Abstract
Biological invasions may expose populations to strong selection for local adaptation along geographical gradients in climate. However, evolution during contemporary timescales can be constrained by low standing genetic variation and genetic correlations among life-history traits. We examined limits to local adaptation associated with northern migration of the invasive wetland plant purple loosestrife (Lythrum salicaria) using a selection model incorporating a trade-off between flowering time and size at reproduction, and common garden experiments of populations sampled along a latitudinal transect of approximately 1200 km in eastern North America. A strong trade-off between flowering time and size at reproduction caused early-flowering plants to be smaller with reduced seed production in northern populations. Northward spread was associated with a decline in genetic variance within populations and an increase in genetic skew for flowering time and size, with limited genetic variation for small, early-flowering genotypes. These patterns were predicted by our selection model of local adaptation to shorter growing seasons and were not consistent with expectations from non-adaptive processes. Reduced fecundity may limit population growth and rates of spread in northern populations. Identifying genetic constraints on key life-history traits can provide novel insights into invasion dynamics and the causes of range limits in introduced species.
Keywords: genetic constraints, invasive species, latitudinal clines, Lythrum salicaria, rapid evolution
1. Introduction
Genetic constraints have long been recognized as limiting factors in adaptive evolution (Darwin 1859; Fisher 1930; Lande 1979; Maynard Smith et al. 1985), but their influence on responses to selection in natural populations is not well understood (Antonovics 1976; Blows & Hoffman 2005). Biological invasions provide opportunities for investigating limits to contemporary evolution (Lankau et al. 2009), because rapid range expansion can expose populations to strong selection for local adaptation (Maron et al. 2004; Xu et al. 2010), particularly along geographical gradients in climate and seasonality where responses to selection should be predictable. Theory suggests that the rate of evolutionary response to selection can directly affect the speed of biological invasion (García-Ramos & Rodríguez 2002), but will be limited by the availability of standing genetic variation for ecologically relevant traits.
Fitness trade-offs may also constrain adaptive differentiation across ecological gradients, even when genetic variation for life-history traits exists (e.g. Etterson & Shaw 2001; Griffith & Watson 2006). A fitness trade-off between two traits occurs if natural selection cannot simultaneously improve both traits (Lande 1979; Lande & Arnold 1983). Constraints on adaptation imposed by fitness trade-offs represent a lack of genetic variation for particular combinations of traits, and this can inhibit populations from adapting to novel environments (Blows & Hoffman 2005). Genetic constraints could potentially influence species distributions by preventing local adaptation at range margins, but this has rarely been investigated empirically (reviewed in Eckert et al. 2008) and to our knowledge no study has done so for an invasive species.
Variation in growing season is often associated with changes in the timing of reproduction in a wide variety of plants and animals (Bradshaw & Holzapfel 2001; Parmesan 2006). Indeed, genetically based latitudinal or altitudinal clines in traits related to growth, phenology and life history appear to be common in introduced plants (reviewed in Colautti et al. 2009; see also Alexander et al. 2009). Invasive species that spread from equatorial to polar latitudes experience a decrease in season length and this is likely to result in selection for a shorter time to reproductive maturity. However, life-history theory predicts that early maturation will come at a cost of reduced size (Roff 1992; Stearns 1992). In plants, this results from a positive genetic correlation between flowering time and size at maturity. This trade-off constrains an evolutionary response to selection on flowering time, because reduced size often translates into lower reproductive output. A gradual change in selection for earlier flowering time in invasive plant populations should therefore result in predictable changes to seed production along latitudinal gradients. Lowered fecundity in geographically marginal populations may limit population growth and reduce rates of spread.
Gradual changes in the strength of stabilizing selection along a latitudinal gradient in climate should alter standing genetic variation for ecologically relevant traits. To investigate this process, we implemented a Lande–Arnold model of selection with a genetic constraint on flowering time and size at maturity, to quantify how changes in selection would affect the mean, variance and skew of the frequency distribution of flowering time and size. Using field surveys and common garden studies of populations of the invasive plant Lythrum salicaria (Lythraceae, purple loosestrife) sampled along a latitudinal gradient in climate in eastern North America we then evaluated the model by investigating patterns of quantitative genetic variation. Specifically, we addressed the following questions: (i) Is there evidence that a response to selection during colonization is constrained by a trade-off between flowering time and size at maturity? (ii) Are the patterns of standing genetic variation, including the mean, variance and skew for these traits, those predicted by our selection model?
2. Material and methods
(a). Model of nonlinear selection on flowering time and size at reproduction
We assume quantitative genetic control of flowering time in our model (see Koornneef et al. 1998; Buckler et al. 2009) and used the Lande–Arnold (1983) fitness model to investigate natural selection on flowering time and size at reproduction along a latitudinal gradient of season length (for a detailed model description, see ‘nonlinear selection model’ in the electronic supplementary material). We assumed that (i) selection acts directly on flowering time, but (ii) plants that are larger at flowering have more resources available for reproduction, and that (iii) fitness effects for these two traits are additive, which has been verified in studies of selection in L. salicaria (O'Neil 1997). We assumed a trade-off between flowering time and size at maturity and modelled it as a positive, linear genetic correlation, such that plants allocate limited resources to either growth or reproduction. Variance in these correlated traits can be conveniently represented as the first principal component of the correlation between them (PC1).
If we assume that plants can begin flowering at any time, the phenotypic distribution of PC1 can be modelled as a Gaussian distribution with mean equal to the optimum phenotype (θpc) and variance inversely proportional to the strength of stabilizing selection. However, two factors make this assumption unlikely. First, there is probably a developmental threshold for flowering time, which is the size at which a plant has enough resources to flower and mature seed (Lacey 1986), resulting in a right skew to the distribution of PC1 as small, early-flowering phenotypes are absent. Second, some invasive populations may lack sufficient genetic variation for small, early-flowering phenotypes. If northward migration is rapid, then selection should increase the frequency of alleles for early flowering but a response to selection may be limited by the rate of new mutations, also resulting in a right skew to the distribution of phenotypes. Both scenarios can be modelled as a threshold value of PC1; values below this threshold represent an area of phenotypic space (i.e. early reproduction at small size) that cannot be occupied due to the lack of appropriate genetic variation.
(b). Field survey and common garden experiments
To evaluate the assumptions and predictions of our model we examined variation in life-history traits among L. salicaria populations in the field and in three common gardens. Natural populations (n = 25) were sampled along a 1200 km latitudinal gradient from southern Maryland, USA (38.75° N) to northern Ontario, Canada (42.12° N). Detailed methods and results of the field survey are provided in Montague et al. (2008). Here, we focus on field measurements of the 22 populations in which seed production was estimated (hereafter ‘field survey’), and a subset of these populations that were compared in three separate common garden experiments using open-pollinated seed families collected during the field survey. We used controlled crosses to verify genetic correlations and evaluate the possibility of maternal effects (see ‘controlled crosses’ in the electronic supplementary material). We found no evidence that non-genetic maternal effects, or any form of trans-generational plasticity, has contributed to the pattern of geographic variation in trait mean, variance and skew presented below. In each experiment, populations were chosen to best represent the latitudinal clines in growth and phenology reported in Montague et al. (2008).
In all population comparisons, we measured flowering time as the number of days to first flower, using the Julian Date standardized to the earliest-flowering plant in each experiment. We measured size at maturity as the height of the non-flowering portion of the primary stem (from the soil surface) at the start of flowering (hereafter ‘vegetative size’). To allow a test of a null model (i.e. no latitudinal clines), we added a third life-history trait, ‘inflorescence length’, measured at the time of first flower from the top of the vegetative portion of the primary stem to the inflorescence apex. Inflorescence length correlates with flower production in the greenhouse (ln-transformed flower number r = +0.719, n = 75, p < 0.001) and is under strong directional selection in natural populations (O'Neil 1997). We chose inflorescence length because there was no reason to expect a change in the strength of selection on this trait along a latitudinal gradient. To ensure that our measurements of vegetative size and inflorescence length were good predictors of above-ground growth, we measured total dry biomass of vegetative and reproductive structures and the number of secondary stems at the end of the growing season in all experiments.
(i). Field survey
We used data from 22 populations to determine if vegetative size correlated with other measures of plant size and fitness: total number of secondary stems, dry biomass of vegetative growth, total fruit production and total dry biomass. These data were presented as population means in Montague et al. (2008) and were analysed for phenotypic correlations here. We used GLM and MIXED procedures of SAS 9.1 (SAS Institute Inc., Cary, NC) to fit an overall regression of vegetative size against each correlate of fitness, ignoring population of origin (i.e. ‘all phenotypes’ in table 1). We then performed a mixed model analysis, with population as a random effect, to estimate the average within-population correlations between vegetative size and each of the fitness measurements (i.e. ‘within populations’ in table 1). Population × vegetative size interactions were not significant for any trait (all p > 0.16) except total fruits (p = 0.046), and are not reported in the table because we were interested in the average effect of vegetative size, rather than differences among populations in the slope of the relations.
Table 1.
Significance of phenotypic correlations between vegetative size at first flower and four fitness components measured in a field survey of 22 Lythrum salicaria populations. All measurements were made in natural populations, analysed in a single model (all phenotypes), or as the average within-population correlation from an analysis of covariance with population as a random factor (within populations). Transformations were chosen to best meet assumptions of bivariate normality; for this reason, the predictor variable (vegetative size) was not transformed.
| trait | n | all phenotypes |
within populations | |
|---|---|---|---|---|
| F | r | F | ||
| number of secondary stems | 590 | 509.50*** | +0.681 | 313.43*** |
| ln vegetative dry biomass | 590 | 387.95*** | +0.630 | 4.52* |
| ln total fruits | 590 | 137.54*** | +0.435 | 6.83** |
| ln total dry biomass | 590 | 337.71*** | +0.604 | 46.23*** |
*p < 0.05.
** p < 0.01.
*** p < 0.001.
(ii). Greenhouse experiment
We tested the assumption of a fitness trade-off between time to first flower and vegetative size at flowering, and measured phenotypic and genetic variation for both traits, using 20 populations in a greenhouse experiment at the University of Toronto (hereafter ‘greenhouse experiment’). In June 2004, we randomly transplanted seedlings from 17 families from each population individually into 10 cm pots arranged in a randomized block design with two individuals per family per population per block (four blocks total). Pots contained Pro-Mix peat saturated with standing water. Plants were grown under long days (16 h), at 20–25°C, with supplementary light provided by high-pressure sodium lamps to keep day length constant. We also recorded date of germination to examine its relation with growth and reproduction. Of 2720 plants transplanted, 20 died and 144 did not flower within the duration of the experiment (170 days) and were excluded from analysis.
We used principal components analysis (PCA) on standardized covariances of population means for three correlated traits (days to flower, vegetative size and inflorescence length) to derive two uncorrelated PC axes (see Blows et al. 2004; Hine et al. 2009). Together, PC1 and PC2 accounted for 97.5 per cent of phenotypic variance and covariance among population means (PC1 = 85.0% and PC2 = 12.5%). Factor loadings (i.e. PC coefficients) indicate that PC1 correlates positively with vegetative size (+0.770) and days to first flower (+0.633), but not inflorescence length (+0.081), while PC2 correlates positively with inflorescence length (+0.826), negatively but weakly with days to flower (−0.475) and positively but weakly with vegetative size (+0.304). Because PC1 and PC2 are orthogonal they can be treated as independent quantitative traits allowing for a test of both model (PC1) and null (PC2) predictions.
To measure standing genetic variation, we used factor loadings to calculate principal component scores for each individual and estimated mean, variance and skew for each PC ‘trait’ in each population. By ‘skew’ we mean the coefficient of skew rather than the third central moment because the latter results in a correlation between the second (i.e. variance) and third moments that is not biologically meaningful (Sokal & Rohlf 1995). Before analysis, 12 outliers were removed owing to their undue influence on estimates of population means. Our model assumes a positive genetic correlation between days to flower and vegetative size. To test this we used best linear unbiased predictors (BLUPs) of family means, calculated from the statistical mixed model:
where T is the trait (days to flower or vegetative size) and Bl the fixed effect of experimental block. Population (p) i and seed family (F) j (nested within population i) are random factors and ε is the error term of individual k. The linear correlation between BLUPs of days to flower and vegetative size represents the average genetic correlation within populations and is not affected by differences among population means. We also tested whether populations differed in the amount of standing genetic variation using a similar model that estimated among-family variances for each population, rather than a single ‘average’ variance component. Variance components were estimated by restricted maximum likelihood (ReML) and tested by hierarchical likelihood ratio test (LRT).
To investigate model predictions concerning latitudinal gradients in genetic variance and skew, we present phenotypic rather than genetic estimates. However, the latter are presented as electronic supplementary material (see ‘genetic estimates of variance and skew’ and figure S1) and exhibit similar clines. We present phenotypic estimates because there is an eightfold reduction in sample size when estimating genetic variance (i.e. 17 families instead of 136 individuals per population) and because open-pollinated seeds are maternal half-sibs, so up to 3/4 of the total genetic variation within a population is distributed as error variance among individuals within a seed family (Lynch & Walsh 1998).
We used least-squares quadratic regressions in GLM to test for latitudinal trends in the mean, variance and skew of PC1. We tested the overall significance of the quadratic function because our selection model predicts nonlinear relations with latitude that may have weak curvature; we did not test whether the quadratic term fits the data better than a linear function because either is possible in our model. Positive correlations between the mean and central moments (i.e. variance, skew and kurtosis) of morphological and life-history traits are a well-known phenomenon (Sokal & Rohlf 1995; Lynch & Walsh 1998), but do not appear to have influenced our results (see ‘scaling effects’ in the electronic supplementary material).
(iii). Mid-latitude field experiment
We compared our greenhouse results to growth and phenology under field conditions across multiple growing seasons (2005–2007) in a randomized common garden experiment with two blocks in a tilled marsh at the Koffler Scientific Reserve (hereafter KSR) in central Ontario (44.03° N, 79.54° W). This site lies at the geographical centre of the latitudinal gradient and has an intermediate season length. In May 2005, we transplanted into each block two individuals from 15 to 20 families from each of 13 populations spanning the latitudinal range. Analysis of life-history traits in this experiment included 934 individuals from 237 seed families. Of these, 121 plants died by the second year and 778 and 775 flowered in the second and third years, respectively.
The PCA of phenotypic traits obtained from the KSR experiment was similar to that obtained from the greenhouse experiment but combined into a single principal component two years of data (i.e. days to flower, vegetative size and inflorescence length × 2 years = 6 measurements) rather than one. Therefore, the PCA of the KSR experiment accounts for both correlations among traits and among years.
(iv). Southern latitude field experiment
The last prediction of our model was a decrease in fecundity in northern populations relative to southern populations because of differences in size at maturity. We tested this by comparing the fecundity of northern and southern populations in a common garden experiment at the University of Virginia's Blandy Experimental Farm (hereafter BEF). This site is located near the southern edge of the latitudinal gradient (39.06° N, 78.06° W) and plants at this site experience a growing season almost twice as long as our northern populations (see Montague et al. 2008). We used a single block of 77 individuals from 40 families, representing two northern and two southern populations. Seedlings were planted in May 2007 and allowed to establish for two growing seasons. In November 2008, we measured fecundity as the dry biomass of reproductive structures. To determine if there were differences in fecundity between northern and southern populations we analysed inflorescence biomass using maximum likelihood in the GENMOD procedure (Poisson distribution with log-link). We tested the significance of region (i.e. north versus south) in a model that included population (nested within region) and seed family (nested within population) using a hierarchical likelihood ratio test.
3. Results
(a). Selection model
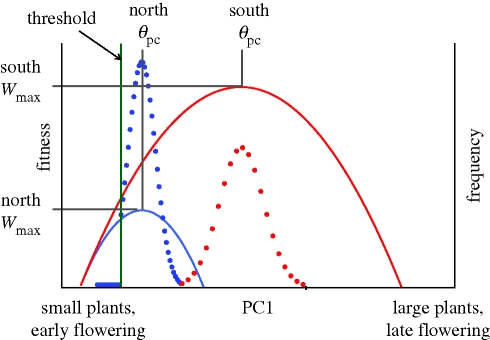
Our model indicated that an increase in the strength of selection on flowering time and size in a northern versus a southern population, resulted in a correlated reduction in the mean and variance and an increase in the skew of PC1 in the northern population (figure 1). The maximum fitness value (i.e. the fecundity of an individual with phenotype θpc) was lower for the northern population, as a result of a trade-off between flowering time and size at maturity.
Figure 1.
Model of nonlinear selection on PC1—the first principal component of time to flowering and size at reproduction. Solid curves contrast the change in absolute fitness (e.g. per-capita seed production) between hypothetical populations at a northern location (blue) and southern location (red). Stronger selection against late-flowering genotypes in the north, or stronger selection for larger genotypes in the south (or both) are sufficient to shift the optimum trait value (θpc) to earlier flowering at a smaller size in the northern population. Absolute fitness (W) is, on average, higher in the southern population due to a longer growing season, allowing for larger plants that flower later. The green line represents a developmental or genetic constraint that precludes phenotypes from attaining smaller PC1 values. The dotted curves contrast the trait frequency distributions that result from the difference in the fitness function between northern and southern populations. Wmax is the maximum fitness, measured as reproductive output of an individual with PC1 = θpc, the optimum phenotype.
We extended the model in figure 1 to represent a latitudinal gradient in climate by considering a gradual change in the strength of selection on flowering time and size at maturity (see ‘nonlinear selection model’ in the electronic supplementary material for details). This resulted in four predicted clines in PC1 with increasing latitude: (i) a decrease in the mean, (ii) a decrease in the variance, (iii) an increase in the skew, and (iv) a decrease in fecundity. These latitudinal clines were robust to the specific parameter values chosen for figure 1.
(b). Testing model assumptions
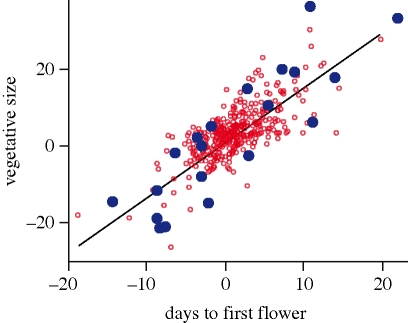
Our model assumed a trade-off between flowering time and size at maturity. Consistent with this, days to flower and vegetative size correlated positively among populations in the greenhouse (figure 2) and among F2 families from controlled crosses (n = 34, r = +0.638, p < 0.001; see ‘controlled crosses’ section in the electronic supplementary material for details). Population means fell along the major axis of covariance within populations, consistent with a constraint on population divergence (figure 2). Population mean days to flower also correlated strongly with vegetative size in the KSR field experiment (second year: r = +0.921, p < 0.001; third year: r = +0.939, p < 0.001), demonstrating that the genetic constraint on population divergence is manifested under both greenhouse (figure 2) and field conditions (table 2). The strong latitudinal clines in days to flower and vegetative size were not the result of differences in germination date, given the weak correlations between germination date and vegetative size (Spearman's ρ = −0.173, p < 0.001) or days to flower (Spearman's ρ = −0.0211, p = 0.294).
Figure 2.
Correlation between days to first flower and vegetative size at first flower in 20 populations of Lythrum salicaria sampled from across the latitudinal range in eastern North America and grown under greenhouse conditions. Blue filled circles are best linear unbiased predictors (BLUPs) of population means and red open circles are BLUPs of 340 seed families, corrected for differences in population means. The line denotes the principal component (PC1) of flowering time and size at first flower. BLUPs are standardized means (i.e. mean = 0) from linear mixed models with vegetative size at first flower or days to flower as the response variable, and population and seed family (nested within population) as random factors (r = +0.740, p < 0.001).
Table 2.
Latitudinal clines in population means, variances and coefficients of skew for the first principal component of time to flowering and vegetative size at flowering in Lythrum salicaria grown under uniform conditions in separate experiments in the greenhouse and the KSR field experiment. Values are Pearson correlation coefficients between latitude (°N) and the population mean, variance and skew of the first principal component (PC1) of days to flower, vegetative size, and inflorescence length; measured in populations from eastern North America, grown under both greenhouse and field conditions. A large value for PC1 represents a plant that flowers later at a larger size. npop is the number of populations in each test.
| Pearson product-moment correlations |
||||
|---|---|---|---|---|
| experiment | npop | mean | variance | skew |
| genetic estimates | ||||
| University of Toronto (greenhouse) | 20 | −0.766*** | −0.602** | +0.491* |
| phenotypic estimates | ||||
| University of Toronto (greenhouse) | 20 | −0.768*** | −0.743*** | +0.741*** |
| Koffler Scientific Reserve (field) | 13 | −0.864** | −0.820*** | +0.610* |
*p < 0.05.
**p < 0.01.
*** p < 0.001.
We also assumed that vegetative size correlates with resource acquisition and therefore fitness under field conditions. Measurements of 22 populations in the field survey supported this assumption; vegetative size (untransformed) correlated linearly and positively with ln-transformed total dry biomass, vegetative dry biomass, total number of secondary stems and total fruit production (table 1).
(c). Latitudinal clines revealed by common garden comparisons
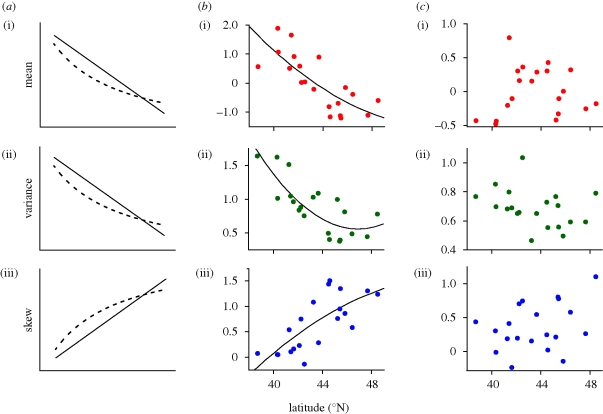
The selection model predicted latitudinal clines in the mean, variance and skew of standing genetic variation for PC1 (figure 3a). Data from both the greenhouse (figure 3b) and the KSR field experiment (table 2) supported these predictions: the mean and variance decreased while the skew increased with increasing latitude. In addition, variance components estimated by ReML for PC1 in the greenhouse experiment were highly significant, both among populations (LRT: d.f. = 1, χ2=369.6, p < 0.001), and among seed families (LRT: d.f. = 1, χ2 = 251.0, p < 0.001). In addition, populations differed significantly in the amount of standing genetic variation (LRT: d.f. = 19, χ2 = 56.4, p < 0.001). Like PC1, PC2 also varied significantly among populations (LRT: d.f. = 1, χ2 = 171.5, p < 0.001) and among families within populations (LRT: d.f. = 1, χ2 = 10.3, p < 0.01). However, in contrast to PC1, the mean, variance and skew of PC2 did not correlate with latitude (figure 3c), and populations did not differ significantly in the amount of standing genetic variation (LRT: d.f. = 19, χ2 = 15.6, p = 0.684).
Figure 3.
(a) Predicted latitudinal trends in the (i) mean, (ii) variance and (iii) coefficient of skew of phenotype frequency distributions for a hypothetical principal component describing flowering time and size at maturity. If the strength of selection on flowering time increases gradually with latitude, the predicted regression is approximately linear (solid line), while a steeper change yields a nonlinear relation (dashed line). (b) Observed mean and central moments and least-squares quadratic regression lines for 20 Lythrum salicaria populations grown under greenhouse conditions for the observed first principal component (PC1) describing days to first flower and vegetative size. Results for PC1 are consistent with predictions of the selection model. (i) r2 =0.600, p < 0.001; (ii) r2 =0.653, p < 0.001 and (iii) r2 =0.557, p < 0.001. (c) Observed mean and central moments and least-squares quadratic regression lines of the second principal component (PC2). Genetic (linear mixed model) estimates are in table 1. (i) r2 =0.274, p < 0.066; (ii) r2 =0.157, p < 0.234 and (iii) r2 =0.157, p < 0.235.
As predicted by our model (figure 1), average fruit production per plant in the field declined with increasing latitude (ln-transformed: n = 23, r = −0.654, p < 0.001) with a striking 23-fold difference between northern (Timmins: average 57 fruits per plant) and southern (Easton: average 1325 fruits per plant) populations. In the BEF field experiment, reproductive biomass was approximately 2.5-fold higher for southern (mean = 34.8 g ± 1.2 s.e.m.) than northern (mean = 17.0 g ± 1.2 s.e.m.) populations (d.f. = 1, χ2 = 20.4, p < 0.001).
4. Discussion
We investigated the effect of genetic constraints on life-history evolution using a nonlinear selection model and common garden studies of invasive populations of L. salicaria sampled along a latitudinal gradient in eastern North American. Measurements of quantitative genetic variation and the analysis of genetic correlations allowed us to validate our model assumptions and test its main predictions. We found evidence for a strong genetic correlation between time to first flower and vegetative size (figure 2). Given this finding and the assumption of a minimum size threshold for flowering, our model predicted that selection against late-flowering genotypes towards the northern range limit would result in latitudinal clines in the mean, variance and skew of the first principal component of flowering time and size at maturity. These predictions were supported in experiments conducted in both the greenhouse (figure 3) and field (table 2).
An important prediction of our selection model is that when the evolution of earlier flowering is constrained by a positive genetic correlation between days to flower and size, stronger stabilizing selection on flowering time will reduce fecundity. Data from a common-garden experiment at BEF strongly supported this prediction. Moreover, size at flowering correlated strongly with predictors of fecundity under field conditions (table 1), a result consistent with our model assumption of selection for larger size. The dramatic reduction in fecundity in northern populations identified in our field survey may play an important role in limiting the northward migration of L. salicaria, at least over the short term.
A variety of deterministic and stochastic evolutionary processes can give rise to latitudinal clines in life-history traits (Endler 1977; Gockel et al. 2001; Vasemagi 2006; Keller et al. 2009). We therefore evaluate alternative hypotheses that could potentially explain the latitudinal clines revealed by our study (and see the electronic supplementary material). We argue that the patterns predicted by our selection model, and supported by our common garden studies, are unlikely to be explained by any of these alternative processes. We conclude our discussion by considering the implications of our results for the study of range limits in introduced species.
(a). Alternative hypotheses for clines
Invasion history, stochastic forces (e.g. genetic drift and founder effects) and migration–selection balance (MSB—Kirkpatrick & Barton 1997; Lenormand 2002) could also influence the mean, variance and skew of phenotypic traits measured in our greenhouse and field experiments. MSB predicts a quadratic relationship between variance and latitude, with less variance in marginal populations due to smaller population size and spatial isolation causing reduced gene flow (Lenormand 2002). Instead, we found a linear relation for PC1 with highest variances at the southern edge of the range (figure 3). Changes in variance and skew due to migration alone should also scale with each other, resulting in a relatively constant coefficient of skew that is contrary to the latitudinal cline we found (figure 3).
Multiple introductions during invasion should increase genetic variation whereas bottlenecks and founder effects reduce it, but this should be true of all traits, resulting in a correlation in variance of PC1 and PC2 across populations. Genetic drift and MSB would also cause a correlation between PC1 and PC2 because genetic variance for all phenotypic traits should generally be lower in small populations where rare alleles are more easily lost. Contrary to these predictions the variance of PC1 and PC2 were not correlated among populations (Pearson correlation: r = +0.320, p = 0.169). Estimated population sizes in the field survey ranged between 300 and 1 000 000 among the 20 populations used in the greenhouse experiment, but variance and coefficient of skew of PC1 correlated more strongly with population means (partial correlation: ρVar = +0.776, ρSkew = −0.847) than population sizes, (ln abundance ρVar = −0.275, ρSkew = +0.120). In contrast, PC2 correlated only weakly with population means (ρVar = −0.078, ρSkew = +0.007) and somewhat more strongly with population size (ρVar = −0.252, ρSkew = −0.295), although in the opposite direction to what would be caused by genetic drift. Thus, population size is a poor correlate of variance and coefficient of skew, contrary to the prediction of genetic drift and MSB.
(b). Implications for species distributions and range limits
Genetic constraints on local adaptation have often been invoked to explain geographical range limits, but there are few empirical tests of this hypothesis (Bradshaw 1991; Davis & Shaw 2001; Blows & Hoffman 2005). Constraints on traits that determine the timing of reproduction may be particularly important in preventing range expansion to environments with shorter growing seasons typical of high latitudes and altitudes (Griffith & Watson 2006), and also in response to anthropogenic changes in climate and seasonality (Bradshaw & Holzapfel 2001; Franks et al. 2007). Studies on a wide range of plant and animal species have compared levels of diversity at neutral maker loci between geographically central versus peripheral populations and many have found evidence of a decline in genetic variation towards range margins (reviewed in Eckert et al. 2008). However, for most studies it is unclear whether these patterns are also associated with a reduction in genetic variation for ecologically relevant traits. Moreover, the few studies that have investigated the genetic basis of range limits in plants (e.g. Etterson & Shaw 2001; Griffith & Watson 2006) have sampled relatively few populations, limiting opportunities to detect strong geographical patterns.
Our study is novel because we examined quantitative genetic variation in a large sample of populations spanning much of the geographical range of L. salicaria in eastern North America. The close match between model predictions and empirical results strongly supports the hypothesis that fitness trade-offs between life-history traits compromise local adaptation in northern populations of L. salicaria. Indeed, per capita fecundity declined sharply with increasing latitude (see also Montague et al. 2008), and this may set the northern range limit by reducing persistence following disturbance (Tilman 1997), and limiting further spread to unoccupied sites (García-Ramos & Rodríguez 2002; Holt et al. 2005). Further studies of invasive populations of L. salicaria that have spread to higher latitudes in other parts of its introduced range would be valuable to corroborate our findings. Native populations of L. salicaria in northern Europe exhibit similar latitudinal clines in flowering time and size at reproduction (Olsson & Ågren 2002) but it is not known whether genetic constraints influence range limits. Our results highlight the importance of estimating the genetic variance and covariance of ecologically relevant traits for evaluating evolutionary constraints, natural selection and non-adaptive processes on range expansion in invasive species.
Acknowledgements
We thank J. Montague and S. Yakimowski for field assistance; S. Yakimowski, M. Blows and K. Samis for comments on the manuscript; the Ontario Government and the University of Toronto for scholarship support to RIC; the Canada Research Chair programme and an Ontario Premier's Discovery Award for funding to S.C.H.B.; and the Natural Sciences and Engineering Research Council of Canada (NSERC) for a graduate scholarship to R.I.C. and Discovery Grants to C.G.E. and S.C.H.B.
References
- Alexander J. M., Edwards P. J., Poll M., Parks C. G., Dietz H.2009Establishment of parallel altitudinal clines in traits of native and introduced forbs. Ecology 90, 612–622 (doi:10.1890/08-0453.1) [DOI] [PubMed] [Google Scholar]
- Antonovics J.1976The nature of limits to natural selection. Ann. Mo. Bot. Gard. 63, 224–247 (doi:10.2307/2395303) [Google Scholar]
- Blows M. W., Hoffman A. A.2005A reassessment of genetic limits to evolutionary change. Ecology 86, 1371–1384 (doi:10.1890/04-1209) [Google Scholar]
- Blows M. W., Chenoweth S., Hine E.2004Orientation of the genetic variance–covariance matrix and the fitness surface for multiple male sexually-selected traits. Am. Nat. 163, 329–340 (doi:10.1086/381941) [DOI] [PubMed] [Google Scholar]
- Bradshaw A. D.1991Genostasis and the limits to evolution. Phil. Trans. R. Soc. Lond. B 333, 289–305 (doi:10.1098/rstb.1991.0079) [DOI] [PubMed] [Google Scholar]
- Bradshaw W. E., Holzapfel C. M.2001Genetic shift in photoperiodic response correlated with global warming. Proc. Natl Acad. Sci. USA 98, 14 509–14 511 (doi:10.1073/pnas.241391498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S., et al. 2009The genetic architecture of maize flowering time. Science 325, 714–718 (doi:10.1126/science.1174276) [DOI] [PubMed] [Google Scholar]
- Colautti R. I., Maron J. L., Barrett S. C. H.2009Common garden comparisons of native and introduced plant populations: latitudinal clines can obscure evolutionary inferences. Evol. Appl. 2, 187–199 (doi:10.1111/j.1752-4571.2008.00053.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C.1859On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- Davis M. B., Shaw R. G.2001Range shifts and adaptive responses to Quaternary climate change. Science 292, 673–679 (doi:10.1126/science.292.5517.673) [DOI] [PubMed] [Google Scholar]
- Eckert C. G., Samis K. E., Lougheed S. C.2008Genetic variation across species' geographical ranges: the central–marginal hypothesis and beyond. Mol. Ecol. 17, 1170–1188 (doi:10.1111/j.1365-294X.2007.03659.x) [DOI] [PubMed] [Google Scholar]
- Endler J. A.1977Geographic variation, speciation and clines Princeton, NJ: Princeton University Press; [PubMed] [Google Scholar]
- Etterson J. R., Shaw R. G.2001Constraint on adaptive evolution in response to global warming. Science 294, 151–154 (doi:10.1126/science.1063656) [DOI] [PubMed] [Google Scholar]
- Fisher R. A.1930The genetical theory of natural selection Oxford, UK: Oxford University Press [Google Scholar]
- Franks S. J., Sim S., Weis A. E.2007Rapid evolution of flowering time by an annual plant in response to climate fluctuation. Proc. Natl Acad. Sci. USA 104, 1278–1282 (doi:10.1073/pnas.0608379104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ramos G., Rodríguez D.2002Evolutionary speed of species invasions. Evolution 56, 661–668 (doi:10.1554/0014-3820(2002)056[0661:ESOSI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Gockel J., Kennington W. J., Hoffman A., Goldstein D. B., Partridge L.2001Nonclinality of molecular variation indicates selection in maintaining a morphological cline of Drosophila melanogaster. Genetics 158, 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith T. M., Watson M. A.2006Is evolution necessary for range expansion? Manipulating reproductive timing of a weedy annual transplanted beyond its range. Am. Nat. 167, 153–164 (doi:10.1086/498945) [DOI] [PubMed] [Google Scholar]
- Hine E., Chenoweth S. F., Rundle H. D., Blows M. W.2009Characterizing the evolution of genetic variance using genetic covariance tensors. Phil. Trans. R. Soc. B 364, 1567–1578 (doi:10.1098/rstb.2008.0313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R. D., Barfield M., Gomulkiewicz R.2005Theories of niche conservatism and evolution. Could exotic species be potential tests? In Species invasions: insights into ecology, evolution, and biogeography (eds Sax D. F., Stachowicz J. J., Gaines S. D.), pp. 259–290 Sunderland, MA: Sinauer [Google Scholar]
- Keller S. R., Sowell D. R., Neiman M., Wolfe L. M., Taylor D. R.2009Adaptation and colonization history affect the evolution of clines in two introduced species. New Phytol. 183, 678–690 (doi:10.1111/j.1469-8137.2009.02892.x) [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N. H.1997Evolution of a species' range. Am. Nat. 150, 1–23 (doi:10.1086/286054) [DOI] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Peeters A. J. M., Soppe W.1998Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370 (doi:10.1146/annurev.arplant.49.1.345) [DOI] [PubMed] [Google Scholar]
- Lacey E. P.1986Onset of reproduction in plants: size- versus age-dependency. Trends Ecol. Evol. 1, 72–75 (doi:10.1016/0169-5347(86)90021-2) [DOI] [PubMed] [Google Scholar]
- Lande R.1979Quantitative genetic analysis of multivariate evolution, applied to brain:body size allometry. Evolution 33, 402–416 (doi:10.2307/2407630) [DOI] [PubMed] [Google Scholar]
- Lande R., Arnold S. J.1983The measurement of selection on correlated characters. Evolution 37, 1210–1226 (doi:10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- Lankau R. A., Nuzzo V., Spyreas G., Davis A. S.2009Evolutionary limits ameliorate the negative impact of an invasive plant. Proc. Natl Acad. Sci. USA 106, 15 362–15 367 (doi:10.1073/pnas.0905446106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T.2002Gene flow and the limits of natural selection. Trends Ecol. Evol. 17, 183–189 (doi:10.1016/S0169-5347(02)02497-7) [Google Scholar]
- Lynch M., Walsh B.1998Genetics and analysis of quantitative traits Sunderland, MA: Sinauer [Google Scholar]
- Maron J. L., Vilà M., Bommarco R., Elmendorf S., Beardsley P.2004Rapid evolution of an invasive plant. Ecol. Monogr. 74, 261–280 (doi:10.1890/03-4027) [Google Scholar]
- Maynard Smith J., Burian R., Kauffman S., Alberch P., Campbell J., Goodwin B., Lande R., Raup D., Wolfpert L.1985Developmental constraints on evolution. Q. Rev. Biol. 60, 265–287 [Google Scholar]
- Montague J. L., Barrett S. C. H., Eckert C. G.2008Re-establishment of clinal variation in flowering time among introduced populations of purple loosestrife (Lythrum salicaria, Lythraceae). J. Evol. Biol. 21, 234–245 (doi:10.1111/j.1420-9101.2007.01456.x) [DOI] [PubMed] [Google Scholar]
- O'Neil P.1997Natural selection on genetically correlated phenological characters in Lythrum salicaria L. (Lythraceae). Evolution 51, 267–274 (doi:10.2307/2410980) [DOI] [PubMed] [Google Scholar]
- Olsson K., Ågren J.2002Latitudinal population differentiation in phenology, life history and flower morphology in the perennial herb Lythrum salicaria. J. Evol. Biol. 15, 983–996 (doi:10.1046/j.1420-9101.2002.00457.x) [Google Scholar]
- Parmesan C.2006Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669 (doi:10.1146/annurev.ecolsys.37.091305.110100) [Google Scholar]
- Roff D. A.1992The evolution of life histories: theory and analysis New York, NY: Chapman and Hall [Google Scholar]
- Sokal R. R., Rohlf F. J.1995Biometry, 3rd edn.New York, NY: Freeman [Google Scholar]
- Stearns S. C.1992The evolution of life histories Oxford, UK: Oxford University Press [Google Scholar]
- Tilman D.1997Community invasibility, recruitment limitation, and grassland biodiversity. Ecology 78, 81–92 [Google Scholar]
- Vasemagi A.2006The adaptive hypothesis of clinal variation revisited: single-locus clines as a result of spatially restricted gene flow. Genetics 173, 2411–2414 (doi:10.1534/genetics.106.059881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. Y., Julien M. H., Fatemi M., Girod C., Van Klinken R. D., Gross C. L., Novak S. J.2010Phenotypic divergence during the invasion of Phyla canescens in Australia and France: evidence for selection-driven evolution. Ecol. Lett. 13, 32–44 (doi:10.1111/j.1461-0248.2009.01395.x) [DOI] [PubMed] [Google Scholar]