Abstract
The geographical region between mainland Asia and New Guinea is characterized by numerous small islands with isolated human populations. Phenotypically, groups in the west are similar to their neighbours in mainland Southeast Asia, eastern groups near New Guinea are similar to Melanesians, and intervening populations are intermediate in appearance. A long-standing question is whether this pattern primarily reflects mixing between groups with distinct origins or whether natural selection has shaped this range of variation by acting differentially on populations across the region. To address this question, we genotyped a set of 37 single nucleotide polymorphisms that are evolutionarily independent, putatively neutral and highly informative for Asian–Melanesian ancestry in 1430 individuals from 60 populations spanning mainland Asia to Melanesia. Admixture analysis reveals a sharp transition from Asian to Melanesian genetic variants over a narrow geographical region in eastern Indonesia. Interestingly, this admixture cline roughly corresponds to the human phenotypic boundary noted by Alfred Russell Wallace in 1869. We conclude that this phenotypic gradient probably reflects mixing of two long-separated ancestral source populations—one descended from the initial Melanesian-like inhabitants of the region, and the other related to Asian groups that immigrated during the Paleolithic and/or with the spread of agriculture. A higher frequency of Asian X-linked markers relative to autosomal markers throughout the transition zone suggests that the admixture process was sex-biased, either favouring a westward expansion of patrilocal Melanesian groups or an eastward expansion of matrilocal Asian immigrants. The matrilocal marriage practices that dominated early Austronesian societies may be one factor contributing to this observed sex bias in admixture rates.
Keywords: admixture, sex-biased, ancestry, Indonesia, Austronesian
1. Introduction
Two broad spheres of cultural influence dominate Indo-Pacific pre-history. Modern humans first settled in the area around 45 thousand years ago (O'Connell & Allen 2004; Barker 2005). Melanesians, who probably resembled these earliest settlers (Brothwell 1960; Howells 1973; Krigbaum & Datan 1999; Bellwood 2007) and are today largely restricted to New Guinea and its surrounding islands, are thought to be their direct descendents. However, Melanesian groups may once have ranged more widely across Island Southeast Asia (ISEA) (Howells 1973). Conversely, populations to the west and north (e.g. in western Indonesia, Borneo, Sulawesi, the Philippines and mainland Southeast Asia) (figure 1) are characterized by Asian features. The arrival time of these populations in ISEA is not well known, nor is the extent to which they contributed genetically to the then-isolated Melanesian populations. Certainly, the spread of some Asian groups was relatively recent, coinciding with the first agricultural settlements in ISEA (Bellwood 2005). This process is often termed the Austronesian expansion, and is putatively linked to demic dispersals from mainland China during the Mid-Holocene.
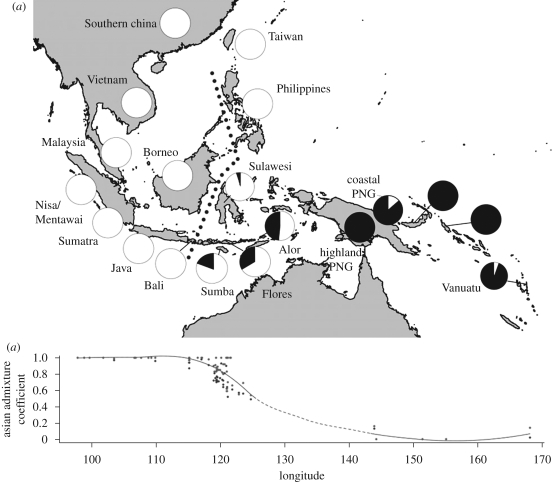
Figure 1.
Local admixture rates across the Indo-Pacific region. (a) Pie charts showing mean regional admixture rates (Asian component in white; Melanesian component in black). Wallace's biogeographic line is shown as a dotted line. Regional admixture rates are shown for data reduction purposes; admixture rates for all 60 populations (with confidence intervals) are listed in the electronic supplementary material. (b) Change in Asian admixture rates calculated from all SNPs combined (black line). Regions with no data indicated by a dashed line (exact gradient unknown). Asian admixture estimated from autosomal and X chromosomal SNPs are indicated by black and grey points, respectively. Note the decline in Asian admixture beginning in eastern Indonesia, as well as preferential retention of X chromosomal (grey) versus autosomal (black) diversity.
Mitochondrial DNA (mtDNA) and Y chromosome (NRY) data suggest that the Austronesian expansion differentially affected populations in ISEA. Broadly speaking, populations in the west have considerable Asian ancestry (Karafet et al. 2005); Asian lineages occur less frequently further east (Friedlaender et al. 2007); and in the remote highlands of New Guinea, where Austronesian languages and cultural items are absent (Bellwood 2007), Asian lineages have not been found at all. This pattern has been attributed to groups expanding recently out from mainland Asia/Taiwan and mixing with pre-existing populations in Melanesia. Furthermore, haploid data are discordant with respect to the relative frequencies of lineages with putatively Asian and Melanesian origins, which has been attributed to sex-biased admixture. For instance, mtDNA lineages with Asian affinity are often found at higher frequency than Asian Y chromosomes in eastern Indonesia and Oceania (Mona et al. 2009), thereby suggesting that the admixture process favoured Asian women (Hage & Marck 2003). Because the entire haploid mtDNA and NRY are strongly affected by genetic drift and founder events owing to their small effective size (Cox 2006), and possibly by natural selection that may have acted on functional sites anywhere within these non-recombining systems, some caution is needed when interpreting this result. Studies of multiple, putatively neutral regions of the nuclear genome have substantially more power to address questions of this nature (Ellegren 2009); however, only three such published studies have relevance for the Indo-Pacific region (Friedlaender et al. 2008; Kayser et al. 2008; Kimura et al. 2008). While these three studies differ in the kind of marker genotyped (autosomal STRs or single nucleotide polymorphisms (SNPs)) and the particular population(s) sampled (mostly Pacific Islanders from different island groups), they tend to agree that approximately 80 per cent of the Polynesian autosomal gene pool is of East Asian origin and approximately 20 per cent is of Melanesian origin. The largest of the three studies also inferred that Taiwanese aboriginals have a predominantly (approx. 100%) Asian origin, while island Melanesian groups share fewer than 20 per cent of Asian markers (Friedlaender et al. 2008). Because each of these studies targeted individual populations or small geographical regions, we still do not have a good understanding of the spatial distribution of Asian–Melanesian ancestry across the Indo-Pacific region. Important questions remain. Does the extent of Asian–Melanesian ancestry really differ across the Indo-Pacific region? Do patterns of ancestry in ISEA mirror those in flanking regions (i.e. mainland Asia and Oceania)? Has incursive Asian gene flow produced a recognizable geographical pattern? And do these admixture rates have a sex-specific bias?
To address these questions, we adopted a different strategy to previous studies. Rather than sampling a large number of randomly identified genomic markers, most of which contain little information about admixture, we assembled a relatively small panel of SNPs (n = 37) that exhibit high FST between representative populations in Asia (southern Han Chinese) and Melanesia (Papua New Guinea (PNG) highlanders). These two groups were chosen because (i) their nuclear genetic diversity has been at least partially characterized (Jakobsson et al. 2008; Li et al. 2008), (ii) they represent two extremes of population differentiation among Southeast Asian populations (Bellwood 2007), and (iii) there is little evidence for pre-historic contact between them—PNG highland populations were largely insulated from Asian advances into the Pacific during the Holocene (evidence reviewed in Cox 2008). We then genotyped these markers in the largest panel of ISEA samples studied to date: 1430 individuals from 60 populations spanning mainland Asia to Melanesia. This approach—using targeted ancestry informative markers (AIMs) chosen from putative source populations outside our study range—allows us to use a smaller panel of markers without significant loss of power. Furthermore, we chose SNPs equally from the autosomes and the X chromosome to address the question of sex-specific admixture. This is possible because the X chromosome spends two-thirds of its time in females and only one-third of its time in males, whereas the autosomes spend equal time in males and females. Therefore, sex-biased migration and mixture processes can, in principle, be detected by examining patterns of diversity on the autosomes and the X chromosome (Hedrick 2007). We also chose our SNPs so that they were effectively unlinked (i.e. statistically independent) and are located in intergenic regions of the genome to minimize the confounding effects of natural selection.
2. Material and methods
(a). Ancestry informative markers
We identified SNPs that show high FST between southern Han Chinese and PNG highlanders. FST was calculated as described previously (Cox et al. 2008). SNPs were chosen from two sources: the HOMINID dataset, a collection of resequenced putatively neutral regions distributed across the human genome (Wall et al. 2008), and the Jakobssen dataset, a collection of 500 000 SNPs typed in the HGDP-CEPH panel (Jakobsson et al. 2008). To address the question of sex-specific admixture, SNPs (n = 37) were selected from both the autosomes and the X chromosome (electronic supplementary material, table S1). All SNPs showed high FST between the two populations (autosome mean = 0.76, X chromosome mean = 0.72; a difference of less than 6%). Moreover, to minimize the effects of natural selection, all SNPs were located away from genes (including introns, UTRs and immediate flanking regions) and are more than 1 cM distant from other SNPs in the panel (i.e. they are evolutionarily independent).
(b). Samples
We assayed 1430 DNA samples from 60 populations across the Indo-Pacific region (mainland China to Vanuatu). Sample sizes varied, but averaged to 33 genotypes per SNP per population (electronic supplementary material, table S2). Sample details are available elsewhere (Karafet et al. 2005; Lansing et al. 2007, 2008), with the exception of New Britain, which is a composite sample of individuals from two inland, Papuan-speaking regions in eastern New Britain—Mali (Marabu) and Kaket (Rangulit and Malasait); Long Gi-Berau, which consists mostly of ethnic Dayak Kenyah; Long Soloy-Berau, which consists mostly of ethnic Dayak Punan; and Timur Hadakewa, which has been included in our greater Flores group, although these individuals were sampled on the neighbouring (but culturally related) island of Lembata.
(c). Data generation
We generated more than 76 000 genotypes for the sample panel. SNPs were genotyped using two different methods: TaqMan, a probe-based PCR assay, and Sequenom, a multiplexed single-base primer extension platform. All genotyping was performed by the University of Arizona Genetics Core facility (http://uagc.arl.arizona.edu/). Appropriate controls were run for both methods, and a subset of SNPs was validated against DNA sequences generated previously (Wall et al. 2008).
(d). Admixture estimates
Because we ascertained SNPs using specific criteria rather than sampling an unbiased range of SNP diversity (i.e. we did not draw from the full site frequency spectrum), many methods of inferring admixture could not be applied here. This includes recently developed coalescent approaches (Bertorelle & Excoffier 1998; Chikhi et al. 2001; Wang 2003). Instead, we inferred admixture rates using a modified weighted least-squares estimator (Chakraborty et al. 1992). This method has the advantage of computational speed, and is therefore readily amenable to resampling approaches and power analyses. We scripted the algorithm in R (code available on request) and validated it against the original test datasets (Chakraborty et al. 1992). The method was modified to account for the sampling error in each of the ‘parental’ (P1 and P2) and ‘hybrid’ (H) populations by inferring a frequency density for P1, P2 and H at each SNP (i.e. a weighted histogram that returns the observed allele frequency). Drift, a stochastic process, was addressed by examining a large number of unlinked (i.e. statistically independent) SNPs. Using a computationally intensive resampling approach, random variables were drawn from the three frequency distributions, admixture was calculated using the least-squares estimator and the process was repeated 105 times. The median admixture rate with 95 per cent confidence intervals (i.e. 0.025 and 0.975 quantiles) was calculated from the distribution of resampled admixture rates. Code was parallelized and run on a UNIX-based high-performance computing grid at the University of Arizona.
3. Results
To obtain Asian admixture rates, we typed 37 SNPs: 18 from the autosomes and 19 from the X chromosome (electronic supplementary material, table S1). All SNPs have FST > 0.50 between southern Han Chinese and PNG highlanders, which places them in the top 2 per cent of all genomic polymorphisms (Jakobsson et al. 2008). Autosomal SNPs had an average FST of 0.76 (range: 0.51–0.95); X chromosomal SNPs had an average FST of 0.72 (range: 0.50–0.85). All SNPs are from intergenic regions (i.e. map far from genes and known functional regions) and are effectively unlinked (i.e. each SNP is more than 1 cM away from any other marker in the panel). These 37 markers were typed in 1430 individuals from 60 Indo-Pacific populations (i.e. approx. 33 genotypes per SNP per population) (electronic supplementary material, table S2). Most differences in these SNP frequencies help to distinguish between Asian and Melanesian populations; for instance, over half of the observed variance of our dataset is carried on the first principal component (electronic supplementary material, figure S1). These AIMs are thus well chosen to capture the major variation along the Asian–Melanesian gradient, although we emphasize that other important components of variation are likely to exist in ISEA.
We assembled a dataset of 37 allele frequencies for 60 populations (i.e. a matrix containing 2220 entries). To reduce this dataset to more manageable proportions, we used the allele frequencies to estimate admixture rates for each population. Admixture rates were calculated using an approach modified from Chakraborty et al. (1992) (see §2 for details). We defined two representative ‘parental’ populations: one for Asia (southern Han Chinese) and one for Melanesia (PNG highlanders). SNP frequencies are similar in all seven Chinese populations (southern Han, Taiwanese Hakka, Miao, She, Tujia, Yao and Yi), and indeed, most of our mainland Asian populations. Admixture estimates (see subsequently) reinforce this shared history. Similarly, no Asian admixture was inferred for the ancestors of all the Papuan-speaking populations in this study (indigenous groups from PNG highlands, New Britain and Bougainville). Consequently, the choice of populations selected to represent the extremes of Asian–Melanesian diversity has little effect on admixture estimates.
We estimated admixture rates for the autosomal and X chromosomal SNPs combined (electronic supplementary material, table S3), as well as for the autosomes (electronic supplementary material, table S4) and X chromosome separately (electronic supplementary material, table S5). For ready comparison, regional admixture rates (i.e. summarized for major island groups) are listed in table 1. In brief, populations from Borneo, Bali and westward exhibit extremely high rates of Asian ancestry (effectively fixed at 100%). While Asian ancestry still predominates across Wallace's biogeographic line in Sulawesi (approx. 97%), the extent of the Asian contribution drops off rapidly in the islands further east: approximately 81 per cent on Sumba, approximately 66 per cent on Flores and approximately 51 per cent on Alor (figure 1a). Further east in Melanesia, only the Austronesian-speaking regions of coastal PNG (approx. 14%) and Vanuatu (approx. 6%) show any evidence of Asian admixture. Asian admixture was not observed in our Papuan-speaking populations from New Britain and Bougainville (i.e. Nasioi). The change from predominantly Asian to predominantly Melanesian genomic ancestry occurs rapidly within a relatively small area of eastern Indonesia that falls roughly between 120 and 145 east longitude (figure 1b) (Cox 2008). We have no samples from 125 to 145 east longitude, but based on published haploid loci from these regions (Mona et al. 2007), the Melanesian genomic component is probably dominant beyond 130 east longitude (i.e. from New Guinea and further east).
Table 1.
Regional admixture rates. See electronic supplementary material, tables S3–S6, for detailed information on individual populations.
| Asian admixture |
|||||||
|---|---|---|---|---|---|---|---|
| region | location | population | all | A | X | difference | female preference |
| mainland Asia | China | Chinese | 1.00 | 1.00 | 1.00 | 0 | 0 |
| Vietnam | Vietnamese | 1.00 | 1.00 | 1.00 | 0 | 0 | |
| Malaysia | Malay | 1.00 | 0.97 | 0.99 | 0.03 | + | |
| ISEA | Taiwan | Aboriginal | 1.00 | 1.00 | 1.00 | 0 | 0 |
| Philippines | Aeta | 1.00 | 1.00 | 0.83 | −0.17 | − | |
| Philippines | Filipino | 1.00 | 1.00 | 1.00 | 0 | 0 | |
| Indonesia | Sumatra | Toba | 1.00 | 1.00 | 1.00 | 0 | 0 |
| Nias | Nias | 1.00 | 1.00 | 1.00 | 0 | 0 | |
| Mentawai | Mentawai | 1.00 | 1.00 | 1.00 | 0 | 0 | |
| Java | Dieng | 0.98 | 0.96 | 1.00 | 0.04 | + | |
| Java | Javanese | 1.00 | 1.00 | 1.00 | 0 | 0 | |
| Borneo | Borneo | 1.00 | 0.99 | 1.00 | 0.01 | + | |
| Bali | Bali | 0.99 | 0.95 | 1.00 | 0.04 | + | |
| Sulawesi | Sulawesi | 0.97 | 0.91 | 0.99 | 0.08 | + | |
| Flores/Lembata | Flores/Lembata | 0.66 | 0.62 | 0.69 | 0.08 | + | |
| Sumba | Sumba | 0.81 | 0.74 | 0.86 | 0.12 | + | |
| Alor | Alor | 0.51 | 0.49 | 0.54 | 0.04 | + | |
| Melanesia | PNG | Coastal | 0.14 | 0.13 | 0.16 | 0.03 | + |
| Bismarck Archipelago | New Britain | 0 | 0 | 0 | 0 | 0 | |
| Bougainville | Nasioi | 0 | 0 | 0 | 0 | 0 | |
| Vanuatu | Maewo | 0.06 | 0.02 | 0.14 | 0.12 | + | |
Admixture rates also differ between the autosomes and the X chromosome (figure 1b, black and grey dots, respectively). We observe higher mean rates of Asian admixture on the X chromosome, which is consistent with approximately 7 per cent greater contribution on average from Asian women during the admixture process (electronic supplementary material, table S6). This difference in admixture rates reaches statistical significance for only a few populations because the confidence intervals on our admixture rates are generally large. (The Aetas of the Philippines are an important exception.) To check whether admixture rates are, in a broad sense, higher on the X chromosome, we instead compared our data with expectations under the binomial distribution. Across 43 Indonesian populations (where we have greater control of sampling quality and coverage), we observe 31 cases where admixture is higher on the X chromosome than on the autosomes. This outcome is statistically highly unlikely (p = 0.0027) if this admixture ratio were fluctuating by chance alone. When we exclude cases where Asian ancestry reaches 100 per cent on both the autosomes and the X chromosome (i.e. there is zero difference in admixture rates), the observed pattern is even less likely (p < 0.00001). We conclude that Asian women made a larger contribution than Asian men to the ancestors of modern ISEA populations during the admixture process.
Finally, we explored whether this bias in admixture rates is structured spatially across Indonesia. Visual inspection of the data suggests a higher level of bias in the southeast (electronic supplementary material, figure S2). To check whether this pattern differs statistically from a uniform expectation, we selected the k nearest neighbours of each sampling location, and determined whether neighbouring points exhibited lower (−1), identical (0) or greater (+1) admixture on the X chromosome relative to the autosomes. We also set the number of cases where Asian ancestry reaches 100 per cent on both the autosomes and the X chromosome to the observed level (n = 7). The empirical dataset was then compared with simulations where admixture rates were permitted to fluctuate randomly (i.e. higher or lower in each sampling location). We found that the observed distribution was an outlier under all of these simulations, regardless of the choice of k (all p < 0.002). Therefore, the bias towards Asian women is significantly structured across Indonesia; neighbouring populations have similar admixture biases, and this effect is increasingly prevalent towards the southeast.
4. Discussion
An Asian–Melanesian phenotypic gradient across the Indo-Pacific region has long been recognized (Wallace 1869); however, the proportion of nuclear loci with Asian and Melanesian ancestry in populations across this region has not been well studied. Here, we perform the first survey of multiple, putatively neutral DNA polymorphisms in a large set of populations to infer admixture dynamics across ISEA. We first genotyped a series of AIMs that distinguish Asian and Melanesian diversity, and then determined the geographical pattern and extent of Asian–Melanesian ancestry. Rather than appearing as a broad cline across Indonesia, we find that a dramatic change from Asian to Melanesian ancestry occurs within a relatively narrow geographical window in the far east—essentially within the Indonesian province of Nusa Tenggara Timur. In particular, to the west of Sumba and Flores, Asian ancestry approaches 100 per cent; while east of Alor, evidence of Asian ancestry diminishes dramatically (see below). Alor is among the most westerly locations where Papuan languages are spoken, and Melanesian Y chromosome lineages are common on all of these islands (Lansing et al. 2007, 2008), but not further west (Karafet et al. 2005). Interestingly, this transition is shifted eastward relative to Wallace's line—a boundary that separates the biogeographic regions of Asia and Wallacea. At its southern limit, Wallace's line falls between the islands of Bali and Lombok (figure 1), which are separated by a deep-water sea channel that marks the southern edge of the Sunda Shelf. During ice-age glacial advances, the Sunda land mass included Borneo, Bali, Java and Sumatra, together with mainland Southeast Asia. However, even in periods of low sea level, deep water in Wallacea separated the Sunda shelf from the eastern landmass of Sahul (connecting New Guinea and Australia). While the distribution of many flora and fauna conforms to Wallace's line, the seafaring capabilities of human settlers to this region undoubtedly overcame this barrier to dispersal. Indeed, Asian ancestry exceeds 50 per cent as far east as the island of Alor, which is well within Wallacea and approximately 1000 km east of Bali, as well as on the island of Sulawesi, which is located east of Wallace's line in the north (figure 1). Curiously, Wallace himself noted this difference, positing a second line in eastern Indonesia corresponding to changes in human phenotype (Wallace 1869; Cox 2008). Wallace's second ‘phenotypic’ line broadly parallels the rapid decline in Asian admixture identified here.
The historical processes underlying this sharp transition from Asian to Melanesian ancestry are not completely clear. Human genetic diversity is typically partitioned over geography in more gradual clines observed at the level of continents (Serre & Paabo 2004). Steep and narrow clines are more unusual (Novembre & Di Rienzo 2009), partly because long-term stability requires large initial gene frequency differences between source populations, and repeated gene flow tends to destablize them (Wijsman & Cavalli-Sforza 1984). A major question emerging from this study relates to the age of the cline in eastern Indonesia. Was it established in the Paleolithic by the encounter of genetically differentiated hunter–gatherer groups (Hill et al. 2007), or did it arise more recently with the mixing of Austronesian farmers and local populations in eastern Indonesia? Climatic changes following the last glacial maximum (approx. 18 kya; Mulvaney & Kamminga 1999) may have spurred expansions of Asian hunter–gatherers into ISEA from further north on the mainland (Soares et al. 2008). Indeed, the spread of the Southeast Asian Hoabinhian culture into Sumatra is one tangible marker of these movements (Bellwood 2007). Dispersals of Asian hunter–gatherers radiating over an extended period of time during the Paleolithic (e.g. 35–8 kya) may have introduced a proportion of the Asian alleles that we detect in western ISEA.
Alternatively, as pointed out in many previous studies based on both genetic (Cox 2005, 2006, 2008; Karafet et al. 2005; Hill et al. 2007; Lansing et al. 2007; Mona et al. 2009) and archaeological data (Bellwood 2005, 2007), there is good evidence that significant Asian contact occurred in eastern Indonesia and Melanesia during the Austronesian expansion. Indeed, much of the pattern of admixture we observe in this study, especially in Wallacea and Near Oceania, may well reflect cultural processes associated with the expansion of Austronesian farmers into the territory previously occupied by Melanesian hunter–gatherers (given subsequently). A third alternative involves a combination of Paleolithic and Neolithic migrational processes. Despite the utility of this set of highly informative autosomal and X-linked SNPs for obtaining admixture rates, more extensive genetic data distributed across the genome would be required to infer the timing of different waves of migration (Hellenthal et al. 2008). Furthermore, the observed variance in admixture rates among individual communities may well be caused by a variety of demographic factors, such as genetic drift and repeated founder events, during the admixture process. The exact nature and mode of action of these factors at the community level remain unclear.
A major finding of this study is that mean rates of Asian admixture are higher on the X chromosome than on the autosomes, suggesting that Asian women made a approximately 7 per cent greater contribution on average during the admixture process(es). This is consistent with previous studies noting a higher proportion of Asian mtDNA versus Y chromosome lineages in many Indo-Pacific populations from eastern Indonesia (Mona et al. 2009). Our results, derived from unlinked and highly informative nuclear markers, are concordant with this finding. In our study, this sex bias in admixture rate appears more prevalent towards the southeast. What cultural processes might underlie this pattern? Further east in Oceania, it has been attributed to the role of matrilocal communities during the Austronesian expansion (Hage 1999; Hage & Marck 2003; Kayser et al. 2008). Matrilocal residence has been inferred as the ancestral state in early Austronesian societies (Jordan et al. 2009), including many whose descendent communities are analysed here. In matrilocal groups, husbands live with their wife's kin, and therefore local Melanesian men would be preferentially incorporated into expanding Asian groups. Over time, this process tends to increase Asian maternal versus paternal ancestry (i.e. increase the frequency of Asian X-linked and mtDNA markers, and decrease the frequency of Asian autosomal and Y chromosomal markers), and probably underpins the bias towards higher rates of Asian admixture on the X chromosome relative to the autosomes. Conversely, an unbiased admixture process would not cause an imbalance in autosome and X chromosome admixture rates.
Other evidence also links the observed pattern in the eastern portion of this range to the Austronesian expansion. The only Melanesian groups in our study that speak Papuan languages—that is, New Britain and Bougainville—show no evidence of Asian admixture, in accordance with earlier studies (Friedlaender et al. 2007). Asian admixture is also infrequent in the Austronesian-speaking populations of Melanesia; that is, coastal PNG and Vanuatu. Indeed, Maewo islanders are one of only two Vanuatu populations where Austronesian Y chromosome lineages have been detected (at a frequency of approx. 10%; Cox 2006). Even the low rates of Asian admixture that we infer for these Austronesian-speaking communities (approx. 10%) may therefore be elevated relative to surrounding groups. The Aeta, a hunter–gatherer Negrito group from the Philippines, is a key exception to general trends. Here, Asian admixture is significantly lower on the X chromosome relative to the autosomes (p > 0.05; electronic supplementary material, tables S4 and S5), suggesting that Asian admixture in the Aeta was biased towards males. This deviation in autosome and X chromosome admixture rates (approx. 17%) is the most extreme of any in our study (electronic supplementary material, table S6). However, Negrito populations probably experienced the flip side of the Austronesian expansion; older communities such as the Aeta adopted Austronesian languages only recently, and local hunter–gatherer women may have preferentially married men from neighbouring agricultural communities. For completeness, we note that an alternative hypothesis for the admixture pattern we observe is a westward expansion of Melanesians practising patrilocality into a territory previously occupied by populations of predominantly Asian ancestry. Some evidence for such a westward expansion comes from archaeobotanical and botanical data supporting a pre-Austronesian dispersal of banana cultivars from New Guinea into eastern Indonesia (Denham & Donohue 2009).
A remaining question is why Asian–Melanesian ancestry changes over such a small area in eastern Indonesia. It may mark the region where indigenous Papuan groups were large enough to resist incursive Austronesian populations during the Mid-Holocene. Such demographic resistance may have its foundation in pre-existing agricultural traditions related to those found in New Guinea (Denham 2005). While the only clear evidence of pre-Austronesian agriculture in the region occurs in the highlands of eastern New Guinea (Denham et al. 2003), indigenous agriculture may have been practiced more widely than current archaeological evidence suggests. Patterns of Y chromosome diversity have been presented to support this model (Mona et al. 2007). Alternately, the rapid phenotypic cline may instead result from changes in Austronesian agricultural practices (Cox 2008). The domestication of rice has long been credited as a presumptive trigger of the Austronesian expansion (Bellwood 1978), but rice agriculture becomes less important from west to east across Indonesia. The current eastward limit of rice falls in eastern Indonesia, where the seasonal tropical climate of more northerly latitudes is transformed into the ‘season-less’ monotony of the equatorial zone (Dewar 2003). This natural climatic variation probably underpins the change from rice agriculture to tuber-based economies (Spriggs 2000). Therefore, the rapid admixture gradient that we observe may reflect the decreasing ability of rice agriculture to propel the Austronesian expansion into the long occupied territory of indigenous Papuan groups (Cox 2008).
Here, we present the first regional picture of Asian admixture rates across the Indo-Pacific. Rates of Asian ancestry vary both geographically and in a sex-specific manner: we infer Asian ancestry to be approximately 100 per cent for most ISEA populations in the west and north, whereas most Melanesian groups show little Asian ancestry—an absence that is especially notable in Papuan-speaking groups. Instead of a gradual cline, we show that the relative contributions of these neutral markers change rapidly over a small area of eastern Indonesia (i.e. near the lesser Sunda islands of Flores, Sumba, Lembata and Alor). Because our markers are distributed across the human genome, the observed cline better fits a demographic scenario, and the primary cause of the phenotypic boundary first identified by Alfred Wallace is most probably the mixing of two phenotypically distinct populations, not natural selection. Additionally, we show that admixture is biased towards the X chromosome relative to the autosomes, which indicates that Asian women were probably favoured in this admixture process. The same general pattern dominates ISEA, with increasing prevalence towards the southeast. Here, in eastern ISEA at least, it may reflect the matrilocal residence system of ancestral Austronesian societies.
Acknowledgements
Indonesian samples were obtained in collaboration with the Eijkman Institute for Molecular Biology, Jakarta, Indonesia, with the assistance of Indonesian Public Health clinic staff, following protocols for the protection of human subjects established by both the Eijkman Institute and the University of Arizona Institutional Review Boards. Permission to conduct research in Indonesia was granted by the Indonesian Institute of Sciences, and informed consent was obtained from all project participants.
We thank Kevin Keyes, Daniel Teberg and Meryanne Tumonggor for assistance in analysing genetic data in the Hammer Laboratory at the University of Arizona. We also thank Jonathan Friedlaender (Temple University) for providing samples from New Britain, as well as for helpful discussion regarding their provenance; Brian Hallmark (University of Arizona) for GIS support; and Lisa Kent (University of Arizona) for data entry. Indonesian samples were obtained by J.S.L. and H.S., and by Golfiani Malik, Wuryantari Setiadi, Loa Helena Suryadi and Meryanne Tumonggor of the Eijkman Institute for Molecular Biology, Jakarta, Indonesia, with the assistance of Indonesian Public Health clinic staff. This research was supported by grants from the National Science Foundation, the James McDonnell Foundation Robustness Programme at the Santa Fe Institute and the Eijkman Institute, Jakarta, Indonesia.
Author contributions: M.P.C. designed research; J.S.L. and H.S. collected samples; M.P.C. and T.M.K. performed research; M.P.C. analysed data; and M.P.C. and M.F.H. wrote the paper.
The authors declare no conflicts of interest.
References
- Barker G.2005The archaeology of foraging and farming at Niah Cave, Sarawak. Asian Perspect. 44, 90–106 (doi:10.1353/asi.2005.0004) [Google Scholar]
- Bellwood P.1978Man's conquest of the Pacific: the prehistory of Southeast Asia and Oceania Auckland, New Zealand: Collins [Google Scholar]
- Bellwood P.2005The first farmers: the origins of agricultural societies Oxford, UK: Blackwell Publishing [Google Scholar]
- Bellwood P.2007Prehistory of the Indo-Malaysian Archipelago Canberra, Australia: ANU E Press, Australian National University [Google Scholar]
- Bertorelle G., Excoffier L.1998Inferring admixture proportions from molecular data. Mol. Biol. Evol. 15, 1298–1311 [DOI] [PubMed] [Google Scholar]
- Brothwell D. R.1960Upper Pleistocene human skull from Niah caves, Sarawak. Sarawak Mus. J. 15–16, 323–349 [Google Scholar]
- Chakraborty R., Kamboh M. I., Nwankwo M., Ferrell R. E.1992Caucasian genes in American Blacks: new data. Am. J. Hum. Genet. 50, 145–155 [PMC free article] [PubMed] [Google Scholar]
- Chikhi L., Bruford M. W., Beaumont M. A.2001Estimation of admixture proportions: a likelihood-based approach using Markov Chain Monte Carlo. Genetics 158, 1347–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. P.2005Indonesian mitochondrial DNA and its opposition to a Pleistocene era origin of proto-Polynesians in Island Southeast Asia. Hum. Biol. 77, 179–188 (doi:10.1353/hub.2005.0037) [DOI] [PubMed] [Google Scholar]
- Cox M. P.2006Extreme patterns of variance in small populations: placing limits on human Y-chromosome diversity through time in the Vanuatu Archipelago. Ann. Hum. Genet. 71, 390–406 (doi:10.1111/j.1469-1809.2006.00327.x) [DOI] [PubMed] [Google Scholar]
- Cox M. P.2008The genetic environment of Melanesia: clines, clusters and contact. In Population genetics research progress (ed. Koven V. T.), pp. 45–83 New York, NY: Nova Science Publishers [Google Scholar]
- Cox M. P., Woerner A. E., Wall J. D., Hammer M. F.2008Intergenic DNA sequences from the human X chromosome reveal high rates of global gene flow. BMC Genet. 9, e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham T.2005Envisaging early agriculture in the highlands of New Guinea: landscapes, plants and practices. World Archaeol. 37, 290–306 (doi:10.1080/00438240500095447) [Google Scholar]
- Denham T., Donohue M.2009Pre-Austronesian dispersal of banana cultivars West from New Guinea: linguistic relics from eastern Indonesia. Archaeol. Oceania 44, 18–28 [Google Scholar]
- Denham T. P., Haberle S. G., Lentfer C., Fullager R., Field J., Therin M., Porch N., Winsborough B.2003Origins of agriculture at Kuk Swamp in the Highlands of New Guinea. Science 301, 189–193 (doi:10.1126/science.1085255) [DOI] [PubMed] [Google Scholar]
- Dewar R. E.2003Rainfall variability and subsistence systems in Southeast Asia and the western Pacific. Curr. Anthropol. 44, 369–388 (doi:10.1086/368348) [Google Scholar]
- Ellegren H.2009The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 25, 278–284 (doi:10.1016/j.tig.2009.04.005) [DOI] [PubMed] [Google Scholar]
- Friedlaender J. S., Friedlaender F. R., Hodgson J. A., Stoltz M., Koki G., Horvat G., Zhadanov S., Schurr T. G., Merriwether D. A.2007Melanesian mtDNA complexity. PLoS ONE 2, e248 (doi:10.1371/journal.pone.0000248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlaender J. S., et al. 2008The genetic structure of Pacific Islanders. PLoS Genet. 4, e19 (doi:10.1371/journal.pgen.0040019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage P.1999Reconstructing ancestral Oceanic society. Asian Perspect. 38, 200–277 [Google Scholar]
- Hage P., Marck J.2003Matrilineality and the Melanesian origin of Polynesian Y chromosomes. Curr. Anthropol. 44, S121–S127 (doi:10.1086/379272) [Google Scholar]
- Hedrick P. W.2007Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution 61, 2750–2771 (doi:10.1111/j.1558-5646.2007.00250.x) [DOI] [PubMed] [Google Scholar]
- Hellenthal G., Auton A., Falush D.2008Inferring human colonization history using a copying model. PLoS Genet. 4, e1000078 (doi:10.1371/journal.pgen.1000078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., et al. 2007A mitochondrial stratigraphy for Island Southeast Asia. Am. J. Hum. Genet. 80, 29–43 (doi:10.1086/510412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howells W. W.1973The Pacific islanders New York, NY: Charles Scribner's Sons [Google Scholar]
- Jakobsson M., et al. 2008Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451, 998–1003 (doi:10.1038/nature06742) [DOI] [PubMed] [Google Scholar]
- Jordan F. M., Gray R. D., Greenhill S. J., Mace R.2009Matrilocal residence is ancestral in Austronesian societies. Proc. R. Soc. B 276, 1957–1964 (doi:10.1098/rspb.2009.0088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T. M., et al. 2005Balinese Y-chromosome perspective on the peopling of Indonesia: genetic contributions from pre-Neolithic hunter–gatherers, Austronesian farmers, and Indian traders. Hum. Biol. 77, 93–114 (doi:10.1353/hub.2005.0030) [DOI] [PubMed] [Google Scholar]
- Kayser M., Lao O., Saar K., Brauer S., Wang X., Nürnberg P., Trent R. J., Stoneking M.2008Genome-wide analysis indicates more Asian than Melanesian ancestry of Polynesians. Am. J. Hum. Genet. 82, 194–198 (doi:10.1016/j.ajhg.2007.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R., Ohashi J., Matsumura Y., Nakazawa M., Inaoka T., Ohtsuka R., Osawa M., Tokunaga K.2008Gene flow and natural selection in oceanic human populations inferred from genome-wide SNP typing. Mol. Biol. Evol. 25, 1750–1761 (doi:10.1093/molbev/msn128) [DOI] [PubMed] [Google Scholar]
- Krigbaum J., Datan I.1999The deep skull of Niah. Borneo 5, 13–17 [Google Scholar]
- Lansing J. S., et al. 2007Coevolution of languages and genes on the island of Sumba, eastern Indonesia. Proc. Natl Acad. Sci. USA 104, 16 022–16 026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing J. S., Watkins J. C., Hallmark B., Cox M. P., Karafet T. M., Sudoyo H., Hammer M. F.2008Male dominance rarely skews the frequency distribution of Y chromosome haplotypes in human populations. Proc. Natl Acad. Sci. USA 105, 11 645–11 650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., et al. 2008Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (doi:10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- Mona S., Tommaseo-Ponzetta M., Brauer S., Sudoyo H., Marzuki S., Kayser M.2007Patterns of Y-chromosome diversity intersect with the trans-New Guinea hypothesis. Mol. Biol. Evol. 24, 2546–2555 (doi:10.1093/molbev/msm187) [DOI] [PubMed] [Google Scholar]
- Mona S., et al. 2009Genetic admixture history of eastern Indonesia as revealed by Y-chromosome and mitochondrial DNA analysis. Mol. Biol. Evol. 26, 1865–1877 (doi:10.1093/molbev/msp097) [DOI] [PubMed] [Google Scholar]
- Mulvaney D. J., Kamminga J.1999Prehistory of Australia Washington, DC: Smithsonian Institution Press [Google Scholar]
- Novembre J., Di Rienzo A.2009Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 10, 745–755 (doi:10.1038/nrg2632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell J. F., Allen J.2004Dating the colonization of Sahul (Pleistocene Australia–New Guinea): a review of recent research. J. Archaeol. Sci. 31, 835–853 (doi:10.1016/j.jas.2003.11.005) [Google Scholar]
- Serre D., Paabo S.2004Evidence for gradients of human genetic diversity within and among continents. Genome Res. 14, 1679–1685 (doi:10.1101/gr.2529604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P., et al. 2008Climate change and postglacial human dispersals in Southeast Asia. Mol. Biol. Evol. 25, 1209–1218 (doi:10.1093/molbev/msn068) [DOI] [PubMed] [Google Scholar]
- Spriggs M.2000Out of Asia: the spread of southeast Asian Pleistocene and Neolithic maritime cultures in Island Southeast Asia and the western Pacific. Mod. Quat. Res. SE Asia 16, 51–75 [Google Scholar]
- Wall J. D., Cox M. P., Mendez F. L., Woerner A., Severson T., Hammer M. F.2008A novel DNA sequence database for analyzing human demographic history. Genome Res. 18, 1354–1361 (doi:10.1101/gr.075630.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A. R.1869The Malay Archipelago: the land of the orang-utan, and the bird of paradise London, UK: Macmillan and Company [Google Scholar]
- Wang J.2003Maximum-likelihood estimation of admixture proportions from genetic data. Genetics 164, 747–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman E. M., Cavalli-Sforza L. L.1984Migration and genetic population structure with special reference to humans. Ann. Rev. Ecol. Syst. 15, 279–301 (doi:10.1146/annurev.es.15.110184.001431) [Google Scholar]