Abstract
The transmission dynamics of influenza in tropical regions are poorly understood. Here we explore geographical variations in the reproduction number of influenza across equatorial, tropical and subtropical areas of Brazil, based on the analysis of weekly pneumonia and influenza (P&I) mortality time series in 27 states. The reproduction number (R) was low on average in Brazil (mean = 1.03 (95% CI 1.02–1.04), assuming a serial interval of 3 days). Estimates of the reproduction number were slightly lower for Brazil than for the USA or France (difference in mean R = 0.08, p < 0.01) and displayed less between-year variation (p < 0.001). Our findings suggest a weak gradient in the reproduction number with population size, where R increases from low population in the North to high population in the South of Brazil. Our low estimates of the reproduction number suggest that influenza population immunity could be high on average in Brazil, potentially resulting in increased viral genetic diversity and rate of emergence of new variants. Additional epidemiological and genetic studies are warranted to further characterize the dynamics of influenza in the tropics and refine our understanding of the global circulation of influenza viruses.
Keywords: influenza, reproduction number, tropics, geography, Brazil, mortality
1. Background
Influenza remains a major public health issue globally, despite the availability of vaccines and anti-viral treatments (WHO et al. 2003). A better understanding of the disease transmission dynamics is crucial for improving the existing control strategies (Chowell et al. 2008b). Influenza has marked winter seasonal patterns in temperate areas of the world, where viruses are reintroduced every winter and cause large and intense outbreaks, followed by fade-out periods in warmer months where little influenza activity is detected (Nelson 2006; Viboud et al. 2006a). By contrast, there is a great diversity of influenza seasonal patterns in the tropics and timing of peak virus activity varies between locales (Alonso et al. 2007; Russell 2008; Laguna-Torres 2009). A number of recent epidemiological and phylogenetic studies have explored the dynamics of influenza in the tropics, with a specific focus on disease burden, seasonality and circulation patterns (Wong et al. 2004, 2006; Alonso et al. 2007; Rambaut et al. 2008; Russell 2008; Laguna-Torres 2009). Most recently, a source-sink model of influenza virus evolution has been proposed, whereby new virus variants preferentially emerge out of the tropics, in particular from East–SouthEast Asia, and seed epidemics in temperate areas (Rambaut et al. 2008; Russell 2008).
A largely unexplored aspect of the global epidemiology of influenza is how infection transmission and population immunity patterns differ between temperate and tropical regions, and how these differences, if any, fit with the proposed source-sink model of influenza evolution. One possible mechanism could involve latitudinal differences in population immunity, where high immunity in the tropics could increase rates of emergence of new virus variants in this region. Alternatively, rates of emergence of new virus variants may be homogeneous globally, but most variants originating from temperate areas could be purged at the end of each winter season owing to strong seasonal bottlenecks. By contrast, new variants could persist in a meta-population of highly connected and seasonally diverse epidemics in tropical regions (Russell 2008). These mechanisms could be tested by comparative analysis of disease dynamics in tropical and temperate locations, in particular studies of transmissibility, immunity and attack rates.
The transmissibility of a pathogen is a central quantity in epidemiology and disease control and is measured by the basic reproduction number (R0), defined as the average number of secondary cases generated by a primary case during the infectious period in an entirely susceptible population (Anderson & May 1991; Diekmann & Heesterbeek 2000). In the case of seasonal influenza epidemics, R0 cannot be estimated owing to partial immunity in individuals infected in previous years with antigenically related strains, and annual vaccination of a fraction of the high-risk population (Chowell et al. 2008b). However, we can estimate a different reproduction number, R, which measures the transmission potential at the beginning of a seasonal epidemic, combining the intrinsic transmissibility of the influenza virus and the average level of immunity in the population. Comparisons of R estimates between tropical and temperate locations could shed light on potential differences in population immunity and transmissibility globally.
Although there are estimates of the reproduction number from a few temperate countries for seasonal influenza (mean R ∼ 1.1–1.3; Chowell et al. 2008b) and for the devastating influenza pandemic of 1918–1919 (range: 1.5–5.4, depending on the serial interval, location and pandemic wave considered (Vynnycky et al. 2007; Andreasen et al. 2008; Chowell & Nishiura 2008; Chowell et al. 2008a)), no estimate exists for tropical regions. Here we took advantage of the unique epidemiology of seasonal influenza in Brazil, which has been described in a recent study (Alonso et al. 2007), to explore geographical differences in the reproduction number across a wide range of latitudes and correlate these differences with geographic, socio-economic and population factors.
2. Material and methods
(a). Data sources
In a previous study exploring Brazilian vital statistics by month and state from 1979 to 2001, an annual wave of influenza-related mortality was shown to originate in the sparsely populated equatorial North, and travel to highly populous states in the subtropical South over a three-month period (Alonso et al. 2007). The amplitude of seasonal mortality was low in the northern parts of Brazil and became more pronounced in the southern regions (Alonso et al. 2007; de Mello 2009; Moura 2009). Here, we further analysed the Brazilian mortality dataset to derive reproduction number estimates over a wide range of latitudes (+5° N to −35° S), taking advantage of the more refined spatial and temporal resolution available for recent years.
Brazil's Ministry of Health provided daily mortality records from 1996 to 2006 for all districts of Brazil. We selected deaths from pneumonia and influenza (P&I), which are considered robust indicators of timing and severity of influenza epidemics (Simonsen 1999; Viboud et al. 2006b) using codes 480–487 from the International Classification of Diseases (ICD) 9th revision and codes J10.0–J18.9 from ICD-10. We aggregated deaths by week and state, including the 26 Brazilian states and the federal district (referred to as ‘27 states’ for simplicity—see figure 1 for selected time series).
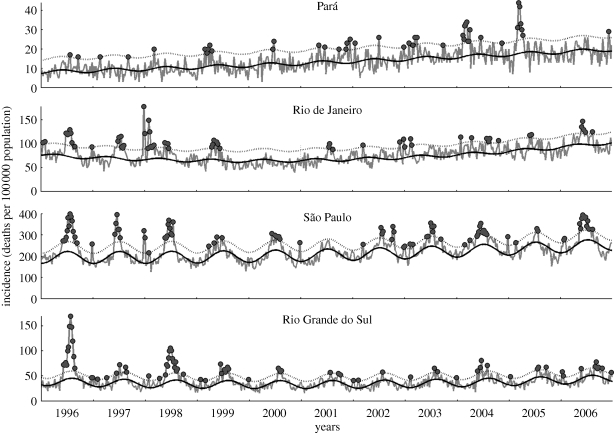
Figure 1.
P&I weekly mortality rate per 100 000 population in four Brazilian states, 1996–2006 (Pará, equatorial; Rio de Janeiro and São Paulo, tropical; and Rio Grande do Sul, subtropical). Grey dots represent weeks where observed mortality exceeds the upper limit of the 95% CI (dotted curve) of the seasonal baseline (solid curve; see electronic supplementary material).
The Brazilian Institute of Geography and Statistics (IBGE) (2005) provided administrative unit boundaries, and geographic and demographic indicators at the state and microregion levels (see maps in figure 2). We used an urbanization index defined as the proportion of the population living in rural areas (figure 2a), population density (estimated as the ratio between population size and surface area, figure 2b), population size (figure 2c), as well as latitude and longitude coordinates of population centres. Indicators of population density and urbanization were well correlated during 1996–2006 (Spearman ρ = 0.53, p < 0.0001). We also calculated the average infant mortality rate as an indicator of socio-economic status (defined as the total annual death rate from all-causes in children under 1 year of age, averaged across 1996–2006).
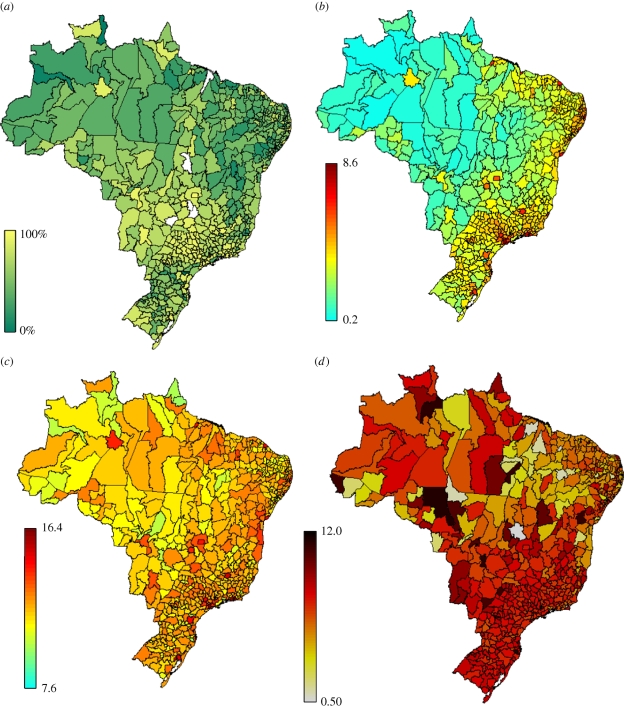
Figure 2.
Maps of urbanization, population size and population density, and P&I mortality rates in Brazilian microregions (n = 554). (a) Urbanization index (0–100%), measured by the percentage of the population living in rural areas across Brazil's microregions; (b) population density in 2000 (log-scale, people/km2); (c) population size in 2000 (log-scale); and (d) average seasonal P&I mortality rates per 100 000 (1996–2006).
(b). Reproduction number for seasonal influenza in Brazil
(i). Transmission model
We used a compartmental transmission model previously developed to estimate the reproduction number of seasonal influenza epidemics based on mortality data from temperate countries (Chowell et al. 2008b). We provide a brief description of the estimation procedure below and the reader can refer to Chowell et al. (2008b) for more details. The population is classified into five disease categories: susceptible (S), exposed (E), infectious (I), recovered/protected (P) and dead (D), where the total population size at time t is given by N(t) = S(t) + E(t) + I(t) + P(t). We assume homogeneous mixing, that is, each individual has the same probability of having contact with any other individual in the population. For each seasonal epidemic, the total population is assumed constant according to the population size estimate for a given location and year. Susceptible individuals infected with the virus enter the latent period (category E) at the rate β I/N, where β is the mean transmission rate per day and I/N is the probability of contacting an infected individual out of the total population size N. Latent individuals progress to the infectious class at the rate κ (1/κ is the mean latent period). Infectious individuals either recover or die from influenza at the mean rates γ and δ, respectively. Recovered individuals are assumed protected for the duration of the influenza season. The mortality rate is given by δ = γ [CFP/(1 − CFP)], where CFP is the mean case fatality proportion. The system of differential equations that describes the above epidemic process is given by:
and
The initial number of P&I deaths D(0) is set to be the number of P&I deaths in the first epidemic week. Furthermore, using the case fatality percentage (CFP), we also approximate the number of recovered individuals in the first epidemic week as P(0) = D(0)/CFP − D(0). Thus, the initial susceptible population, S(0), can be roughly estimated by S(0) = N − P(0) − D(0) − E(0) − I(0). The reproduction number, R, is given by the product of the mean transmission rate β and the mean infectious period 1/(γ + δ), that is, R = β/(γ + δ); it is estimated for each state and season. Hence, the estimates of β and R capture spatial and temporal variations in partial population immunity and virus transmissibility. The formulation is equivalent to assuming a partially susceptible population at the beginning of the epidemic, and estimating R0, the basic reproduction number for influenza (Earn et al. 2000).
(ii). Parameter estimation
The mean latent and infectious periods were each fixed to 1.5 days (mean serial interval of 3 days) and the CFP of influenza was fixed to 0.2 per cent according to previous studies (Cauchemez et al. 2004; Ferguson et al. 2005; Chowell et al. 2008a). We also present a sensitivity analysis with a mean serial interval of 6 days, since the value of this parameter is debated (Andreasen et al. 2008; Chowell et al. 2008b). We also assessed the sensitivity of our estimates to assuming a longer interval between infection and death of about 7 days (Mills et al. 2004). Parameters β, E(0) and I(0) were estimated by least squares fitting of the model to the cumulative number of weekly P&I deaths during the initial ascending phase of the epidemic. The advantage of using the cumulative over the weekly number of new deaths is that the former somewhat smoothes out known reporting delays (Ferrari et al. 2005; Chowell et al. 2008a). A sensitivity analysis using the weekly rather than the cumulative number of deaths to estimate the reproduction number was also carried out. We estimated the reproduction number for epidemics with an ascending phase comprising at least four weeks immediately preceding the epidemic peak, where the peak week is the week with highest death rate in a given year. When the initial epidemic phase was comprised by more than four epidemic weeks, we used the ascending phase period that gave the best fit to our SEIR model using the χ2 goodness-of-fit statistic (Chowell et al. 2008a). Because of the short latent period for influenza, we assumed E(0) = I(0); this simplification allowed us to estimate only two parameters from the exponential growth phase of the epidemic.
(iii). Uncertainty of reproduction number estimates
We estimated the uncertainty of reproduction number estimates via parametric bootstrap as in previous studies (Chowell & Nishiura 2008; Chowell et al. 2008b). For each epidemic, we simulated 200 alternate realizations of the epidemic trajectory, by perturbation of the best-fit solution of the cumulative weekly epidemic curve. We added to the best-fit curve a simulated Poisson error structure computed using the increment in the ‘true’ number of deaths from week t to week t + 1 as the Poisson mean for the number of new deaths observed in the t to t + 1 interval.
(iv). Sensitivity analyses
In prior work (Chowell et al. 2008b), we showed that the estimation algorithm was robust to the choice of the error distribution for bootstrap resampling, the distribution of the latent and infectious periods, the method for estimating confidence intervals, the number of weeks used in the estimations, and the CFP values (two-fold increase or decrease). In this study, we performed various additional sensitivity analyses, as detailed below.
Sensitivity to choice of mortality indicator. Since this study was focused on Brazil, a tropical/subtropical country where the seasonality of influenza is less defined than in temperate areas, we needed to assess the robustness of R estimates to the use of various mortality indicators. In particular, we tested the impact of using excess P&I deaths above a seasonal baseline (Chowell et al. 2008b) in the R estimation procedure, rather than using crude P&I deaths (electronic supplementary material, figures S1–S5). To estimate excess P&I mortality, we expanded on a classical approach developed by Serfling in 1963 and previously applied to data from a variety of temperate countries (Serfling 1963; Viboud et al. 2004; Simonsen et al. 2005; Chowell et al. 2008b). Specifically, we iteratively applied a linear regression model with harmonic terms to Brazilian P&I mortality to identify epidemic weeks and produce an expected level of mortality in the absence of influenza virus activity (electronic supplementary material). Excess P&I mortality was defined as observed minus expected mortality, which is thought to produce a highly specific indicator of influenza disease patterns (Serfling 1963; Viboud et al. 2004; Simonsen et al. 2005; Chowell et al. 2008b).
Sensitivity to R estimation method. We conducted additional sensitivity analyses to take into account a possible delay between infection and death, test the impact of using weekly death counts instead of cumulative counts, and the robustness to various R estimation algorithms. In particular, we estimated R with an entirely different method that does not rely on a SEIR transmission model, and combines estimates of the initial exponential growth rate with the theoretical distribution of the generation interval (Lipsitch et al. 2003; Wallinga & Lipsitch 2007; Chowell et al. 2008a). We assumed an initial exponential growth and estimated the intrinsic growth rate ‘r’ by fitting a straight line (with slope ‘r’ and intercept b) to the initial increase in weekly deaths (in logarithmic scale). The longest epidemic period consistent with exponential growth was determined by the goodness-of-fit test statistic (Chowell et al. 2008a). The reproduction number was calculated by substituting the estimate for r into an expression derived from the linearization of the classical susceptible–exposed–infectious–recovered transmission model (Lipsitch et al. 2003; Wallinga & Lipsitch 2007):
| 2.1 |
where 1/b1 and 1/b2 are, respectively, the mean latent and infectious periods. This expression for R assumes exponentially distributed latent and infectious periods, where the mean generation interval between two successive cases is given by Tc = 1/b1 + 1/b2. We also obtained an upper bound estimate for the extreme case of a fixed generation interval (delta distribution), using the following expression (Wallinga & Lipsitch 2007):
| 2.2 |
Sensitivity to heterogeneities in influenza transmission and death by age. Since we relied on aggregated time series of influenza-related deaths, combining all ages, we explored the effects of age differences in transmission and case fatality rates on reproduction number estimates. We simulated an age-structured ordinary differential equation model, as in Chowell et al. (2009), using realistic age-specific contact rates recently estimated from surveys conducted in Utrecht, the Netherlands, for six age groups (0–5, 6–12, 13–19, 20–39, 40–59, ≥60; Wallinga et al. 2006). We incorporated in the model age variation in severity of disease by setting age-specific values for hospitalization given clinical infection Weycker et al. (2005) and three broad values of the case fatality rate given hospitalization (1% (0–5 years), 3% (6–59 years) and 15% (less than or equal to 60 years).
Sensitivity to spatial heterogeneities. Most of our analyses were done at the week and state level, because a weekly time resolution was essential to estimate R and there were too few deaths at lower levels of spatial aggregation. However, using coarse state-level mortality data can lead to underestimation of R, if local epidemics within a state lack synchrony. To address this potential issue, we compiled more spatially refined mortality time series for the greater metropolitan areas of São Paulo and Rio de Janeiro.
(c). Comparison of reproduction number estimates (R) for Brazil, France, Australia and the USA
Since we were interested in comparisons of the reproduction number between tropical and temperate regions, we compared estimates obtained for Brazil with estimates for the USA, France and Australia, using epidemiological data previously published in Chowell et al. (2008b). In this comparison, the same R estimation approach was applied to weekly national P&I mortality time series from all four countries. Confidence intervals calculated for summary national R estimates combined statistical uncertainty in the estimation procedure of individual R estimates, and variability from taking an average over multiple states and/or influenza seasons.
3. Results
(a). Estimates of influenza reproduction number in Brazilian states and sensitivity analyses
In most of the 297 epidemics studied in 27 Brazilian states during 1996–2006, the initial ascending phase lasted for four weeks or more, so that robust estimates of the reproduction number R could be obtained for 67–85% of the states each season. The average reproduction number across all states and seasons was 1.03 (95% CI 1.02–1.04) with a serial interval of 3 days and 1.04 (95% CI 1.02, 1.06) with a serial interval of 6 days. There was no time trend in the annual time series of R estimates averaged for the 27 states (p = 0.65; electronic supplementary material, figure S6). A map of average state-level estimates is provided in figure 3, while the distributions of R estimates for selected states are shown in figure 4.
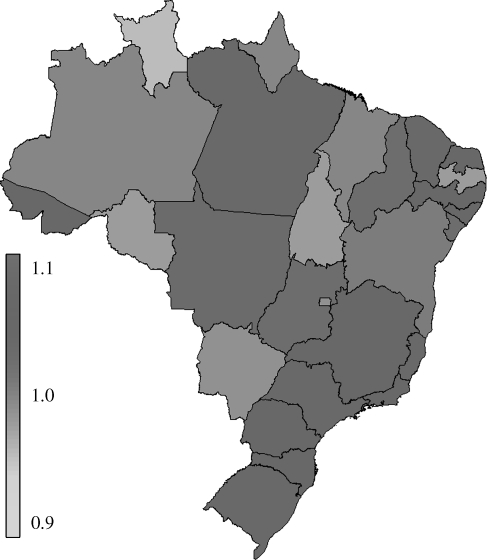
Figure 3.
Map of mean reproduction number estimates during seasons 1996–2006 in 27 Brazilian states.
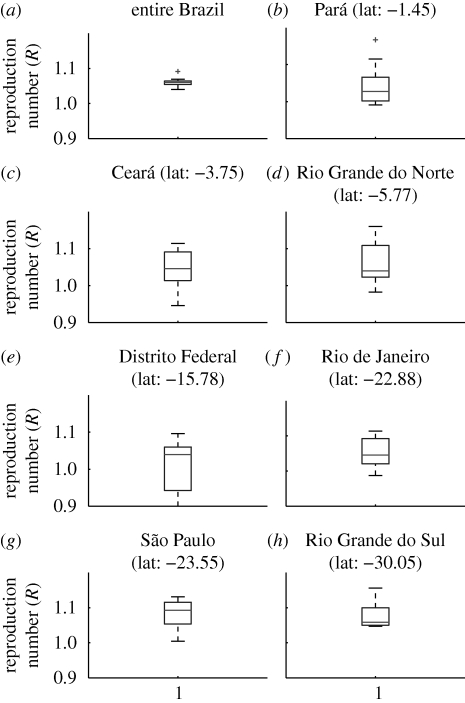
Figure 4.
(a–h) Boxplots of the mean estimates of the reproduction number R for seasons 1996–2006 for selected states of Brazil representing a range of latitudes. Boxes have lines at the 25, 50 and 75 percentiles and whiskers show the rest of the data extending to a maximum of 1.5 times the interquartile range. Points outside the ends of the whiskers are indicated with a grey cross.
Overall, our estimates of the reproduction number were robust to a number of assumptions. First, we selected four geographically distant states with large enough population sizes to conduct a sensitivity analysis on the type of mortality indicator used to estimate R. We calculated P&I mortality in excess of a seasonal baseline and ran R estimation algorithms for São Paulo and Rio de Janeiro (tropical states), Rio Grande do Sul (subtropical) and Pará (equatorial). We found that R estimates were robust to using crude P&I or excess P&I mortality (electronic supplementary material, table S1), as the overall mean R for all four states and seasons (n = 37) was 1.06 (95% CI 1.05, 1.08) using crude P&I mortality and 1.09 (1.0, 1.18) using excess P&I mortality (n = 20) (t-test for differences in mean R, p = 0.35). Because of substantial reduction in the duration of the ascending phase when using excess mortality, it was not possible to estimate R for all seasons in the P&I excess mortality method.
Second, we tested the robustness of estimates to the choice of the R estimation algorithms. There was little difference in using weekly or cumulative mortality epidemic curves, with an increase in R estimates by about 2 per cent when using the weekly number of P&I deaths. When considering a longer interval between infection and death (7 days), our estimates increased slightly by about 7 per cent. Estimates were also robust to realistic age variations in transmission and disease severity. Simulations using an age-structured transmission model that accounts for age-specific rates of contacts, hospitalization and death, indicated that our methodology slightly underestimates the reproduction number by 3–8% when the theoretical reproduction number is between 1.1 and 1.3.
To test the effect of the spatial level of aggregation, we also estimated the reproduction number for the greater metropolitan areas of Rio de Janeiro, where R = 1.07 (95% CI 1.03, 1.11) and São Paulo, where R = 1.06 (95% CI 1.03, 1.1). These results were within 3 per cent of our estimates derived from mortality aggregated at the state level.
Finally, we carried out a sensitivity analysis relying on an entirely different method to estimate R, which combines estimates of the initial exponential growth rate with the assumed distribution of the generation interval (Lipsitch et al. 2003; Wallinga & Lipsitch 2007; Chowell et al. 2008a). Estimates changed by less than 0.1 on average (absolute difference) when assuming exponential or fixed latent and infectious periods, which provide lower and upper bounds on R for a given value of the growth rate and the mean generation interval (Wallinga & Lipsitch 2007). Overall, these various sensitivity analyses suggest that our estimates are within 10 per cent or less of their true values, despite using a simple transmission model and state-level epidemiological data.
(b). Comparison of the reproduction number estimates between Brazil and temperate countries
To conduct a fair comparison of the reproduction number between Brazil and the USA, France and Australia, for which we had national data, we also compiled national mortality time series for Brazil and derived annual national R estimates. National estimates were consistent with state-level estimates in Brazil, with an average R = 1.06 (95% CI 1.05, 1.07) and little inter-annual variation (range of R 1.04–1.09; electronic supplementary material, table S1).
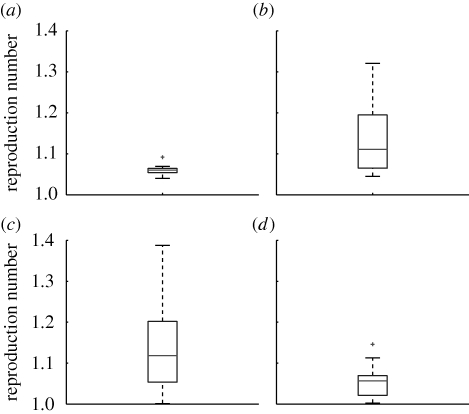
Box plots illustrating the distribution of R estimates for Brazil, France, USA and Australia are provided in figure 5 for a serial interval of 3 days. The average R estimate for Brazil was lower than that in the USA (mean R = 1.14, 95% CI 1.11, 1.17; p = 0.01) and France (mean R = 1.14, 95% CI 1.10, 1.19; p = 0.04) but there was no difference with Australia (mean R = 1.06, 95% CI 1.04, 1.08; p = 0.5). Moreover, there was less between-year variation in estimates from Brazil than from the USA, France and Australia (p < 0.001; see also estimates for individual seasons and different serial interval assumptions in electronic supplementary material, table S2).
Figure 5.
Boxplots of the mean estimates of the reproduction number R for Brazil (a), the USA (b), France (c), and Australia (d). Boxes have lines at the 25, 50 and 75 percentiles and whiskers show the rest of the data extending to a maximum of 1.5 times the interquartile range. Points outside the ends of the whiskers are indicated with a grey cross.
To account for a potential bias owing to unequal length of the epidemiological time series in the four countries (nine estimates were obtained for Brazil and 25–30 estimates for the temperate countries), we simulated shorter time series for temperate countries by randomly sampling nine estimates from the empirical distribution of R in temperate countries. Results for 100 randomized samples confirmed that the observed range of R in Brazil was significantly lower than in the USA (maximum R = 1.28, p < 0.01) and France (maximum R = 1.31, p < 0.01). By contrast, the difference in the range of R estimates disappeared with Australia (maximum R = 1.11, p = 0.32).
(c). Reproduction number estimates and geographic, socio-economic and population factors
Finally, we explored the association between the reproduction number of influenza and various covariates (table 1 and electronic supplementary material, figure S7). The mean R across seasons 1996–2006 increased moderately with the population size of each state (Spearman correlation ρ = 0.53, p = 0.004) and the distance from the Equator (Spearman ρ = 0.46, p = 0.02). Population size was the only factor to remain statistically significant after adjusting for all other variables and applying Bonferroni correction for multiple comparisons.
Table 1.
Relationship between influenza reproduction number and population size, population density, urbanization index (defined as the proportion of the population living in rural areas) and latitude of Brazilian states (n = 27) for the period 1996–2006. The strength of the relationship is given by the Spearman univariate correlation coefficient and the corresponding p-value as well as the partial rank (Spearman) correlation coefficient after adjusting for all other variables. The threshold p value after the Bonferroni correction for multiple comparisons is 0.005 (10 comparisons).
| variable | univariate correlation | partial correlation |
|---|---|---|
| population size | 0.53** | 0.43* |
| population density | 0.32 | −0.09 |
| all-cause infant mortality rate | 0.14 | 0.17 |
| urbanization index | −0.31 | 0.05 |
| latitude | −0.46* | −0.27 |
*p < 0.05.
**p < 0.005.
4. Discussion
To the best of our knowledge, this is the first study to systematically estimate the reproduction number of influenza across a range of latitudes encompassing equatorial, tropical and subtropical areas, using spatially and temporally refined P&I mortality data spanning multiple influenza seasons. Our estimates of the reproduction number at the level of states in Brazil were very close to 1.0 (mean R = 1.03) and systematically lower than estimates for the USA and France, considering a short serial interval as in recent studies (Cauchemez et al. 2004; Ferguson et al. 2005; Chowell et al. 2008a). We also found significantly less between-year variation in the reproduction number of influenza for Brazil (range 1.04–1.09), when compared with those for the USA and France (range 1.0–1.39). Further, our results suggest a weak gradient in the reproduction number with population size, where R increases from lowest to highly populous states of Brazil.
Our findings suggest that the reproduction number of influenza in Brazil is low and remains just slightly above the epidemic threshold of 1.0, in contrast to temperate locales like the USA or France, where epidemics can be associated with higher transmission potential. Although most of the Australian population lives in temperate zones, a growing fraction of the population lives in the northern and tropical part of this country and could be contributing to the observed similarities in influenza transmission patterns between the two countries. It is also worth noting that the low population size of Australia relative to the USA and France, led to greater uncertainty in reproduction number estimates, as reflected by wide confidence intervals (see electronic supplementary material, table S2, also Chowell et al. (2008b)). Finally, there may be other unidentified factors beyond climate or population size that produce similar patterns in the transmission dynamics of seasonal influenza in Australia and Brazil.
We did not find a temporal trend in the average reproduction number in Brazil over the study period, despite the launch of an influenza immunization programme in 1999 targeting seniors more than 65 years. Although annual influenza vaccination campaigns may be contributing to declining mortality rates in Brazilian seniors, they are unlikely to generate significant reductions in transmission rates, as seniors respond weakly to influenza vaccines (Simonsen et al. 2005) and transmission is primarily driven by school age children and young adults (Weycker et al. 2005; Wallinga et al. 2006).
Our modelling approach retains the minimal complexity necessary to estimate the reproduction number of influenza, similar to recent efforts (Ferguson et al. 2003; Mills et al. 2004; Ferguson et al. 2005; Viboud et al. 2006c; Wallinga & Lipsitch 2007; Andreasen et al. 2008; Chowell et al. 2008a). In this work, we conducted a range of sensitivity analyses to test the robustness of our estimates to various modelling assumptions and found very consistent results across analyses. In particular, because mixing patterns and risk of death is known to vary with age, we undertook simulation studies using an age-structured model of disease transmission for Brazil, including compartments for severe disease outcomes. Results from this more complex model showed that R estimates were robust to age variation in disease patterns, consistent with previous sensitivity analyses (Mills et al. 2004). We also explored the sensitivity of R estimates to assumptions about the time lag between infection and death through two independent algorithms and found that results were robust to such assumptions (see electronic supplementary material; SEIR model versus estimation of the exponential growth rate directly from the mortality curve). Likewise, uncertainties in case fatality rate values did not substantially affect our estimates.
We relied on time series of P&I mortality from Brazilian states to estimate R. These data were derived from the centralized collection of death certificates organized by the Ministry of Health, which are not prone to geographical differences in reporting rates. To ensure the validity of our results, we also estimated the reproduction number using influenza-specific mortality (ICD9 code = 487), a highly conservative and specific mortality outcome, and confirmed our low estimates for Brazil (R = 1.01 (95% CI 0.96, 1.06)). A similar sensitivity analysis was conducted with epidemiological data from temperate countries in a previous study (Chowell et al. 2008b). Furthermore, we tried a less conservative approach to estimate R, by using the P&I excess mortality curves and starting earlier in the epidemic (in the week with 0 excess deaths). In general, we found slightly higher estimates of R by this method, by 0.1 or less, consistently in all four countries studied. We note that the statistical differences between Brazil, France and the USA disappeared with this less conservative approach. Since R estimates relied on total P&I deaths, of which only a fraction is attributable to influenza virus activity, we also checked that estimates were independent of baseline seasonal P&I death rates across all states (Spearman ρ < 0.39; p > 0.06 for partial correlation adjusting for population size and latitude, using R estimates derived from both the SEIR and growth rate methods). Together with the reassuring results of our sensitivity analysis using excess P&I deaths above a seasonal baseline in four large states, we conclude that our estimates are robust to variation in baseline non-influenza mortality rates across states.
Even though we did not explicitly consider in our model a realistic contact network with high levels of clustering related to age, geography or other factors, theoretical network-based models can be used to gauge the impact of heterogeneities on the transmission dynamics of infectious diseases. Simulations have shown that a high level of population clustering has negligible impact on the epidemic growth rate when transmissibility is low (Miller 2009), which is the case for seasonal influenza.
Overall, while our modelling approach is subject to assumptions and limitations, we believe that these caveats should not bias our main results highlighting geographical differences in the transmission potential of influenza, given that the same methodology was applied to similar data from different locations.
Estimates of the reproduction number for seasonal influenza epidemics combine the intrinsic transmissibility of the influenza virus with population immunity levels, and it is difficult to disentangle the role of each factor. However, the most parsimonious explanation for the observed differences in reproduction number estimates between countries (and seasons) is perhaps differences in population immunity. In the context of the global circulation of influenza virus, our results suggest that population immunity could be high on average in Brazil, and perhaps in other tropical settings. If population immunity were higher on average in the tropics, it would be consistent with a sink-source model of influenza virus evolution originating from the tropics. We cannot rule out that other factors could affect the transmission dynamics of influenza in the studied populations, including differential seasonal forcing across Brazilian states and countries.
It is worth noting a parallel relationship between low reproduction number and high genetic diversity across the three (sub)types of influenza virus, whereby the A/H3N2 subtype is associated with epidemics with higher transmission potential, when compared with the other two subtypes (Chowell et al. 2008b), and displays the least amount of viral diversity at any time point (Ferguson et al. 2003). Indeed, co-circulation of antigenically distinct lineages of influenza A/H3N2 viruses is rare, as illustrated by the highly pruned phylogenetic tree of the haemagglutinin A/H3 surface antigen. By contrast, co-circulation of antigenic variants is more common for the less transmissible A/H1N1 and B viruses (Ferguson et al. 2003). If confirmed, latitudinal differences in influenza transmission and genetic diversity could partially drive a source-sink model of influenza evolution focused on the tropics (Rambaut et al. 2008; Russell 2008). This mechanism could also explain why in the case of Brazil, epidemics seem to first arise in the equatorial zone and gradually spread southward to more temperate locales (Alonso et al. 2007), which is consistent with an equatorial origin of new virus variants.
We found weak associations between reproduction number estimates at the state level and population sizes. For comparison, there was no association between R and population size during the 1918–1919 influenza pandemic in England and Wales (Chowell et al. 2008a). An analysis of the 1918–1919 pandemic in 45 US cities found that R was not correlated with population size and weakly correlated with population density in the fall wave (Mills et al. 2004). Overall, the reproduction number of seasonal and pandemic influenza appears nearly invariant across a wide range of population sizes and densities at the level of states or cities, despite differences in social connectivity patterns in urban and rural settings.
Several limitations of our study are worth noting. First, we assumed that P&I mortality was a reliable proxy for influenza incidence, as demonstrated in several previous studies (Ferguson et al. 2003; Mills et al. 2004; Viboud et al. 2004, 2006c). Second, we assumed that reporting and coding of death certificates were homogeneous across Brazil and over the study period (1996–2006). One may expect that more rural locations may have poorer reporting or coding of deaths. However, a previous study reported higher influenza-related death rates in rural areas of England and Wales with lower population sizes, which argues against such bias (Chowell et al. 2008a), and is consistent with mortality patterns in the Brazilian dataset (not shown). Finally, we relied on administrative regions of Brazil, which are not necessarily the most meaningful spatial units for disease dynamics (Chowell et al. 2008a). Despite these limitations, our influenza study set in Brazil suggests low levels of transmission potential across Brazilian states, which are statistically associated with population size.
We cannot entirely rule out that reproduction number estimates are prone to increased measurement error in Brazil, particularly in the northernmost equatorial states, because of weaker seasonality in virus activity and lower population size. However, this study is a first step towards quantifying influenza transmission dynamics in tropical settings and providing a better understanding of the global ecology and emergence of new virus variants. Further research will shed light on whether the patterns described here are specific to Brazil or can be generalized to the tropics—a key area for persistence and evolution of influenza virus. Future influenza research in the tropics should focus on collection of morbidity rather than mortality data, quantification of attack rates and evolutionary patterns, and prioritize studies in East–SouthEast Asia, where transmission patterns could differ from those in Brazil (Russell 2008).
References
- Alonso W. J., Viboud C., Simonsen L., Hirano E. W., Daufenbach L. Z., Miller M. A.2007Seasonality of influenza in Brazil: a traveling wave from the Amazon to the subtropics. Am. J. Epidemiol. 165, 1434–1442 (doi:10.1093/aje/kwm012) [DOI] [PubMed] [Google Scholar]
- Anderson R. M., May R. M.1991Infectious diseases of humans New York, NY: Oxford University Press [Google Scholar]
- Andreasen V., Viboud C., Simonsen L.2008Epidemiologic characterization of the summer wave of the 1918 influenza pandemic in Copenhagen: implications for pandemic control strategies. J. Infect. Dis. 97, 270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazilian Institute of Geography and Statistics. 2005 Interactive Maps. Maps of the territorial divisions. See http://mapas.ibge.gov.br/divisao/viewer.htm. (accessed on 12 February 2008) [Google Scholar]
- Cauchemez S., Carrat F., Viboud C., Valleron A. J., Boëlle P. Y.2004A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat. Med. 23, 3469–3487 (doi:10.1002/sim.1912) [DOI] [PubMed] [Google Scholar]
- Chowell G., Nishiura H.2008Quantifying the transmission potential of pandemic influenza. Phys. Life Rev. 5, 50–77 (doi:10.1016/j.plrev.2007.12.001) [Google Scholar]
- Chowell G., Bettencourt L. M., Johnson N., Alonso W. J., Viboud C.2008aThe 1918–1919 influenza pandemic in England and Wales: spatial patterns in transmissibility and mortality impact. Proc. Biol. Sci. 275, 501–509 (doi:10.1098/rspb.2007.1477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G., Miller M. A., Viboud C.2008bSeasonal influenza in the United States, France, and Australia: transmission and prospects for control. Epidemiol. Infect. 136, 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G., Viboud C., Wang X., Bertozzi S., Miller M.2009Adaptive vaccination strategies to mitigate pandemic influenza: Mexico as a case study. PLoS ONE 4, e8164 (doi:10.1371/journal.pone.0008164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann O., Heesterbeek J.2000Mathematical epidmeiology of infectious diseases: model building, analysis and interpretation Chichester, UK: Wiley [Google Scholar]
- Earn D. J., Rohani P., Bolker B. M., Grenfell B. T.2000A simple model for complex dynamical transitions in epidemics. Science 287, 667–670 (doi:10.1126/science.287.5453.667) [DOI] [PubMed] [Google Scholar]
- Ferguson N. M., Galvani A. P., Bush R. M.2003Ecological and immunological determinants of influenza evolution. Nature 422, 428–433 (doi:10.1038/nature01509) [DOI] [PubMed] [Google Scholar]
- Ferguson N. M., Cummings D. A. T., Cauchemez S., Fraser C., Riley S., Meeyai A., Iamsirithaworn S., Burke D. S.2005Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437, 209–214 (doi:10.1038/nature04017) [DOI] [PubMed] [Google Scholar]
- Ferrari M. J., Bjørnstad O. N., Dobson A. P.2005Estimation and inference of R0 of an infectious pathogen by a removal method. Math. Biosci. 198, 14–26 (doi:10.1016/j.mbs.2005.08.002) [DOI] [PubMed] [Google Scholar]
- Laguna-Torres V. A., et al. 2009Influenza-like illness sentinel surveillance in Peru. PLoS ONE 4, e6118 (doi:10.1371/journal.pone.0006118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., et al. 2003Transmission dynamics and control of severe acute respiratory syndrome. Science 300, 1966–1970 (doi:10.1126/science.1086616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mello W. A., de Paiva T. M., Ishida M. A., Benega M. A., Dos Santos M. C., Viboud C., Miller M. A., Alonso W. J.2009The dilemma of influenza vaccine recommendations when applied to the tropics: the Brazilian case examined under alternative scenarios. PLoS ONE 4, e5095 (doi:10.1371/journal.pone.0005095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C.2009Spread of infectious disease through clustered populations. J. R. Soc. Interface 6, 1121–1134 (doi:10.1098/rsif.2008.0524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C. E., Robins J. M., Lipsitch M.2004Transmissibility of 1918 pandemic influenza. Nature 432, 904–906 (doi:10.1038/nature03063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura F. E., Perdigão A. C., Siqueira M. M.2009Seasonality of influenza in the tropics: a distinct pattern in northeastern Brazil. Am. J. Trop. Med. Hyg. 81, 180–183 [PubMed] [Google Scholar]
- Nelson M. I., et al. 2006Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog. 2, e125 (doi:10.1371/journal.ppat.0020125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Pybus O. G., Nelson M. I., Viboud C., Taubenberger J. K., Holmes E. C.2008The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (doi:10.1038/nature06945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. A., et al. 2008The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 (doi:10.1126/science.1154137) [DOI] [PubMed] [Google Scholar]
- Serfling R.1963Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Rep. 78, 494–506 [PMC free article] [PubMed] [Google Scholar]
- Simonsen L.1999The global impact of influenza on morbidity and mortality. Vaccine 17(Suppl. 1), S3–S10 [DOI] [PubMed] [Google Scholar]
- Simonsen L., Reichert T. A., Viboud C., Blackwelder W. C., Taylor R. J., Miller M. A.2005Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch. Intern. Med. 165, 265–272 (doi:10.1001/archinte.165.3.265) [DOI] [PubMed] [Google Scholar]
- Viboud C., Boelle P. Y., Pakdaman K., Carrat F., Valleron A. J., Flahault A.2004Influenza epidemics in the United States, France, and Australia, 1972–1997. Emerg. Infect. Dis. 10, 32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Alonso W. J., Simonsen L.2006aInfluenza in tropical regions. PLoS Med. 3, e89 (doi:10.1371/journal.pmed.0030089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Bjornstad O. N., Smith D. L., Simonsen L., Miller M. A., Grenfell B. T.2006bSynchrony, waves, and spatial hierarchies in the spread of influenza. Science 312, 447–451 (doi:10.1126/science.1125237) [DOI] [PubMed] [Google Scholar]
- Viboud C., Tam T., Fleming D., Handel A., Miller M. A., Simonsen L.2006cTransmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine 24, 6701–6707 (doi:10.1016/j.vaccine.2006.05.067) [DOI] [PubMed] [Google Scholar]
- Vynnycky E., Trindall A., Mangtani P.2007Estimates of the reproduction numbers of Spanish influenza using morbidity data. Int. J. Epidemiol. 36, 881–889 (doi:10.1093/ije/dym071) [DOI] [PubMed] [Google Scholar]
- Wallinga J., Lipsitch M.2007How generation intervals shape the relationship between growth rates and reproductive numbers. Proc. Biol. Sci. 274, 599–604 (doi:10.1098/rspb.2006.3754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J., Teunis P., Kretzschmar M.2006Using data on social contacts to estimate age-specific transmission parameters for respiratory-spread infectious agents. Am. J. Epidemiol. 164, 936–944 (doi:10.1093/aje/kwj317) [DOI] [PubMed] [Google Scholar]
- Weycker D., et al. 2005Population-wide benefits of routine vaccination of children against influenza. Vaccine 23, 1284–1293 (doi:10.1016/j.vaccine.2004.08.044) [DOI] [PubMed] [Google Scholar]
- WHO 2003Influenza—Fact sheet 211. See http://www.who.int/mediacentre/factsheets/fs211/en/index.html (accessed in April 2007) [Google Scholar]
- Wong C. M., Chan K. P., Hedley A. J., Peiris J. S.2004Influenza-associated mortality in Hong Kong. Clin. Infect. Dis. 39, 1611–1617 (doi:10.1086/425315) [DOI] [PubMed] [Google Scholar]
- Wong C. M., Yang L., Chan K. P., Leung G. M., Chan K. H., Guan Y., Lam T. H., Hedley A. J., Peiris J. S.2006Influenza-associated hospitalization in a subtropical city. PLoS Med. 3, e121 (doi:10.1371/journal.pmed.0030121) [DOI] [PMC free article] [PubMed] [Google Scholar]