Abstract
The most rapid species radiations have been reported from ‘evolutionary laboratories’, such as the Andes and the Cape of South Africa, leading to the prevailing view that diversification elsewhere has not been as dramatic. However, few studies have explicitly assessed rates of diversification in northern regions such as Europe. Here, we show that carnations (Dianthus, Caryophyllaceae), a well-known group of plants from temperate Eurasia, have diversified at the most rapid rate ever reported in plants or terrestrial vertebrates. Using phylogenetic methods, we found that the majority of species of carnations belong to a lineage that is remarkably species-rich in Europe, and arose at the rate of 2.2–7.6 species per million years. Unlike most previous studies that have inferred rates of diversification in young diverse groups, we use a conservative approach throughout that explicitly incorporates the uncertainties associated with phylogenetic inference, molecular dating and incomplete taxon sampling. We detected a shift in diversification rates of carnations coinciding with a period of increase in climatic aridity in the Pleistocene, suggesting a link between climate and biodiversity. This explosive radiation suggests that Europe, the continent with the world's best-studied flora, has been underestimated as a cradle of recent and rapid speciation.
Keywords: diversification rate, species radiation, Europe, Dianthus, phylogeny
1. Introduction
Biodiversity can arise at spectacular rates. Unusually rapid episodes of species diversification have recently been reported in several megadiverse lineages from equatorial regions and island-like environments (Baldwin & Sanderson 1998). At the same time, most research to date seems to suggest that diversity in temperate northern regions has arisen at moderate speeds, owing to higher rates of extinction and/or lower rates of speciation (Ricklefs 2006; Mittelbach et al. 2007; Schemske 2009). Indeed, in plants, episodes of hyperdiversification have so far only been documented in oceanic archipelagos (Baldwin & Sanderson 1998; García-Maroto et al. 2009) and megadiverse floristic hotspots, such as the tropics (Richardson et al. 2001; Kay et al. 2005; Hughes & Eastwood 2006) and the Cape of South Africa (Verboom et al. 2003; Klak et al. 2004). Perhaps for this reason, studies of plant diversification have focused on ‘exotic’ regions where the chances of detecting explosive radiations are assumed to be higher. However, Northern Hemisphere biomes such as the Mediterranean region of Europe are floristically unique, constituting unorthodox and compelling systems on which to test species radiations.
For historical reasons, Europe is undoubtedly the world's best-studied continent, both botanically and geologically. Surprisingly, to date, there has been no attempt to analyse rates of diversification in genera with exceptional endemic species richness (more than 100 species) in Europe. Species-level studies of large clades are often avoided owing to a priori concerns from botanists regarding the low interspecific genetic variation that often characterizes species-rich genera and greatly limits phylogenetic resolution. Paradoxically, this very feature constitutes one of the most striking signatures of recent rapid radiation. A recent study comparing diversification patterns in Eurasian plant groups revealed a mixed picture—some lineages have originated quite rapidly, whereas others have diversified considerably more slowly (Linder 2008). Rapid diversification in European angiosperms has so far only been detected in relatively small clades from the Mediterranean basin or alpine regions (Kadereit et al. 2004; Linder 2008; Bittkau & Comes 2009; Guzmán et al. 2009), the sizes of which cannot match the scope and scale of large radiations recorded in other parts of the globe. Nevertheless, the rates of origination of these clades suggest that evolutionary processes in the region may have been considerably rapid.
One of the most diverse plant groups in Europe is Dianthus (carnations and pinks), an extremely popular genus in horticulture, as carnations are the world's second most important flower crop (AIPH/Union Fleurs 1997). Dianthus is distributed throughout Eurasia and Africa (approx. 300 species), but is almost exclusively a temperate taxon, with the exception of six tropical African representatives. Over 100 species of carnations occur in Europe (more than 70 endemic), raising the question of how and when such remarkable diversity arose. Dianthus is a taxonomically difficult clade (Tutin & Walters 1993) characterized by large numbers of endemic species with small geographically restricted ranges, suggesting that diversity has originated only recently. Here, we investigate the tempo of evolution in Dianthus using a conservative approach designed to incorporate the uncertainty associated with reconstructing evolutionary history from extant species data. We compare the speed of diversification of Dianthus with the most rapid radiations documented in the globe and investigate whether rates of diversification in the genus have been constant through time.
2. Material and methods
(a). Sampling strategy and phylogenetic analyses
We sampled 104 species of Dianthus, covering all previously recognized sections and subsections and all major geographical regions (Pax & Hoffmann 1934). Representatives of Velezia, Petrorhagia and Saponaria, which are believed to be closely related to Dianthus (Williams 1893; Fior et al. 2006), were also included in the analyses. Voucher information and GenBank accession records are provided in the electronic supplementary material, table S1. We sequenced the nuclear region ITS1-5.8S-ITS2 and the plastid regions trnK-matK, psbA-trnK and trnH-psbA for all accessions (dataset A). A matrix of indel characters was produced using the ‘simple indel coding’ approach (Simmons & Ochoterena 2000) as implemented in SeqState (Müller 2005). For the molecular dating analysis, we sequenced the matK gene for 25 species of Dianthus, one species of Velezia and two species of Petrorhagia, which were then added to Fior and colleagues’ (Fior et al. 2006) family-wide Caryophyllaceae matK dataset (dataset B).
Bayesian and maximum-likelihood (ML) phylogenetic analyses based on the combined dataset A were run on the Vital-IT cluster of the Swiss Institute of Bioinformatics. A general time-reversible (GTR) model with gamma-distributed rate variation was chosen as the optimal model of evolution for all partitions using jModelTest (Posada 2008). Bayesian analyses were conducted in MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003). The indel-character matrix was included and analysed under a binary model. We ran two sets of four Markov chains for 40 million generations, sampling every 1000 generations. Trees from the first 25 per cent of generations were discarded based on an assessment of convergence in Tracer 1.4 (Rambaut & Drummond 2007). For comparison, we performed ML analyses using RAxML 7.2.1 alpha (Stamatakis 2006). A total of 20 runs with a random starting seed were used to infer the best topology, employing a GTRCAT approximation to the nucleotide data and a BINCAT approximation to the indel data.
(b). Divergence time estimation
The age of Dianthus was obtained by applying the Bayesian uncorrelated lognormal method implemented in BEAST 1.4.8 (Drummond & Rambaut 2007) and the non-parametric smoothing method implemented in PATHd8 (Britton et al. 2007) to dataset B. The Caryophyllaceae tree was calibrated using the age of a recently described fossil inflorescence from the Middle–Late Eocene (Jordan & Macphail 2003). Following a detailed phylogenetic study (Jordan & Macphail 2003), the fossil was placed within or as sister to subfamilies Alsinoideae and Caryophylloideae (Frajman et al. 2009). Given the uncertainty regarding the positioning of the fossil with respect to these subfamilies, we chose the most conservative approach (the one that produces the oldest ages) and applied an age constraint at the crown node of Alsinoideae and Caryophylloideae. Divergence dating was repeated using the two age extremes of the fossil (34 and 45 Myr before present; Frajman et al. 2009) as constraints. In BEAST, these constraints were applied as minimum age constraints by use of a lognormal prior (zero offset of 34 or 45, standard deviation of 1). In PATHd8, we fixed the node age at 34 or 45 Myr old. To further incorporate the uncertainty associated with ages derived from paleontological data (Ho & Phillips 2009), we repeated the analyses by assigning an age of 55 Myr before present to the fossil (i.e. increasing the maximum estimated age of the fossil by 10 Myr). For comparison, we conducted an alternative dating analysis using the maximum age for the split between Caryophyllaceae and Amaranthaceae (40 Myr before present) obtained from a study that calculated the ages of several angiosperm families and that is widely used as a source of calibration points for dating molecular phylogenies of plants (Wikström et al. 2001).
Dataset B did not provide sufficient resolution within Dianthus to accurately estimate the divergence times of each of the main subgeneric lineages. Therefore, we conducted a new set of BEAST and PATHd8 analyses using dataset A. We applied age constraints to the split of Dianthus and Petrorhagia and to the crown node of Dianthus, based on the estimates for these nodes in the dated Caryophyllaceae trees.
The ML trees obtained in RAxML were used as the input for PATHd8. The BEAST runs were performed using the optimal molecular models determined in jModelTest (Posada 2008). Runs were repeated using either a birth–death or a Yule speciation prior for the branching rates. We report the results based on runs using the birth–death prior, as the ages were very similar between both methods. Five independent runs of 5 million to 10 million generations, sampling every 1000 generations, were performed for each of the analyses. Adequacy of sampling and run convergence were assessed using the effective sample size diagnostic in Tracer 1.4 (Rambaut & Drummond 2007). Maximum clade credibility trees were built using TreeAnnotator 1.4.8 (Drummond & Rambaut 2007) after removal of the appropriate number of burn-in generations. We describe the results based on the Bayesian analyses, as these yielded older ages, which are more conservative for the purposes of this paper.
(c). Diversification rates
We estimated absolute net diversification rates for Dianthus lineages and compared them with the most rapid episodes of hyper-diversification reported to date. Recent studies comparing published diversification rates of recent radiations have done so without taking into account that the rates reported in previous publications were obtained using a variety of estimators (e.g. Hughes & Eastwood 2006; Moyle et al. 2009). We therefore recalculated rates for each radiation using a standardized methodology. Diversification rates were estimated using Magallón and Sanderson's whole-clade method (Magallón & Sanderson 2001), which does not assume complete taxon sampling. Rates were calculated for both crown and stem groups, at two extremes of the relative extinction rate (k = 0, no extinction; and k = 0.9, high rate of extinction; where k = speciation rate/extinction rate), as implemented in the R package Geiger (Harmon et al. 2008). For each radiation, we obtained total species richness and both crown and stem ages from the original publications (with the exception of the ages of Lupinus, which were kindly provided by Colin Hughes). When available, we used the 95 per cent highest posterior density intervals for the age of each radiation; otherwise, we took the absolute minimum and maximum ages reported.
The most recent systematic treatment of Dianthus estimated the number of species to be close to 300 (Pax & Hoffmann 1934). In order to account for uncertainty in species boundaries, we revised these estimates using a synthetic taxonomical criterion based on morphology and allopatry, and obtained a conservative estimate of 260 species of Dianthus and 6 species of Velezia. We assigned missing species to a particular lineage based on morphology and/or geography. Our method for allocation of missing taxa relies on synapomorphies that had previously been identified as useful in taxonomic treatments (Boissier 1867; Williams 1893) and floristic keys of the genus (Hooper 1959; Reeve 1967).
(d). Diversification dynamics
We used the birth–death likelihood (BDL) method (Rabosky 2006a) implemented in the R package Laser (Rabosky 2006b) to test a null model of constant diversification rates against several rate-variable models. To assess the effect of incomplete taxon sampling in the BDL analyses, we simulated trees including missing species using a Perl script (Day et al. 2008) that adds taxa to a tree randomly along the branches of the clade to which the species are believed to belong. This procedure was repeated for each of 1000 randomly sampled Bayesian trees (obtained from the BEAST analysis of dataset A, excluding outgroup taxa) that reflect topological and branching-time variation, generating a new set of trees that also reflects uncertainty regarding the placement of missing taxa within the clades to which they were assigned.
Akaike information criterion (AIC) scores were calculated for each of the two rate-constant models—pure birth (constant speciation, no extinction) and birth–death (constant speciation and extinction, extinction greater than 0)—and for each of the three rate-variable models—density-dependent speciation with logistic component, density-dependent speciation with exponential component and yule2rate (a multi-rate variant of the pure birth model, allowing one shift in speciation rate). The best-fit model (yule2rate) was selected by comparing the difference in AIC between the best rate-constant model (birth–death) and each of the rate-variable models. The critical value of ΔAICRC (the ΔAIC between the best rate-constant and the best rate-variable model) was calculated based on a distribution of ΔAICRC values under the null hypothesis of rate constancy. This distribution was generated by fitting each of the candidate models to a set of 1000 phylogenies simulated with the same number of taxa as the original chronogram and the same speciation rate as estimated under the pure birth model.
In order to test whether yule2rate was consistently selected as the best model, we fitted BDL models (Rabosky 2006a) to each of the 1000 resampled Bayesian chronograms (with and without missing taxa), generating a posterior distribution of the difference in AIC scores between the birth–death and the yule2rate model (ΔAICRC). This distribution was then compared with the null distribution of ΔAICRC. By repeating BDL analyses across a large sample of chronograms, we were also able to obtain confidence intervals on parameter estimates. In order to visualize the temporal dynamics of diversification in Dianthus, we produced lineage-through-time (LTT) plots using the R package Ape (Paradis et al. 2004). Plots were produced for the original trees without missing species or for trees simulated including missing species.
3. Results
(a). Phylogenetic relationships and divergence times
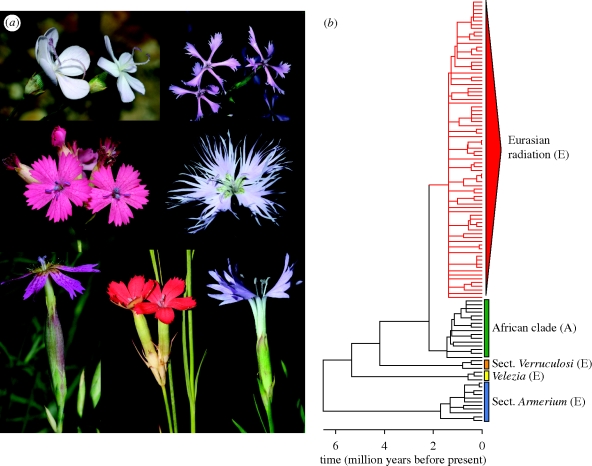
Phylogenetic analyses revealed that Dianthus is monophyletic provided that the small genus Velezia is included (i.e. six species from the Mediterranean basin). Dianthus forms five strongly supported lineages, which are well defined morphologically and geographically. One of these lineages contains the vast majority of Eurasian taxa (over 200 species) and is by far the most diverse in the genus (figure 1a,b; electronic supplementary material, fig. S1). Over 60 per cent of species in this lineage are endemic to the Mediterranean basin and the adjacent Irano-Anatolian biodiversity hotspots, despite the fact that the two regions account for less than 10 per cent of the area of distribution of the clade (electronic supplementary material, fig. S2). This lineage also includes the totality of northern European endemics.
Figure 1.
The radiation of carnations. (a) Floral diversity in European Dianthus. Photographs by P. Vargas, except top left C. Herrera and top right O. Fragman Sapir. (b) Maximum credibility tree of Dianthus (including Velezia) showing times of divergence, obtained from the BEAST analysis of dataset A, calibrated using the oldest age of the Caryophyllaceae fossil. The clades highlighted in the tree represent the five subgeneric Dianthus clades retrieved in the phylogenetic analyses. The outgroup is not shown. Branches highlighted in red correspond to the Eurasian radiation. All nodes shown in the tree have Bayesian posterior probabilities above 0.95. A, African distribution; E, Eurasian distribution.
Dating analyses suggest that the genus first evolved in the Late Miocene/Early Pliocene (electronic supplementary material, fig. S3). The crown age of Dianthus was estimated to be 1.9–7.0 Myr old (95% highest posterior density interval) following the conservative fossil-based approach, or 1.2–3.4 Myr old following the more liberal secondary calibration approach based on Wikström et al. (2001).
(b). Diversification rates
Given our conservative estimate of diversity (200 species) and the oldest crown age estimate for the large Eurasian lineage of carnations (0.9–2.1 Myr), we obtained a net diversification rate of 2.2–5.4 species per million years (sp Myr−1). Using the youngest age of the fossil, this estimate increases to 2.9–7.6 sp Myr−1. Both these estimates exceed or closely match those of the most rapid radiations reported to date in any part of the globe (table 1; electronic supplementary material, table S2). Using the alternative calibration based on the angiosperm-wide study (Wikström et al. 2001), the diversification rate estimate of Eurasian carnations would be even higher (4.2–16.4 sp Myr−1).
Table 1.
Rates of diversification of Dianthus in comparison to the other most rapid radiations documented to date.
| taxon | number of species in clade | diversification rate (crown group, sp Myr−1) | diversification rate (stem group, sp Myr−1) | geographical region |
|---|---|---|---|---|
| plants | ||||
| Cistus (Guzmán et al. 2009) | 10 | 1.46–2.44a | 1.78–2.91a | Mediterranean |
| Costus (Kay et al. 2005) | 51 | n.a. | 0.55–2.62 | Neotropics |
| Echium (García-Maroto et al. 2009) | 19 | 0.35–1.53a | 0.36–1.40a | subtropical islands |
| Gentianella (von Hagen & Kadereit 2001) | 170 | 1.48–2.78 | 1.71–3.21 | Neotropics |
| Hebe complex (Wagstaff et al. 2002) | 100 | n.a. | 1.18 | New Zealand |
| Inga (Richardson et al. 2001) | 300 | 0.37–2.50 | n.a. | Neotropics |
| Lupinus (Hughes & Eastwood 2006) | 81 | 1.30–3.78a | 1.17–3.23a | tropical Andes |
| Neo-Astragalus (Scherson et al. 2008) | 15 | 1.64–2.55 | 1.44–1.82 | South America |
| Primula (Kadereit et al. 2004) | 25 | 1.05 | n.a. | alpine Europe |
| Ruschioideae (Klak et al. 2004) | 1563 | 0.77–1.75 | n.a. | Cape of South Africa |
| Soldanella (Kadereit et al. 2004) | 16 | 2.12 | n.a. | alpine Europe |
| vertebrates | ||||
| Lake Malawi cichlids (McCune 1997) | 400 | n.a. | 3.00–5.99 | tropical lakes |
| Lake Victoria cichlids (Verheyen et al. 2003) | 500 | n.a. | 8.29–62.15 | tropical lakes |
| Lake Victoria and Lake Malawi cichlids (Day et al. 2008) | 1135 | n.a. | 1.67–2.71 | tropical lakes |
| Zosterops (birds) (Moyle et al. 2009) | 80 | 1.95–2.63 | 0.79–0.98 | palaeotropics/temperate Asia |
| Dianthus Eurasian radiation (this study) | ||||
| fossil calibration | 200 | 2.21–7.55a | 1.80–6.09a | temperate Eurasia |
| 75b | 1.74–5.94a | 1.47–4.96a | temperate Eurasia | |
| secondary calibration | 200 | 4.19–16.45a | 3.42–13.59a | temperate Eurasia |
aEstimates are based on 95% highest posterior density intervals of the age estimates reported for each taxon. Otherwise, based on minimum and maximum ages reported.
bNumber of species from this clade sampled in this study.
(c). Differential diversification
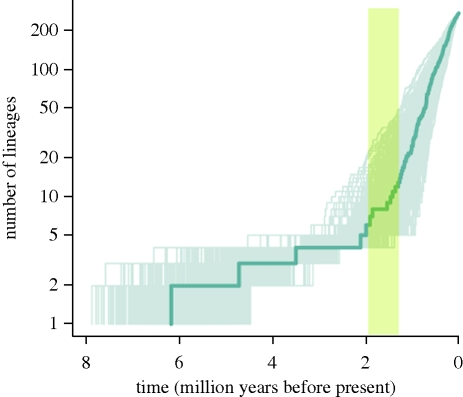
The BDL analyses strongly rejected the null hypothesis that diversification rates in Dianthus have been constant through time (electronic supplementary material, table S3). The same best-fit model was selected whether missing species were added or not, and we therefore describe results including missing species. The best-fit model was the rate-variable yule2rate model (p < 0.005), which assumes a pure-birth process with a shift in speciation rates at a particular point in time. Acceptance of this model was not conditional on branch lengths and topology of the Dianthus chronogram, as revealed by the posterior distribution of model-fit statistics (figure 2). According to the scenario proposed by our analyses, carnations diversified at a constant rate (0.3 ± 0.07 sp Myr−1) until 2.0–1.3 Myr ago, when a dramatic sevenfold to eightfold shift to a new rate (2.2 ± 0.34 sp Myr−1) took place (electronic supplementary material, table S4). This acceleration is shown in the lineage-through-time plots (figure 3; electronic supplementary material, fig. S4), which display a sudden increase in rate with no plateau, a typical pattern found in explosive radiations (Crisp & Cook 2009).
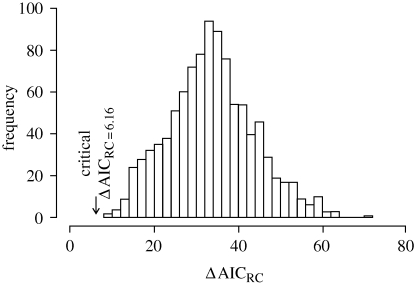
Figure 2.
Posterior distribution of the difference in AIC scores (ΔAICRC) between the best rate-constant model (birth–death) and the best rate-variable model (yule2rate) in the BDL analysis. This distribution was generated after fitting BDL models to each of 1000 Bayesian chronograms of Dianthus (including Velezia) with missing species added. All values of the distribution are above the critical ΔAICRC (6.16; arrow). The critical ΔAICRC was estimated by tabulating ΔAICRC statistics for 1000 phylogenies simulated under the rate-constant birth–death model. The null hypothesis of rate constancy is therefore rejected for all chronograms in favour of the yule2rate model.
Figure 3.
Log-lineage-through-time plot for genus Dianthus (including Velezia), based on 1000 resampled trees with missing taxa added using the method of Day et al. (2008). The line in bold corresponds to the maximum credibility tree from the Bayesian dating analysis (conservative calibration). The shaded panel highlights the time window in which a dramatic increase in diversification rates took place, coinciding with a period of profound climatic transformations in the Pleistocene.
4. Discussion
Our study has revealed that the vast majority of carnations belong to a hyperdiverse Eurasian lineage (approx. 200 species) that diversified very recently and rapidly in the last 1–2 Myr. Contrary to expectations, our conservative upper estimates of the diversification rate of Eurasian carnations surpass those of the most rapid radiations documented in plants from tropical regions (Inga, Lupinus, Gentianella), islands (Echium, Hebe) and the Cape of South Africa (Ruschioideae ‘ice plants’). The rate of origination in carnations is also comparable with that reported for the most rapid radiation of terrestrial vertebrates (Zosterops birds) and is only surpassed by the rates of lacustrine radiations, as in cichlid fish from Lake Victoria (McCune 1997). The finding of rapid diversification in Dianthus is particularly striking given that it concerns one of the most well-known groups of plants from Europe, the continent with the world's best-studied flora. The fact that three other European clades (Cistus, Soldanella, Primula; table 1) showed a diversification rate above 1.0 sp Myr−1 confirms that this continent has been overlooked as a natural laboratory of recent species origination.
Our method was carefully designed to ensure that the diversification rates presented for Dianthus are not overestimates. First, our molecular dating approach explicitly incorporated the uncertainty associated with paleontological data, yielding conservative (old) ages for all nodes in the phylogenetic tree. Even if the Caryophyllaceae fossil was 10 Myr older than currently thought, the upper estimate of diversification rate of Eurasian carnations (4.2 sp Myr−1) would still exceed previous upper estimates in any other group of plants. Second, to account for the fact that taxonomic oversplitting may lead to artificially elevated diversification rate estimates, we used a minimum estimate of diversity of Dianthus in all our analyses. As with other recent radiations (Turner et al. 2001), species boundaries in Dianthus are often unclear as hybrids can occur in the wild, and we were therefore particularly cautious when conducting species diagnosis, revising diversity values downwards. The synthetic taxonomical approach we employed has typically been used for groups that have diversified extensively and recently, such as cichlid fish (Turner et al. 2001) and legumes (Lupinus; Hughes & Eastwood 2006). Most importantly, our results are robust to the level of taxon sampling in Dianthus: in the unlikely case that all of the species not included in this study belong to a clade other than the Eurasian lineage, the absolute net diversification rate for this clade, given the species we sampled, is still 19–66 times faster than the average background rate in angiosperms (0.09 sp Myr−1; Magallón & Sanderson 2001). Finally, in the BDL analyses, we accounted for incomplete taxon sampling by using the random taxon addition method of Day et al. (2008). This method strongly relies on our criteria for assignation of missing species. Nevertheless, had we used an alternative classification, we would still expect to get an acceleration of rates towards the present, given that the crown nodes of all the subgeneric groups are located very close to the tips of the tree.
The reconstruction of past evolutionary events from extant species data presents additional limitations that are more difficult to account for. Recent studies on large clades have suggested that when a given ‘carrying capacity’ is reached, diversity becomes strongly regulated by ecological limiting factors, rather than by diversification rate per se (Rabosky 2009a,b; Ricklefs 2009). Therefore, comparisons of net diversification rates would tend to encounter higher rates in younger clades, simply because they are young (Rabosky 2009a,b). In our comparison, the clades considered are all of remarkably recent origin (average 2.3 Myr before present; electronic supplementary material, table S2), and it is thus reasonable to assume that they are in early stages of diversification and are all still strongly influenced by speciation and extinction processes. However, if some of these clades have indeed reached a diversity limit, then their rates could have been underestimated. An alternative explanation for the high diversity of Eurasian Dianthus would be that this clade has diversified at a similar rate to the other groups, but presents a higher diversity carrying capacity (Rabosky 2009a,b). This alternative hypothesis would imply that the ecological limits on species richness in temperate Eurasia exceed those in tropical areas, a pattern that is uncommon in the literature (Willig et al. 2003), but which cannot be dismissed for Dianthus without further testing.
Understanding why Eurasian carnations have diversified more rapidly than many megadiverse groups in other parts of the world requires a reconsideration of paradigms concerning global patterns of diversification. Diversity in carnations does not appear to be correlated with latitude or insularity, but is instead strongly associated with a climatic regime characterized by seasonality and summer drought. Our analyses detected an abrupt shift in diversification rates of carnations in the Early–Mid-Pleistocene (figure 3), post-dating the onset of a seasonal summer-dry climate in the circum-Mediterranean region similar to that exhibited today (Suc 1984). The establishment of pronounced seasonality in precipitation levels in the region lead to the development of a novel summer-drought regime in which rapid local adaptation processes could have operated (Thompson 2005). We propose that this unique environmental context triggered the formidable acceleration in diversification of carnations detected in our analyses.
Unlike the vast majority of taxa adapted to summer drought (Thompson 2005), Dianthus is almost exclusively a summer-flowering genus. In the Mediterranean basin, for instance, the flowering period of Dianthus is conspicuously out of synchronization with the remaining members of the flora, which blossom en masse in spring (Thompson 2005). It is tempting to hypothesize that the possession of such an unusual phenology in a context of predictable summer drought may have influenced diversification processes by triggering strong local divergence of floral characters in response to pollinators, which are rare in the summer. An additional force that may have promoted rapid reproductive isolation is the occurrence of polyploidization events, given that genome duplications seem to occur readily in the genus (Carolin 1957; Weiss et al. 2002). However, both these hypotheses raise the question as to why Dianthus has diversified so extensively in Europe but not in the summer-dry Cape of South Africa, despite the fact that African carnations share similar phenology and climate, and should be as prone to polyploidization as their Northern Hemisphere relatives. The asymmetry in species richness between the two regions is particularly striking given that the genus has been present in southern Africa for a significant amount of time (1–2 Myr)—a region that, unlike Europe, is well known for its outstanding floristic diversity (Goldblatt & Manning 2002). A possible explanation would be that such disparity was generated by differences in ecological limits associated with the available geographical area: in Europe, the area dominated by a summer-dry climate is one order of magnitude larger than in southern Africa, and may therefore have enabled diversification on a much greater scale (Cowling et al. 1996).
Clearly, geography seems to have played a prominent role in the evolution of carnations in Eurasia. Dianthus lacks major interspecific ecological differentiation, but contains large numbers of narrow endemics (i.e. over 50% of European taxa), a strong signal that the prevailing model of speciation has been geographical. Our data therefore challenge the view that diversification is faster in adaptive radiations (Rundell & Price 2009) and support recent hypotheses proposing that non-adaptive, allopatric diversification can proceed remarkably rapidly (Kozak et al. 2006; Moyle et al. 2009). The circum-Mediterranean region, where most species of carnations occur, is one of the most topographically complex regions of the globe (with hundreds of islands, mountains and peninsulas) and constitutes an ideal setting for processes conducive to allopatric speciation. The combination of terrain complexity and eco-climatic novelty seems to explain why the Mediterranean basin contains a flora of over 24 000 species of angiosperms (10% of the world's total), despite being the youngest of all Mediterranean-type climate biomes (Thompson 2005). In this context of recent biota assembly, it is likely that the explosive radiation of Dianthus was not an exception. We propose that studies of other taxa with outstanding endemic species richness in this region, such as Astragalus (133 spp.), Centaurea (221 spp.) or Silene (194 spp.), will also produce exceptional rates of diversification.
Acknowledgements
We thank Tim Barraclough, Mark Chase, Dan Rabosky, Susanne Renner and one anonymous reviewer for comments; Jan Schnitzler, John Manning, Samantha Wilkinson, Emilio Cano, Lazlo Csiba, Francisco Balao, Hanno Schaefer, Susanne Fritz, Juanjo Aldasoro, Cajsa Anderson, Isabel Sanmartín and Félix Muñoz for assistance; Ori Fragman Sapir and Carlos Herrera for permission to use photographs; James Cotton for permission to use his computer script; Colin Hughes for providing data; Alexandros Stamatakis and Simon Berger for help with analyses; and the Swiss Institute of Bioinformatics for use of their cluster. This work was funded by the European Commission (Marie Curie EST ‘HOTSPOTS’). V.S. was supported by a Royal Society Wolfson Research Merit Award.
References
- AIPH/Union Fleurs 1997Yearbook of the international horticultural statistics, vol. 45 The Hague, The Netherlands: AIPH [Google Scholar]
- Baldwin B. G., Sanderson M. J.1998Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc. Natl Acad. Sci. USA 95, 9402–9406 (doi:10.1073/pnas.95.16.9402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittkau C., Comes H. P.2009Molecular inference of a Late Pleistocene diversification shift in Nigella s. lat. (Ranunculaceae) resulting from increased speciation in the Aegean archipelago. J. Biogeogr. 36, 1346–1360 (doi:10.1111/j.1365-2699.2008.02003.x) [Google Scholar]
- Boissier E.1867Flora orientalis Basel, Switzerland: Basileae et Genevae [Google Scholar]
- Britton T., Anderson C. L., Jacquet D., Lundqvist S., Bremer K.2007Estimating divergence times in large phylogenetic trees. Syst. Biol. 56, 741–752 (doi:10.1080/10635150701613783) [DOI] [PubMed] [Google Scholar]
- Carolin R. C.1957Cytological and hybridization studies in the genus Dianthus. New Phytol. 56, 81–97 (doi:10.1111/j.1469-8137.1957.tb07451.x) [Google Scholar]
- Cowling R. M., Rundel P. W., Lamont B. B., Arroyo M. K., Arianoutsou M.1996Plant diversity in Mediterranean-climate regions. Trends Ecol. Evol. 11, 362–366 (doi:10.1016/0169-5347(96)10044-6) [DOI] [PubMed] [Google Scholar]
- Crisp M. D., Cook L. G.2009Explosive radiation or cryptic mass extinction? Interpreting signatures in molecular phylogenies. Evolution 63, 2257–2265 (doi:10.1111/j.1558-5646.2009.00728.x) [DOI] [PubMed] [Google Scholar]
- Day J., Cotton J., Barraclough T.2008Tempo and mode of diversification of Lake Tanganyika cichlid fishes. PLoS ONE 3, e1730 (doi:10.1371/journal.pone.0001730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A.2007BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior S., Karis P. O., Casazza G., Minuto L., Sala F.2006Molecular phylogeny of the Caryophyllaceae (Caryophyllales) inferred from chloroplast matK and nuclear rDNA ITS sequences. Am. J. Bot. 93, 399–411 (doi:10.3732/ajb.93.3.399) [DOI] [PubMed] [Google Scholar]
- Frajman B., Eggens F., Oxelman B.2009Hybrid origins and homoploid reticulate evolution within Heliosperma (Sileneae, Caryophyllaceae)—a multigene phylogenetic approach with relative dating. Syst. Biol. 58, 328–345 (doi:10.1093/sysbio/syp030) [DOI] [PubMed] [Google Scholar]
- García-Maroto F., Mañas-Fernández A., Garrido-Cárdenas J. A., López-Alonso D., Guil-Guerrero J. L., Guzmán B., Vargas P.2009Δ6-Desaturase sequence evidence for explosive Pliocene radiations within the adaptive radiation of Macaronesian Echium (Boraginaceae). Mol. Phylogenet. Evol. 52, 563–574 (doi:10.1016/j.ympev.2009.04.009) [DOI] [PubMed] [Google Scholar]
- Goldblatt P., Manning J. C.2002Plant diversity of the Cape region of southern Africa. Ann. Miss. Bot. Gard. 89, 281–302 (doi:10.2307/3298566) [Google Scholar]
- Guzmán B., Lledó M. D., Vargas P.2009Adaptive radiation in Mediterranean Cistus (Cistaceae). PLoS ONE 4, e6362 (doi:10.1371/journal.pone.0006362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon L. J., Weir J. T., Brock C., Glor R. E., Challenger W.2008GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- Ho S. Y. W., Phillips M. J.2009Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380 (doi:10.1093/sysbio/syp035) [DOI] [PubMed] [Google Scholar]
- Hooper S. S.1959The genus Dianthus in central and south Africa. Hooker's Icones Plantarum 37, 1–58 [Google Scholar]
- Hughes C., Eastwood R.2006Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl Acad. Sci. USA 103, 10 334–10 339 (doi:10.1073/pnas.0601928103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan G. J., Macphail M. K.2003A Middle–Late Eocene inflorescence of Caryophyllaceae from Tasmania, Australia. Am. J. Bot. 90, 761–768 (doi:10.3732/ajb.90.5.761) [DOI] [PubMed] [Google Scholar]
- Kadereit J., Griebeler E., Comes H.2004Quaternary diversification in European alpine plants: pattern and process. Proc. R. Soc. Lond. B 359, 265–274 (doi:10.1098/rstb.2003.1389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay K. M., Reeves P. A., Olmstead R. G., Schemske D. W.2005Rapid speciation and the evolution of hummingbird pollination in neotropical Costus subgenus Costus (Costaceae): evidence from nrDNA ITS and ETS sequences. Am. J. Bot. 92, 1899–1910 (doi:10.3732/ajb.92.11.1899) [DOI] [PubMed] [Google Scholar]
- Klak C., Reeves G., Hedderson T.2004Unmatched tempo of evolution in southern African semi-desert ice plants. Nature 427, 63–65 (doi:10.1038/nature02243) [DOI] [PubMed] [Google Scholar]
- Kozak K. H., Weisrock D. W., Larson A.2006Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon). Proc. R. Soc. B 273, 539–546 (doi:10.1098/rspb.2005.3326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder H.2008Plant species radiations: where, when, why? Proc. R. Soc. B 363, 3097–3105 (doi:10.1098/rstb.2008.0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S., Sanderson M. J.2001Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780 [DOI] [PubMed] [Google Scholar]
- McCune A.1997How fast is speciation? Molecular, geological, and phylogenetic evidence from adaptive radiations of fishes. In Molecular evolution and adaptive radiation (eds Givnish T. J., Sytsma K. J.), pp. 585–610 Cambridge, UK: Cambridge University Press [Google Scholar]
- Mittelbach G. G., et al. 2007Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331 (doi:10.1111/j.1461-0248.2007.01020.x) [DOI] [PubMed] [Google Scholar]
- Moyle R., Filardi C., Smith C., Diamond J.2009Explosive Pleistocene diversification and hemispheric expansion of a ‘great speciator’. Proc. Natl Acad. Sci. USA 106, 1863–1868 (doi:10.1073/pnas.0809861105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K.2005SeqState: primer design and sequence statistics for phylogenetic DNA datasets. Appl. Bioinf. 4, 65–69 [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K.2004APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- Pax F., Hoffmann K.1934Caryophyllaceae. In Die natürlichen Pflanzenfamilen (eds Engler A., Prantl K.), pp. 275–364 Leipzig, Germany: Engelmann [Google Scholar]
- Posada D.2008jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- Rabosky D.2006aLASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol. Bioinf. Online 2, 247–250 [PMC free article] [PubMed] [Google Scholar]
- Rabosky D. L.2006bLikelihood methods for detecting temporal shifts in diversification rates. Evolution 60, 1152–1164 [PubMed] [Google Scholar]
- Rabosky D. L.2009aEcological limits on clade diversification in higher taxa. Am. Nat. 173, 662–674 (doi:10.1086/597378) [DOI] [PubMed] [Google Scholar]
- Rabosky D. L.2009bEcological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol. Lett. 12, 735–743 (doi:10.1111/j.1461-0248.2009.01333.x) [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A. J.2007Tracer v1.4. See http://beast.bio.ed.ac.uk/Tracer
- Reeve H.1967Dianthus L. In Flora of Turkey and the East Aegean islands, vol. 2 (ed. Davis P. H.), pp. 99–131 Edinburgh, UK: Edinburgh University Press [Google Scholar]
- Richardson J. E., Pennington R. T., Pennington T. D., Hollingsworth P. M.2001Rapid diversification of a species-rich genus of Neotropical rain forest trees. Science 293, 2242–2245 (doi:10.1126/science.1061421) [DOI] [PubMed] [Google Scholar]
- Ricklefs R. E.2006Global variations in the diversification rate of passerine birds. Ecology 87, 2468–2478 (doi:10.1890/0012-9658(2006)87[2468:GVITDR]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Ricklefs R. E.2009Speciation, extinction and diversity. In Speciation and patterns of diversity (eds Butlin R., Bridle J., Schluter D.), pp. 257–277 Cambridge, UK: Cambridge University Press [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Rundell R., Price T.2009Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol. Evol. 24, 394–399 (doi:10.1016/j.tree.2009.02.007) [DOI] [PubMed] [Google Scholar]
- Schemske D. W.2009Biotic interactions and speciation in the tropics. In Speciation and patterns of diversity (eds Butlin R., Bridle J., Schluter D.), pp. 219–239 Cambridge, UK: Cambridge University Press [Google Scholar]
- Scherson R. A., Vidal R., Sanderson M. J.2008Phylogeny, biogeography, and rates of diversification of New World Astragalus (Leguminosae) with an emphasis on South American radiations. Am. J. Bot. 95, 1030–1039 (doi:10.3732/ajb.0800017) [DOI] [PubMed] [Google Scholar]
- Simmons M. P., Ochoterena H.2000Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 49, 369–381 (doi:10.1093/sysbio/49.2.369) [PubMed] [Google Scholar]
- Stamatakis A.2006RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- Suc J. P.1984Origin and evolution of the Mediterranean vegetation and climate in Europe. Nature 307, 429–432 (doi:10.1038/307429a0) [Google Scholar]
- Thompson J. D.2005Plant evolution in the Mediterranean Oxford, UK: Oxford University Press [Google Scholar]
- Turner G. F., Seehausen O., Knight M. E., Allender C. J., Robinson R. L.2001How many species of cichlid fishes are there in African lakes? Mol. Ecol. 10, 793–806 (doi:10.1046/j.1365-294x.2001.01200.x) [DOI] [PubMed] [Google Scholar]
- Tutin T. G., Walters S. M.1993Dianthus L. In Flora Europaea, vol. 1 (eds Tutin T. G., Burges N. A., Chater A. D., Edmondson J. R., Heywood V. H., Moore D. M., Valentine D. H., Walters S. M., Webb D. A.), pp. 227–246 Cambridge, UK: Cambridge University Press [Google Scholar]
- Verboom G. A., Linder H. P., Stock W. D., Baum D.2003Phylogenetics of the grass genus Ehrharta: evidence for radiation in the summer-arid zone of the South African cape. Evolution 57, 1008–1021 [DOI] [PubMed] [Google Scholar]
- Verheyen E., Salzburger W., Snoeks J., Meyer A.2003Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 300, 325–329 (doi:10.1126/science.1080699) [DOI] [PubMed] [Google Scholar]
- von Hagen K. B., Kadereit J. W.2001The phylogeny of Gentianella (Gentianaceae) and its colonization of the southern hemisphere as revealed by nuclear and chloroplast DNA sequence variation. Org. Div. Evol. 1, 61–79 (doi:10.1078/1439-6092-00005) [Google Scholar]
- Wagstaff S. J., Bayly M. J., Garnock-Jones P. J., Albach D. C.2002Classification, origin, and diversification of the New Zealand hebes (Scrophulariaceae). Ann. Miss. Bot. Gard. 89, 38–63 (doi:10.2307/3298656) [Google Scholar]
- Weiss H., Dobes C., Schneeweiss G. M., Greimler J.2002Occurrence of tetraploid and hexaploid cytotypes between and within populations in Dianthus sect. Plumaria (Caryophyllaceae). New Phytol. 156, 85–94 (doi:10.1046/j.1469-8137.2002.00500.x) [Google Scholar]
- Wikström N., Savolainen V., Chase M. W.2001Evolution of the angiosperms: calibrating the family tree. Proc. R. Soc. Lond. B 268, 2211–2220 (doi:10.1098/rspb.2001.1782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams F. N.1893A monograph of the genus Dianthus. J. Linn. Soc. 29, 346–469 (doi:10.1111/j.1095-8339.1839.tb02037.x) [Google Scholar]
- Willig M. R., Kaufman D. M., Stevens R. D.2003Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Ann. Rev. Ecol. Evol. Syst. 34, 273–309 (doi:10.1146/annurev.ecolsys.34.012103.144032) [Google Scholar]