) |
| plc | 0.39 | 47 | 0.56 | 0.34 (−0.37 to 1.07) |
| pfoS | 0.35 | 22 | 0.48 | −0.10 (−1.15 to 0.95) |
aThe frequency of C. perfringens optimal codons in one copy of the gene sequence.
bThe number of sites polymorphic for optimal and non-optimal codons.
cThe average frequency of optimal codons at sites polymorphic for optimal and non-optimal codons.
dGamma is an estimate (with 95% CI) of the strength of selection on optimal codons (=2Nes) from polymorphism data.
These analyses of the frequency spectrum of optimal codons across polymorphic sites should be taken with caution, since the approach assumes that the sequences are drawn randomly from an interbreeding population at mutation-selection-drift equilibrium (McVean & Charlesworth 1999). It has been shown that a recent change in population size can have an erratic impact on the expected frequencies (Zeng & Charlesworth 2009), but the apparent consistency between the values of 2Nes estimated by gamma and by S suggests that such issues have not been important in the examples considered here. Furthermore, the frequency spectra for the two E. coli datasets analysed here, with 25 sequences and estimated gamma values of 0 and 1.7 (figure 3), appear (qualitatively) remarkably similar to the expected distributions for samples of 20 sequences from a diploid species, with gamma values of 0 and 4, shown by McVean & Charlesworth (1999).
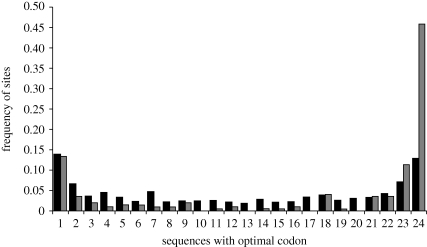
Figure 3.
Distribution of the number of optimal codons at polymorphic sites in 25 strains of E. coli. Data are presented for two sets of genes: 10 genes expressed at low levels (black) and 40 genes expressed at high levels (grey; see also table 2).
The effect of population subdivision on the frequency spectrum, which may be particularly relevant to bacterial species, has not been investigated in detail. In a goodness-of-fit test, the site frequency spectrum for the E. coli low-expression genes differed significantly from that expected (χ2 = 50.1, d.f. = 22, p < 10−3), largely because of the excess of sites with an optimal allele frequency of 7 (figure 3). Such an excess of sites with optimal codons segregating at intermediate frequencies might be expected in samples drawn from a subdivided population. However, the effect is quite small, suggesting again that, for these data, the extent to which the real populations violate the assumptions of the model has had little impact on the results.
The main discrepancy between the observed and expected distributions concerns an excess of sites at extreme optimal codon frequencies in the highly expressed genes; i.e. the leftmost and rightmost grey columns in figure 3 are taller than would be predicted. A possible explanation for the excess of sites with low optimal codon frequencies is that there are certain sites where a codon that is normally optimal is not advantageous. This may be related to the context of the codon: while overall, the frequency of GAA rather than GAG for Glu is only increased a little in highly expressed genes in E. coli (table 1), it has been found that the preference for GAA is strong when the following codon begins with G, but weak in other contexts (Maynard Smith & Smith 1996; Berg & Silva 1997). Also, near the start of highly expressed genes in E. coli, the use of optimal codons is reduced and the frequency of A-ending codons is unusually high, seemingly reflecting conflicting selection pressures (Eyre-Walker & Bulmer 1993). Although we saw no obvious peculiarities to the sites where non-optimal codons were segregating at high frequencies, this merits further investigation.
5. Discussion
The extensive variation in codon usage patterns seen among bacteria is most likely primarily owing to differences in mutation biases. However, in many—but not all—species there is additional variation among genes that is consistent with the action of natural selection. The observation that genes expressed at high levels have increased frequencies of those codons that are expected to be translationally optimal is strongly suggestive that these codons are selectively favoured. The fact that, for some amino acids, the identity of the optimal codons differs among species, coordinated with changes in the population of tRNA genes, reinforces the view that this bias in codon usage is adaptive. However, numerous aspects of how and why natural selection has shaped patterns of codon usage remain unresolved.
To learn more about this we have applied two different approaches to estimate the strength of selection on codon usage in bacteria. Both methods provide estimates of 2Nes, which compounds the effective population size with the selective difference between optimal and non-optimal codons. However, the values (S) from one method reflect very long-term evolution, whereas those (gamma) from the other reflect ongoing selection at polymorphic sites. Gamma values might be expected to be particularly sensitive to the assumption that the sequences analysed came from an idealized population, but the gamma values estimated here were remarkably consistent with estimates of S from the same species. This should not always be the case. Across the phylogeny of bacteria, there have probably been many instances when selection pressures have changed. Then, for example, in a species where codon selection has recently stopped, the gamma value may be close to zero but the S value may still be high because biased codon usage may take a long time to decay (Lawrence & Ochman 1997). In addition, recent demographic changes may impact on the frequency of optimal codons at polymorphic sites and hence gamma (Zeng & Charlesworth 2009), without significant impact on S. At the extreme, Yersinia pestis (the cause of plague) appears to have gone through a very recent severe bottleneck (Achtman et al. 2004) and has such little nucleotide diversity that it would be very difficult to estimate gamma; however, its S value is 1.15 (Sharp et al. 2005), predominantly reflecting selection that occurred in the past in the species from which it was derived, Y. pseudotuberculosis.
Highly expressed genes in many bacteria have S values around 1. The magnitude of the selective difference between optimal and non-optimal codons would then be estimated as on the order of the reciprocal of the effective population size. The effective population sizes for bacterial species are probably not known with any accuracy, but typical values might be in the order of 108 (Lynch 2007), implying that the fitness difference between an optimal and a non-optimal codon may be around 10−8. This is a miniscule value, perhaps reflecting the most subtle form of natural selection known, and only estimable because the selection is repeated over many sites.
This tiny selection coefficient raises a number of issues. One is whether the same form of codon selection can be operating in multicellular eukaryotes. The same approach to estimate S has been adapted for application in eukaryotes (dos Reis & Wernisch 2009). For D. melanogaster and Caenorhabditis elegans, S values of 1.08 and 1.96 were obtained. The same approach to estimate gamma values has been applied to Caenorhabiditis remanei yielding an average value of 0.44 across genes, but with values greater than 1.0 in some genes (Cutter 2008). A number of other analyses have used alternative methods to estimate Nes for codon usage from polymorphism data of Drosophila species. These methods usually require an assignment of the ancestral state at a polymorphic site, which may be difficult in some cases, and especially error prone with bacteria; hence we did not use them here. These analyses have also yielded estimates of the same order of magnitude; for example, Maside et al. (2004) estimated Nes to be around 0.65 in D. americana. Thus, estimates of Nes for Drosophila and Caenorhabditis are of the same order of magnitude as those for bacteria. However, estimates of Ne are typically two orders of magnitude lower than the value given above for bacteria, implying that the fitness difference associated with optimal codons must be two orders of magnitude larger. This has led Lynch (2007) to question whether codon bias in these eukaryotes is caused by some other force, such as biased gene conversion, rather than selection.
A second issue concerns the many sites in the genome where selection on codon usage has occurred. Linkage between sites impairs the efficacy of selection on any one of them, analogous to reducing the effective population size (Hill & Robertson 1966). Bacteria typically have one relatively small chromosome, in which all of the highly expressed genes are linked, and so the strength of selected codon usage bias is expected to be reduced (Li 1987; McVean & Charlesworth 2000). Nevertheless, bacteria have various means of recombination, which vary in frequency among species. This variation in recombination rates could influence the strength of selected codon usage bias, although it has apparently not impacted on H. pylori, which has Nes close to zero, despite perhaps the highest rate of recombination known among bacteria (Suerbaum et al. 1998). If the various sites under codon selection in Drosophila and Caenorhabditis are much less tightly linked than those in bacteria, this could contribute to easing the paradox of similar estimates of Nes in eukaryotes and bacteria (Kaiser & Charlesworth 2009).
The reason why translationally optimal codons are advantageous is also unresolved: it is assumed that they can enhance translational efficiency and/or translational accuracy, but which is more important? The observation that variation among bacteria in the strength of selected codon usage bias is strongly correlated with growth rate (figure 2) may bear on at least one aspect of this debate. In arguing for the accuracy hypothesis, Drummond & Wilke (2008) suggested that non-optimal codons decrease fitness because mistranslated proteins can be toxic. Under this hypothesis, selection against non-optimal codons is stronger in more highly expressed genes because they have more opportunity to be mistranslated. However, it is not clear that the toxic effect of mistranslated proteins would be dependent on rapid growth rate. In contrast, it is obvious that the observed correlation of Nes with growth rate is consistent with the efficiency hypothesis. However, this does not rule out the possibility that inaccuracy of translation is selected against because of its negative impact on the efficiency of translation (Bulmer 1991).
Finally, given the observation that (in many species) codon usage in highly expressed genes is strongly selected and matches tRNA abundance, and yet the identity of the optimal codons can vary among species, there remains an intriguing question: how can this state of co-adaptation between the tRNA gene complement and the codon usage bias in highly expressed genes diverge over time? The observation that the strength of selection varies greatly among contemporary species suggests that there could have been times when ancestral species were subject to relaxed selection, due perhaps to a change of lifestyle or greatly reduced effective population size; selected codon usage bias would then drift and decay. After the re-imposition of selection pressure, the genome could then move to a co-adapted state different from that in the original ancestor. Alternatively, Shields (1990) has suggested that a prolonged influence of mutation bias could provide the impetus for a shift, without the need for a period of drift. Detailed analyses of switches in the identity of optimal codons across the phylogeny of bacteria may provide insights into which, if either, of these processes has played a major role in shaping the patterns of selected codon usage seen in bacteria.
Acknowledgements
We are indebted to Brian Charlesworth for discussion of various aspects of this topic. This work was supported by a studentship from the UK Biotechnology and Biological Sciences Research Council to L.R.E. and a Royal Society of Edinburgh/Caledonian Research Foundation Biomedical Personal Research Fellowship to K.Z.
Footnotes
One contribution of 16 to a Theme Issue ‘The population genetics of mutations: good, bad and indifferent’ dedicated to Brian Charlesworth on his 65th birthday.
References
- Achtman M., et al. 2004Microevolution and history of the pague bacillus, Yersinia pestis. Proc. Natl Acad. Sci. USA 101, 17 837–17 842 (doi:10.1073/pnas.0408026101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H.1994Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S. G. E., Kurland C. G.1990Codon preferences in free-living microorganisms. Microbiol. Rev. 54, 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belozersky A. N., Spirin A. S.1958A correlation between the compositions of deoxyribonucleic and ribonucleic acids. Nature 182, 111–112 (doi:10.1038/182111a0) [DOI] [PubMed] [Google Scholar]
- Berg O. G., Martelius M.1995Synonymous substitution-rate constants in Escherichia coli and Salmonella typhimurium and their relationship to gene expression and selection pressure. J. Mol. Evol. 41, 449–456 (doi:10.1007/BF00160316) [DOI] [PubMed] [Google Scholar]
- Berg O. G., Silva P. J. N.1997Codon bias in Escherichia coli: the influence of codon context on mutation and selection. Nucleic Acids Res. 25, 1397–1404 (doi:10.1093/nar/25.7.1397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G.1986Compositional constraints and genome evolution. J. Mol. Evol. 24, 1–11 (doi:10.1007/BF02099946) [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W.1984The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30, 157–166 (doi:10.1016/0378-1119(84)90116-1) [DOI] [PubMed] [Google Scholar]
- Brewer B. J.1988When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53, 679–686 (doi:10.1016/0092-8674(88)90086-4) [DOI] [PubMed] [Google Scholar]
- Bulmer M.1991The selection-mutation-drift theory of synonymous codon usage. Genetics 129, 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L., Lee W., Hottes A. K., Shapiro L., McAdams H. H.2004Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl Acad. Sci. USA 101, 3480–3485 (doi:10.1073/pnas.0307827100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutourier E., Rocha E. P. C.2006Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol. Microbiol. 59, 1506–1518 (doi:10.1111/j.1365-2958.2006.05046.x) [DOI] [PubMed] [Google Scholar]
- Cutter A. D.2008Multilocus patterns of polymorphism and selection across the X chromosome of Caenorhabditis remanei. Genetics 178, 1661–1672 (doi:10.1534/genetics.107.085803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Charlesworth B.2006Selection intensity on preferred codons correlates with overall codon usage bias in Caenorhabditis remanei. Curr. Biol. 16, 2053–2057 (doi:10.1016/j.cub.2006.08.067) [DOI] [PubMed] [Google Scholar]
- dos Reis M., Wernisch L.2009Estimating translational selection in eukaryotic genomes. Mol. Biol. Evol. 26, 451–461 (doi:10.1093/molbev/msn272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. A., Wilke C. O.2008Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 (doi:10.1016/j.cell.2008.05.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D.1999Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila and Arabidopsis. Proc. Natl Acad. Sci. USA 96, 4482–4487 (doi:10.1073/pnas.96.8.4482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M., Kurland C. G.1984Costs of accuracy determined by a maximal growth rate constraint. Quart. Rev. Biophys. 17, 45–82 (doi:10.1017/S0033583500005254) [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Bulmer M.1993Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 21, 4599–4603 (doi:10.1093/nar/21.19.4599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Bulmer M.1995Synonymous substitution rates in Enterobacteria. Genetics 140, 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.1985Phylogenies and the comparative method. Am. Nat. 125, 1–15 (doi:10.1086/284325) [Google Scholar]
- Fijalkowska I. J., Jonczyk P., Tkaczyk M. M., Bialoskorska M., Schaaper R. M.1998Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl Acad. Sci. USA 95, 10 020–10 025 (doi:10.1073/pnas.95.17.10020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino M. P., Chao L., Riley M. A., Ochman H.1996Asymmetries generated by transcription-coupled repair in Enterobacterial genes. Science 272, 107–109 (doi:10.1126/science.272.5258.107) [DOI] [PubMed] [Google Scholar]
- Galtier N., Lobry J. R.1997Relationships between genomic G + C content, RNA secondary structures, and optimal growth temperatures in prokaryotes. J. Mol. Evol. 44, 632–636 (doi:10.1007/PL00006186) [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C.1982Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10, 7055–7074 (doi:10.1093/nar/10.22.7055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pave A.1980Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 8, r49–r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I., Sharp P. M.2007Predicting gene expression level from codon usage bias. Mol. Biol. Evol. 24, 10–12 (doi:10.1093/molbev/msl148) [DOI] [PubMed] [Google Scholar]
- Herbeck J. T., Degnan P. H., Wernegreen J. J.2005Nonhomogeneous model of sequence evolution indicates independent origins of primary endosymbionts within the Enterobacteriales (gamma-Proetobacteria). Mol. Biol. Evol. 22, 520–532 (doi:10.1093/molbev/msi036) [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A.1966The effect of linkage in limits to artificial selection. Genet. Res. 8, 269–294 (doi:10.1017/S0016672300010156) [PubMed] [Google Scholar]
- Ikemura T.1985Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2, 13–34 [DOI] [PubMed] [Google Scholar]
- Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F. U., Kerner M. J., Frishman D.2008Protein abundance profiling of the Escherichia coli cytosol. BMC Genom. 9, 102 (doi:10.1186/1471-2164-9-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser V. B., Charlesworth B.2009The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25, 9–12 (doi:10.1016/j.tig.2008.10.009) [DOI] [PubMed] [Google Scholar]
- Kanaya S., Yamada Y., Kudo Y., Ikemura T.1999Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238, 143–155 (doi:10.1016/S0378-1119(99)00225-5) [DOI] [PubMed] [Google Scholar]
- Kloster M., Tang C.2008SCUMBLE: a method for systematic and accurate detection of codon usage bias by maximum likelihood estimation. Nucleic Acids Res. 36, 3819–3827 (doi:10.1093/nar/gkn288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A. W., Tollervey D., Plotkin J. B.2009Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (doi:10.1126/science.1170160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe R. G., Huang T. H.1995Generation times and modeling of enterotoxin-positive and enterotoxin-negative strains of Clostridium perfringens in laboratory media and ground beef. J. Food Prot. 58, 1303–1306 [DOI] [PubMed] [Google Scholar]
- Lafay B., Lloyd A. T., McLean M. J., Devine K. M., Sharp P. M., Wolfe K. H.1999Proteome composition and codon usage in spirochaetes, species-specific and DNA strand-specific mutational biases. Nucleic Acids Res. 27, 1642–1649 (doi:10.1093/nar/27.7.1642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafay B., Atherton J. C., Sharp P. M.2000Absence of translationally selected codon usage bias in Helicobacter pylori. Microbiology 146, 851–860 [DOI] [PubMed] [Google Scholar]
- Lawrence J. G., Ochman H.1997Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44, 383–397 (doi:10.1007/PL00006158) [DOI] [PubMed] [Google Scholar]
- Li H.1987Models of nearly neutral mutations with particular implications for the nonrandom usage of synonymous codons. J. Mol. Evol. 24, 337–345 (doi:10.1007/BF02134132) [DOI] [PubMed] [Google Scholar]
- Lobry J. R.1996Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13, 660–665 [DOI] [PubMed] [Google Scholar]
- Lynch M.2007The origins of genome architecture MA, USA: Sinauer Associates [Google Scholar]
- Lynn D. J., Singer G. A. C., Hickey D. A.2002Synonymous codon usage is subject to selection in thermophilic bacteria. Nucleic Acids Res. 30, 4272–4277 (doi:10.1093/nar/gkf546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maside X., Lee A. W., Charlesworth B.2004Selection on codon usage in Drosophila americana. Curr. Biol. 14, 150–154 (doi:10.1016/j.cub.2003.12.055) [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Smith N. H.1996Site-specific codon bias in bacteria. Genetics 142, 1037–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M., Wolfe K. H., Devine K. M.1998Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 47, 691–696 (doi:10.1007/PL00006428) [DOI] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B.1999A population genetic model for the evolution of synonymous codon usage: patterns and predictions. Genet. Res. 74, 145–158 (doi:10.1017/S0016672399003912) [Google Scholar]
- McVean G. A. T., Charlesworth B.2000The effects of Hill–Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155, 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., Wernegreen J. J.2000Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15, 321–326 (doi:10.1016/S0169-5347(00)01902-9) [DOI] [PubMed] [Google Scholar]
- Musto H., Romero H., Zavala A.2003Translational selection is operative for synonymous codon usage in Clostridium perfringens and Clostridium acetobutylicum. Microbiology 149, 855–863 [DOI] [PubMed] [Google Scholar]
- Musto H., Naya H., Zavala A., Romero H., Alvarez-Valin F., Bernardi G.2004Correlations between genomic GC levels and optimal growth temperatures in prokaryotes. FEBS Lett. 573, 73–77 (doi:10.1016/j.febslet.2004.07.056) [DOI] [PubMed] [Google Scholar]
- Muto A., Osawa S.1987The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl Acad. Sci. USA 84, 166–169 (doi:10.1073/pnas.84.1.166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H. E., Moran N. A., Hattori M.2006The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267 (doi:10.1126/science.1134196) [DOI] [PubMed] [Google Scholar]
- Normand P., et al. 2007Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 17, 7–15 (doi:10.1101/gr.5798407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C.1978Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell 14, 179–190 (doi:10.1016/0092-8674(78)90312-4) [DOI] [PubMed] [Google Scholar]
- Post L. E., Nomura M.1980DNA sequences from the str operon of Escherichia coli. J. Biol. Chem. 255, 4660–4666 [PubMed] [Google Scholar]
- Rocha E. P. C.2004Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient coding for translation optimization. Genome Res. 14, 2279–2286 (doi:10.1101/gr.2896904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney A. P., Swezey J. L., Friedman R., Hecht D. W., Maddox C. W.2006Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics 172, 2081–2092 (doi:10.1534/genetics.105.054601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M.1991Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium, codon usage, map position and concerted evolution. J. Mol. Evol. 33, 23–33 (doi:10.1007/BF02100192) [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W.-H.1987The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (doi:10.1093/nar/15.3.1281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Shields D. C., Wolfe K. H., Li W.-H.1989Chromosomal location and evolutionary rate variation in Enterobacterial genes. Science 246, 808–810 (doi:10.1126/science.2683084) [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Bailes E., Grocock R. J., Peden J. F., Sockett R. E.2005Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33, 1141–1153 (doi:10.1093/nar/gki242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C.1990Switches in species-specific codon preferences: the influence of mutation biases. J. Mol. Evol. 31, 71–80 (doi:10.1007/BF02109476) [DOI] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M., Higgins D. G., Wright F.1988‘Silent’ sites in Drosophila genes are not neutral: evidence of selection among alternative synonymous codons. Mol. Biol. Evol. 5, 704–716 [DOI] [PubMed] [Google Scholar]
- Sørensen M. A., Pedersen S.1991Absolute in vivo translation rates of individual codons in Escherichia coli: the two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 222, 265–280 (doi:10.1016/0022-2836(91)90211-N) [DOI] [PubMed] [Google Scholar]
- Stoletzki N., Eyre-Walker A.2006Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 24, 374–381 (doi:10.1093/molbev/msl166) [DOI] [PubMed] [Google Scholar]
- Suerbaum S., Maynard Smith J., Bapumia K., Morelli G., Smith N. H., Kunstmann E., Dyrek I., Achtman M.1998Free recombination within Helicobacter pylori. Proc. Natl Acad. Sci. USA 95, 12 619–12 624 (doi:10.1073/pnas.95.21.12619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N.1962On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl Acad. Sci. USA 48, 582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier E. R. M., Collins R. A.2000The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol. 50, 249–257 [DOI] [PubMed] [Google Scholar]
- Wernegreen J. J., Funk D. J.2004Mutation exposed: a neutral explanation for extreme base composition of an endosymbiont genome. J. Mol. Evol. 59, 849–858 (doi:10.1007/s00239-003-0192-z) [DOI] [PubMed] [Google Scholar]
- Zeng K., Charlesworth B.2009Estimating selection intensity on synonymous codon usage in a non-equilibrium population. Genetics 183, 651–662 (doi:10.1534/genetics.109.101782) [DOI] [PMC free article] [PubMed] [Google Scholar]