Abstract
We examine the application of the maximum entropy production principle for describing ecosystem biogeochemistry. Since ecosystems can be functionally stable despite changes in species composition, we use a distributed metabolic network for describing biogeochemistry, which synthesizes generic biological structures that catalyse reaction pathways, but is otherwise organism independent. Allocation of biological structure and regulation of biogeochemical reactions is determined via solution of an optimal control problem in which entropy production is maximized. However, because synthesis of biological structures cannot occur if entropy production is maximized instantaneously, we propose that information stored within the metagenome allows biological systems to maximize entropy production when averaged over time. This differs from abiotic systems that maximize entropy production at a point in space–time, which we refer to as the steepest descent pathway. It is the spatio-temporal averaging that allows biological systems to outperform abiotic processes in entropy production, at least in many situations. A simulation of a methanotrophic system is used to demonstrate the approach. We conclude with a brief discussion on the implications of viewing ecosystems as self-organizing molecular machines that function to maximize entropy production at the ecosystem level of organization.
Keywords: free energy, living systems, evolution, dissipative systems, self-organization, information
1. Introduction
Many of the chemical transformations that occur on the Earth are catalysed by biological systems, and in most cases these biological machines enhance reaction rates by many orders of magnitude over abiotic processes (Falkowski et al. 2008). Now that human society can match or exceed many of the natural biogeochemical reaction rates, it is critical that we understand how living systems organize to process energy and mass on local, regional and global scales. Given the importance of biogeochemical cycles on climate and ecological processes, it is surprising that we lack any agreed upon theoretical basis for biogeochemistry. Current biogeochemical models largely take an organismal perspective, where emphasis is placed on understanding and modelling the growth and interaction of the various organisms that comprise a given ecosystem. The biogeochemistry is then a secondary consequence of the complex food web (for aquatic systems) or physiological (for terrestrial systems) dynamics. Unfortunately, other than conservation of mass, and implicit conservation of energy, no fundamental rules or theories govern the equations used to model the organisms and their interactions. While the theory of evolution by natural selection provides a mechanism for self-organization of complex biological structures, the theory is indeterminate with regards to the emergent properties biological systems follow, if any (Peters 1976; Murray 2001). Consequently, our biogeochemical models are purely based on imperial observations, where each system is modelled largely as a case study that depends on the organisms present in the particular system under study. As natural or human-induced environmental conditions change, the population of the dominate organisms in an ecosystem shift in response, but we have little understanding or predictive capabilities regarding such system reorganization. Consequently, our biogeochemical models are brittle and of dubious value when predictions are extrapolated beyond observations used to calibrate them. Yet, ironically, extrapolation is often the primary rationale for quantitative modelling.
Dating back to at least Lotka (1922), who proposed that ecosystems organize towards a state of maximum power, there has been a significant amount of work in theoretical ecology that focuses on understanding the governing principles that organize ecosystems. Biological systems are far from equilibrium and contain low entropy ordered structures that are maintained by external energy dissipation (Schrödinger 1944; Morowitz 1968). Accordingly, there has been a great deal of interest in non-equilibrium thermodynamic (or thermodynamically inspired) applications to living systems involving: power (Lotka 1922; Odum & Pinkerton 1955), biomass to maintenance (Margalef 1968), minimum entropy (Prigogine & Nicolis 1971), exergy (Mejer & Jorgensen 1979), ascendancy (Ulanowicz 1986), emergy (Odum 1988), energy dissipation (Schneider & Kay 1994), respiration (Washida 1995; Choi et al. 1999), thermodynamic efficiency (Nielsen & Ulanowicz 2000), constructal theory (Bejan 2007) as well as others (Ulanowicz & Platt 1985; Weber et al. 1988; Toussaint & Schneider 1998; Jorgensen et al. 2000). However, the theories have not gained wide acceptance and are seldom employed to model biogeochemistry. While many of the theories have a similar basis (Jorgensen 1994; Fath et al. 2001), they differ sufficiently to cause confusion.
As evident by this special journal issue, there is a renewed interest in the principle of maximum entropy production (MEP) for non-equilibrium systems owing to both theoretical and observational research. The original application of MEP dates to Paltridge (1975), who demonstrated that if global heat transport between the tropics and the poles operates at MEP, one can accurately predict meridional heat flux, latitudinal temperatures and fractional cloud cover; a similar analysis has also been applied to describe the climatology of Mars and Titan (Lorenz et al. 2001). Others speculated that ecosystems follow MEP (Ulanowicz & Hannon 1987; Swenson 1989), but it garnered little attention without theoretical and experimental support. Dewar's (2003, 2005) analysis now provides a theoretical basis for the MEP principle, as he derives a provisional proof for MEP for non-equilibrium steady-state systems with sufficient degrees of freedom. The general implication of the MEP principle is that systems will organize, within biophysiochemical constraints, so as to maximize the rate of entropy production. MEP is an appealing extension to classic, equilibrium thermodynamics which dictates that systems will move to a state of maximum entropy at equilibrium. In essence, MEP indicates that systems will attempt to achieve equilibrium via the fastest allowable pathways. Equally desirable, the MEP principle does not distinguish between biotic and abiotic systems, so it can be applied generally.
In this manuscript we will examine the application of the MEP principle to describe biogeochemistry. To facilitate this, we will view ecosystems from the perspective of a distributed metabolic network, which will replace the traditional organismal focus. With the new perspective, we will develop, via example with a methanotrophic community, a mathematical framework to describe ecosystem biogeochemistry. We conclude with a discussion on the advantages and limits of the MEP approach.
2. Theoretical framework
(a). Entropy context
In this manuscript, entropy will refer to Clausius' (1850) original definition; the energy dissipated when converting internal energy into work. Under constant temperature and pressure, the energy that can be used for work (or to drive other reactions) is Gibb's free energy, which accounts for losses owing to entropy. Most importantly, if an exergonic chemical reaction occurs but all the energy liberated is dissipated as heat to the surroundings (i.e. energy is not stored), then all the free energy is converted to entropy at the given temperature. Unlike energy, free energy is not conserved. In fact, free energy is more accurately a measure of entropy that can be produced at a given temperature (Lineweaver & Egan 2008). For processes of interest here, entropy production will be equated to chemical or radiative energy dissipation at the temperature of the surroundings, and all discussions refer to internal system entropy production (Meysman & Bruers 2007).
(b). Macrostates, microstates and ecosystems
The MEP principle requires a system to have many degrees of freedom; that is, many different microstate configurations (technically, micropaths) that produce the same entropy-producing macrostate (Dewar 2003). In this paper, we extend the concept of microstates to biological systems. Here, a microstate will refer to the organisms present and their connectivity in an ecosystem. Based on MEP principle, there should exist many different species configurations and trophic connections that give rise to the same entropy-producing macrostate. Over appropriate time scales, species composition and their trophic relationships dramatically change, as has been observed in methanogenic (Fernandez et al. 1999), nitrifying (Graham et al. 2007) and planktonic communities (Beninca et al. 2008) that exhibit dynamics primarily at the species level but maintain similar biogeochemistry (i.e. functional stability). Indeed, multiplicity of biological microstates appears consistent with neutral theory (Pueyo et al. 2007), provided the different configurations are functionally complementary. Because of the large number of microbial species and their high abundances (Gans et al. 2005; Sogin et al. 2006), the microstate analogy is most applicable to microbial systems, which will be our primary focus.
(c). Biophysiochemical constraints
In any implementation of MEP principle, system constraints are critical in determining the relevant solution and the rate of entropy production at MEP (Crooks 2007). As an example, consider a burning mixture of methane and air. If no energy is stored, then all the free energy of combustion is ultimately dissipated as entropy at the temperature of the surroundings. In this case, the process is operating at the MEP state, where the magnitude of entropy production is constrained by the kinetic theory for gasses, which we could calculate a priori. We consider this type of abiotic process operating in a steepest descent mode, where the rate of entropy production is maximized at any instance in time and space, subject to kinetic constraints.
Outside the flammability limits, CH4 and O2 still react in a steepest descent mode, but the reaction proceeds extremely slowly. The reaction rate can be considerably accelerated by the addition of a catalyst, but for self-organizing systems, the catalyst must be created from CH4 and O2 and/or materials contained within the system. The reaction rate, and entropy produced, depends on the effectiveness and quantity of the catalyst. An insufficient amount of catalyst limits the reaction rate, but too much catalyst wastes resources. If the catalyst contains internal energy, which it is likely to, then over-synthesis of catalyst is not consistent with MEP, because entropy could have been produced instead of catalyst. The catalyst's effectiveness (i.e. reaction rate per unit mass) depends on its molecular structure, which in turn depends on the resources available to construct it. Given the available elements, what is the most effective catalyst that can be constructed? Obviously, this is an extremely difficult question to answer given the tremendous number of degrees of freedom. We know the upper bound is the complete removal of the reaction's activation energy, but how much the activation energy can be decreased is constrained by the elemental resources used to build the catalyst. Unlike the relatively simple kinetics of combustion, we cannot determine a priori what the maximum reaction rate is nor what the MEP rate could be. Experiments are necessary.
(d). MEP from individuals or ecosystems?
Of course, the catalysts of interest here are enzymes, but we include the organisms that synthesize enzymes as part of the catalyst. Continuing our example, we know from experiments that methanotrophs catalyse methane oxidation as follows:
| 2.1 |
where methanotrophs, M, both catalyse the reaction and are produced by it if their growth efficiency, ɛM, is greater than 0. The question we address here is how might evolution by natural selection lead to a system that maximizes entropy production? The competitive exclusion principle (Armstrong & McGehee 1980) tells us that the organism with the fastest growth will dominate at the exclusion of all others. To achieve fast growth, an organism must balance efficiency against speed. In equation (2.1) above, if ɛM is close to unity (high efficiency), the reaction will proceed slowly owing to thermodynamic constraints (Gnaiger 1990; Pfeiffer & Bonhoeffer 2002, but also see §3a below) and methanotroph productivity will be low. As ɛM approaches 0 (low efficiency), the reaction can proceed rapidly, but once again with low methanotroph productivity owing to low efficiency. Between 0 and 1 there is an optimum ɛM that maximizes methanotroph productivity, which is selected for by evolution (Ibarra et al. 2002), but this value does not maximize entropy production because the methane that contributed to methanotroph biomass could have been oxidized. We conclude that growth of individual organisms as selected for by evolution does not follow the MEP principle. But ecosystems are not composed of a single species, as grazers, G, are always present and they consume prey, such as methanotrophs, as given by,
| 2.2 |
which is also an autocatalytic reaction that produces exponential growth. Of course, grazers of the methanotroph-grazers are often present, as well as detritivores. Extending this logic naturally leads to food webs observed in nature. A net result of all the predation and recycling is that the total biomass in the system is constantly turned over. It is possible to have all organisms growing near their maximum rates without any biomass accumulation, which can lead to an MEP state. Hence, it is necessary to have a closed food web of prey, predators and detritivores in order to achieve MEP. The predator–prey interactions also ensure that biological structures are continuously and dynamically reallocated to those reactions that can assure MEP under changing conditions.
(e). MEP, information and time scales
Interestingly, the effectiveness of methanotrophs to catalyse equation (2.1) depends on information stored in their genome, which specifies how to construct astoundingly complex organic structures from resources available in the environment. Over evolutionary time, enzymes associated with methanotrophy would presumably improve (Adami 2002), so that less protein is needed by modern methanotrophs than ancient methanotrophs to attain the same reaction rate. Hence, the amount of entropy produced for a given amount of protein changes with evolution, as well as the introduction of new pathways all together. But information embedded in the metagenome makes it difficult to predict MEP in biological systems from first principles as can be done with physical systems and demonstrated by Paltridge (1975). For this paper, we will rely on simple metabolic networks as constraints. Genomic information also permits biological systems to integrate entropy production over time and circumvent the steepest descent pathway of abiotic systems.
As discussed above, MEP rate for a flammable mixture of CH4 and air is dictated by gas kinetics. While combustion is the MEP solution, it tends to destroy ordered structures, so has only short persistence. If a perturbation extinguishes the flame, the CH4 and air mixture will accumulate until a serendipitous spark is reintroduced. Consequently, there can be periods of massive entropy production followed by long periods of no entropy production, and combustion never occurs if the mixture falls outside its flammability limits. If a catalyst is introduced, such as methanotrophs, then entropy can be continuously produced over substantial transients. Even though the instantaneous entropy production will be lower with methanotrophs, the average rate of entropy production can exceed that of the sporadic steepest descent route.
If MEP follows only steepest descent pathways, then not only should CH4 be oxidized, but all biomass as well, as this would produce the greatest instantaneous entropy production. However, if time-averaged entropy production is maximized (Moroz 2008), then allocating some CH4 to methanotrophs and grazers increases entropy production over the integration interval. Unlike physical systems, biological systems have the capability to predict the future, where the information to do so is contained within the system metagenome. For instance, deciduous forests store some resources during the growing season that allows them to maintain dormancy over the winter or dry period, which requires expectations of future conditions. Microbes also exhibit temporal strategies such as spore formation and luxury-uptake mechanisms (Berman-Frank & Dubinsky 1999; Khoshmanesh et al. 2002). When considering biological systems, strategies are not instantaneous, but are integrated over time based on a prediction of the future that has been selected for by evolution. By avoiding the steepest descent pathway, information can allow a system to produce entropy even when confronted with perturbations. Of course, perturbations of sufficient magnitude will disrupt biological systems as well. Nevertheless, we postulate as follows: the difference between abiotic and biotic processes is that the former always follows a pathway of steepest descent, while the latter follows a pathway dictated by information, which leads to greater entropy production when averaged over time. Of course, the two pathways are always competing, such as occurs when fire consumes a forest. Pathways of averaged entropy production may be flanked by pathways of steepest descent.
(f). Distributed metabolic networks
Because of the multitude and redundancy of microstates, understanding flow of energy and mass through ecosystems at the organismal level is problematic (Graham et al. 2007; Beninca et al. 2008). Instead of focusing at the organismal level, we will pursue a functional representation using a metabolic network abstraction. When considering metabolic capabilities of microbial systems, we often find that metabolic function is distributed among all three domains of life: Bacteria, Archaea and Eukaryote. Since the species that comprise metabolic networks can undergo substantial substitution with only minor impact on functional characteristics (Fernandez et al. 2000; Wittebolle et al. 2008), we will view microbial systems as metabolic networks that can be distributed in space and time, but resemble multicellular organisms (Wilson & Sober 1989; Shapiro 1998).
3. Mathematical framework
As defined above, our basic model framework uses the metabolic network perspective that is applied at the ecosystem level. The formulation is similar to how single cells control metabolism by regulating the synthesis and degradation of enzymes associated with pathways necessary for growth. We assume that communities organize such that their combined metabolic expression maximizes entropy production. A methanotrophic microbial community in a batch reactor that is sparged with methane and air will serve as an example to demonstrate the approach.
(a). Optimized metabolic ecosystem network model
In our approach the microbial food web is replaced by a metabolic network that synthesizes generic biological structure, 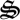 , which consists largely of enzymatic protein, but also represents other macromolecules expressed by microbial communities for growth and form. The biological structure can be allocated to any reaction in the network and serves as the reaction's catalyst. The metabolic network also orchestrates energy and mass acquisition necessary to construct biological structure itself, where the MEP principle governs how biological structure is allocated to any metabolic reaction. The mathematical framework is general and can be extended to any microbial system by expanding the distributed metabolic network based on knowledge from classic microbiology and metagenomics. One can also examine how the introduction or evolution of new metabolic functions alters resource allocation and system dynamics, as well as examine dynamics when metabolic functions are removed.
, which consists largely of enzymatic protein, but also represents other macromolecules expressed by microbial communities for growth and form. The biological structure can be allocated to any reaction in the network and serves as the reaction's catalyst. The metabolic network also orchestrates energy and mass acquisition necessary to construct biological structure itself, where the MEP principle governs how biological structure is allocated to any metabolic reaction. The mathematical framework is general and can be extended to any microbial system by expanding the distributed metabolic network based on knowledge from classic microbiology and metagenomics. One can also examine how the introduction or evolution of new metabolic functions alters resource allocation and system dynamics, as well as examine dynamics when metabolic functions are removed.
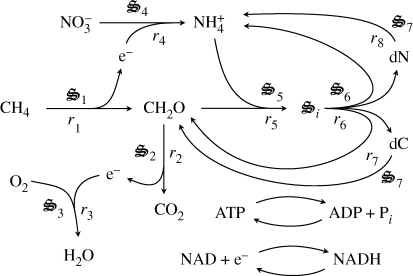
For our aerobic methanotrophic metabolic network example (figure 1), eight half reactions account for methane oxidation (r1, r2, r3), including sugar biosynthesis (r1), nitrate reduction (r4) for biological structure synthesis from 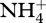 and sugars (r5), and biological structure and detritus degradation (r6, r7, r8) (electronic supplementary material, table S1). Each of the eight reactions has associated biological structure (
and sugars (r5), and biological structure and detritus degradation (r6, r7, r8) (electronic supplementary material, table S1). Each of the eight reactions has associated biological structure (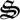 1,…,7), except for reactions 7 and 8, which are both catalysed by the same structure,
1,…,7), except for reactions 7 and 8, which are both catalysed by the same structure, 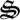 7, to minimize model degrees of freedom. The use of half reactions (electronic supplementary material, table S1) increases the flexibility of the network to use the available electron acceptors or donors, but to ensure electron conservation, all half reactions are coupled to an electron shuttle reaction as follows:
7, to minimize model degrees of freedom. The use of half reactions (electronic supplementary material, table S1) increases the flexibility of the network to use the available electron acceptors or donors, but to ensure electron conservation, all half reactions are coupled to an electron shuttle reaction as follows:
| 3.1 |
where ɛi is the number of electron pairs produced by reaction i. Gibbs free energies are calculated for each reaction (electronic supplementary material, table S1) after coupling to equation (3.1). We use the approach of Alberty (2003, 2006) to calculate the standard Gibbs free energy of reaction, which accounts for proton dissociation equilibria between chemical species (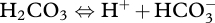 , etc.) at a specified pH and temperature. The overall free energy of reaction, denoted as
, etc.) at a specified pH and temperature. The overall free energy of reaction, denoted as 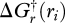 , accounts for the concentration of reactants and products (c(t)), where ionic strength, IS, is used to estimate activity coefficients (Alberty 2003). For the methanotrophic network simulation, we use the following conditions: pH 7, IS 1.92 µM, at 20°C. Free energy, enthalpy and entropy of formation for biological structure is based on Battley (1999, 2003) for bacteria; however, this is not critical as the free energy of living organisms is similar in value to the substrates they are constructed from.
, accounts for the concentration of reactants and products (c(t)), where ionic strength, IS, is used to estimate activity coefficients (Alberty 2003). For the methanotrophic network simulation, we use the following conditions: pH 7, IS 1.92 µM, at 20°C. Free energy, enthalpy and entropy of formation for biological structure is based on Battley (1999, 2003) for bacteria; however, this is not critical as the free energy of living organisms is similar in value to the substrates they are constructed from.
Figure 1.
Simple distributed metabolic network for a methanotrophic-based food web. Seven biological structures, 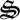 i, catalyse the reactions, including degradation of detrital N (dN) and C (dC).
i, catalyse the reactions, including degradation of detrital N (dN) and C (dC).
Of course, 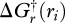 represents the available or needed energy only if the reaction is run reversibly, which is clearly not the case for biological systems. To account for inefficient energy transfer or energy production, all reactions are coupled to an energy source (or sink) reaction in the form of ATP hydrolysis (or synthesis) of the form,
represents the available or needed energy only if the reaction is run reversibly, which is clearly not the case for biological systems. To account for inefficient energy transfer or energy production, all reactions are coupled to an energy source (or sink) reaction in the form of ATP hydrolysis (or synthesis) of the form,
| 3.2 |
Even though we know most ATP reaction couplings from biochemistry, cells have the ability to dissipate ATP (Russell & Cook 1995), and different organisms in a community can have different ATP couplings for the same pathways (Helling 2002). Consequently, we treat energy coupling, ηi(t), as a control variable to be determined by optimization. The combined whole reaction i consists of equations (3.1) and (3.2) and half reaction i (figure 1; electronic supplementary material, table S1). The Gibbs free energy of the combined reaction i, 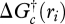 , is then given by
, is then given by
| 3.3 |
where 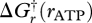 is the Gibbs free energy of equation (3.2). By altering the magnitude and sign of ηi(t), an endergonic reaction can be driven forward, and by inefficient coupling of equation (3.2) with exergonic reactions, energy can be extracted at less than 100 per cent efficiency, allowing the reaction to proceed at higher rates. This is the standard tradeoff between power and efficiency; operating reactions at high throughput necessitates low efficiency, while attaining high efficiency limits reaction rate, which all biotic and abiotic systems must contend with (Gnaiger 1990; Pfeiffer & Bonhoeffer 2002).
is the Gibbs free energy of equation (3.2). By altering the magnitude and sign of ηi(t), an endergonic reaction can be driven forward, and by inefficient coupling of equation (3.2) with exergonic reactions, energy can be extracted at less than 100 per cent efficiency, allowing the reaction to proceed at higher rates. This is the standard tradeoff between power and efficiency; operating reactions at high throughput necessitates low efficiency, while attaining high efficiency limits reaction rate, which all biotic and abiotic systems must contend with (Gnaiger 1990; Pfeiffer & Bonhoeffer 2002).
Metabolic network reaction rates are given by:
| 3.4 |
where νi is an experimentally determined rate constant per unit of biological structure allocated, 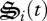 is biological structure allocated to reaction i, and
is biological structure allocated to reaction i, and 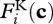 and
and 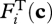 are kinetic and thermodynamic forces, respectively. The kinetic force is given by the general form:
are kinetic and thermodynamic forces, respectively. The kinetic force is given by the general form:
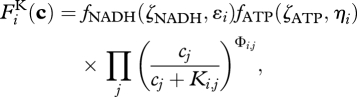 |
3.5 |
where cj is the concentration of substrate j, and the matrix element, Фi,j, equals 1 if reaction i consumes substrate j; otherwise Фi,j is zero (electronic supplementary material, table S1). To ensure electron and energy conservation, we assume that biological structure has a fixed amount of NAD + NADH and ADP + ATP storage per unit of total biological structure, 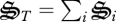 , as specified by the constants
, as specified by the constants  and
and 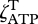 , respectively
, respectively 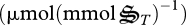 . Consequently, the amounts of NAD and NADH as well as ATP and ADP per biological structure are constrained by,
. Consequently, the amounts of NAD and NADH as well as ATP and ADP per biological structure are constrained by,
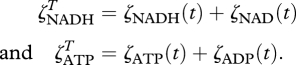 |
3.6 |
We define the functions fNADH and fATP based on 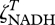 and
and 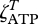 as,
as,
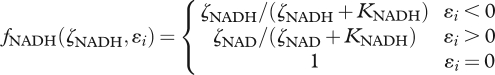 |
3.7 |
and
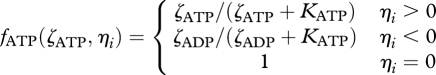 |
3.8 |
Functions (3.7) and (3.8) account for the redox and energy state of the system, analogous to cellular metabolism.
Chemical reaction rates are often limited by kinetics, so the thermodynamic force is often ignored because it is usually close to unity. However, as the reaction approaches equilibrium, 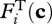 in equation (3.4) approaches 0 and constrains the net reaction rate, no matter how favourable the reaction kinetics may be. It can be shown (Boudart 1976; Jin & Bethke 2003) that the thermodynamic force is related to the Gibbs free energy of the reaction,
in equation (3.4) approaches 0 and constrains the net reaction rate, no matter how favourable the reaction kinetics may be. It can be shown (Boudart 1976; Jin & Bethke 2003) that the thermodynamic force is related to the Gibbs free energy of the reaction, 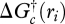 , as follows:
, as follows:
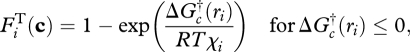 |
3.9 |
where R is the gas constant, T is temperature (K) and χi is the average stoichiometric number for net reaction i (Boudart 1976; Vuddagiri et al. 2000). By adjusting ηi(t), 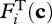 can be driven from 0 to 1 via relationship to equation (3.3). Because of the complexity of community metabolic networks, χi is treated as a tunable parameter for each reaction.
can be driven from 0 to 1 via relationship to equation (3.3). Because of the complexity of community metabolic networks, χi is treated as a tunable parameter for each reaction.
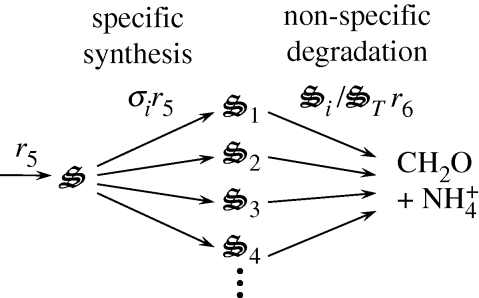
Reactions r5(t) and r6(t) represent the synthesis and degradation of all biological structures, respectively. However, which of the seven biological structures to produce or degrade at time t has not been specified. Consequently, we introduce an additional set of control variables, σi(t), that specify the partitioning of biological structure synthesis, which is analogous to transcription plus translation (figure 2). While we could introduce another set of control variables to determine which biological structures to degrade, we make the assumption that degradation of biological structure is non-specific and depends only on the relative concentration of 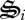 to total biological structure,
to total biological structure, 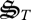 , and the concentration of
, and the concentration of 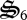 . In addition, biological structure is not perfectly degraded to building block materials (i.e. CH2O and
. In addition, biological structure is not perfectly degraded to building block materials (i.e. CH2O and 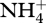 ), but produces some detrital carbon (dC) and nitrogen (dN) (figure 1), as specified by the assimilation parameters faC and faN, respectively (electronic supplementary material, table S1). Breakdown of detrital C and N is controlled by reactions r7(t) and r8(t), but only one biological structure,
), but produces some detrital carbon (dC) and nitrogen (dN) (figure 1), as specified by the assimilation parameters faC and faN, respectively (electronic supplementary material, table S1). Breakdown of detrital C and N is controlled by reactions r7(t) and r8(t), but only one biological structure, 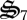 , catalyses both the reactions.
, catalyses both the reactions.
Figure 2.
Illustration of how the partition function (σi) controls synthesis of biological structures, 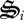 .
.
Based on the metabolic network and reaction rates (figure 1), a mass balance model can be constructed for the state variables, xT(t) = [c(t), 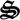 (t), ζ(t)]T, which represent the concentrations of chemical species, c(t), biological structures,
(t), ζ(t)]T, which represent the concentrations of chemical species, c(t), biological structures, 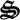 (t), as well as the system's redox and energy states (ζNADH, ζATP), and has the general form,
(t), as well as the system's redox and energy states (ζNADH, ζATP), and has the general form,
| 3.10 |
Once the control variables σ(t) and η(t) are specified over time, the state equation (3.10) can be solved for the state variables (see equations S1–S14 in the electronic supplementary material for the expansion of equation (3.10) in scalar form).
The control variables, [η(t), σ(t)]T, are determined by formulating and solving an interval optimization problem in which average entropy production rate, 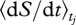 , is maximized over a specified interval of time, δt for each tj interval. Entropy production is given by the negative of a reaction rate multiplied by the Gibbs free energy of the reaction divided by temperature, summed over all reactions (Eu 1992, pp. 131–141), which is readily calculated. The general form of the model is as follows:
, is maximized over a specified interval of time, δt for each tj interval. Entropy production is given by the negative of a reaction rate multiplied by the Gibbs free energy of the reaction divided by temperature, summed over all reactions (Eu 1992, pp. 131–141), which is readily calculated. The general form of the model is as follows:
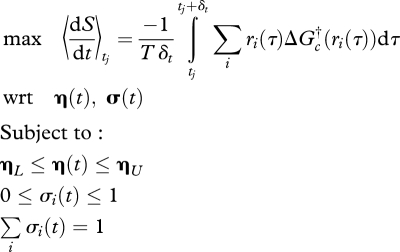 |
3.11 |
which is a class of optimal control problems (Kirk 1970); however, since no constraints are imposed on the state variables, x(t), it is not necessary to include the differential equations (3.10) as part of the constraints, as they are explicitly satisfied. To obtain a solution over a specified time domain, [t0, tf], the optimal control problem (3.11) is solved repetitively over a sufficient number of j intervals, [tj, tj + δt], to cover the entire domain. There are several ways of solving equation (3.11), including linear programming following linearization at time, t (Vallino et al. 1996; Vallino 2003). However, we have implemented an interval optimization method using SNOPT (Gill et al. 2005) that solves the nonlinear programming problem over the interval δt via sequential quadratic programming coupled with block implicit methods to solve the associated differential equations (3.10) (Brugnano & Magherini 2004). In the current implementation, the optimal interval, δt, is subdivided into ng grid points, and the control functions [η(t), σ(t)]T are discretized over the interval as linear piecewise continuous functions. The length of the optimization interval δt, is an interesting aspect of the model, as it reflects the characteristic time scale of environmental variability that a living system has evolved to cope with over a given spatial scale (O'Neill et al. 1986). We expect that microbial systems can be described by short optimization intervals (approx. days), while for forest ecosystems δt would require longer intervals (approx. year). It is also possible to cast the problem as a type of infinite horizon (Carlson et al. 1991) or receding horizon (Ito & Kunisch 2002) optimal control problem, but we are yet to fully investigate that possibility.
4. Simulation results
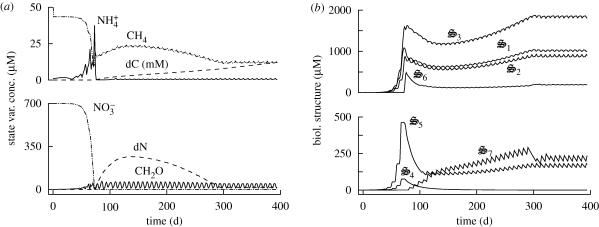
Figures 3–5 show results from a standard simulation based on the above equations for a 18 l methanotrophic microbial community that is sparged with 2.9 per cent methane in air at 20 ml min−1 (STP) starting with initial nutrient concentrations of 700 µM 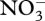 , 70 µM
, 70 µM 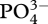 , plus salts and trace elements. The simulation conditions are based on current methanotrophic microcosm experiments that are being used to guide model development and test the MEP principle; however, the model has not been calibrated to the observations yet, but we do provide some preliminary experimental data in the electronic supplementary material for qualitative comparison. For these simulations, an 8-day optimization interval, δt, was employed, which results in
, plus salts and trace elements. The simulation conditions are based on current methanotrophic microcosm experiments that are being used to guide model development and test the MEP principle; however, the model has not been calibrated to the observations yet, but we do provide some preliminary experimental data in the electronic supplementary material for qualitative comparison. For these simulations, an 8-day optimization interval, δt, was employed, which results in 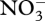 depletion by day 75 (figure 3a), which closely matches observations (electronic supplementary material, figure S2a). Although not evident on the linear scale (figure 3), constant exponential growth begins at time zero but does not result in significant changes in state variables until day 40, which is similarly observed experimentally (electronic supplementary material, figure S2). Because entropy production is maximized over an 8-day interval instead of being instantaneous, entropy production can be increased if some CH4 is allocated to
depletion by day 75 (figure 3a), which closely matches observations (electronic supplementary material, figure S2a). Although not evident on the linear scale (figure 3), constant exponential growth begins at time zero but does not result in significant changes in state variables until day 40, which is similarly observed experimentally (electronic supplementary material, figure S2). Because entropy production is maximized over an 8-day interval instead of being instantaneous, entropy production can be increased if some CH4 is allocated to 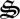 rather than being completely oxidized to CO2. During the first 75 days, biological structure is allocated to respiration (
rather than being completely oxidized to CO2. During the first 75 days, biological structure is allocated to respiration (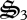 ), CH4 oxidation (
), CH4 oxidation (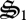 ), CH2O oxidation (
), CH2O oxidation (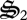 ), biological structure synthesis (
), biological structure synthesis (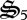 ) and nitrate reduction (
) and nitrate reduction (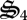 ) in decreasing magnitude (figure 3b). The majority of
) in decreasing magnitude (figure 3b). The majority of 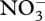 is converted to
is converted to 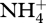 for biological structure synthesis; however, some
for biological structure synthesis; however, some 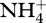 accumulates, but is later consumed after
accumulates, but is later consumed after 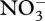 depletion (figure 3a). Ammonium accumulation during the first 75 days occurs to balance excess electrons resulting from CH4 oxidation that is not completely balanced by O2 reduction to H2O. Ammonium accumulation and depletion also occurs in the experimental microcosms (electronic supplementary material, figure S2a), both before and after
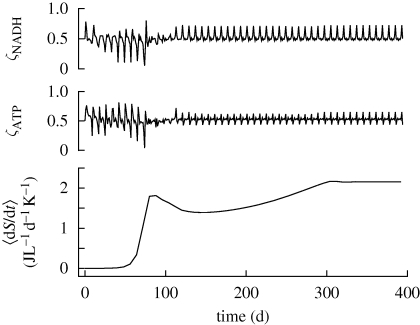
depletion (figure 3a). Ammonium accumulation during the first 75 days occurs to balance excess electrons resulting from CH4 oxidation that is not completely balanced by O2 reduction to H2O. Ammonium accumulation and depletion also occurs in the experimental microcosms (electronic supplementary material, figure S2a), both before and after 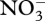 depletion. Entropy production rate increases rapidly during the first 75 days, while both the ‘intracellular’ redox (ζNADH) and energy (ζATP) states oscillate around their balance state of 0.5 µmol mmol−1 (figure 4). Because ‘intracellular’ ζNADH and ζATP have faster characteristic time scales than other state variables (see electronic supplementary material, equations S10 and S11), balancing internal NADH/NAD and ATP/ADP ratios severely constrains reaction rates and may account for energy spilling observed in bacterial cultures (Dauner et al. 2001).
depletion. Entropy production rate increases rapidly during the first 75 days, while both the ‘intracellular’ redox (ζNADH) and energy (ζATP) states oscillate around their balance state of 0.5 µmol mmol−1 (figure 4). Because ‘intracellular’ ζNADH and ζATP have faster characteristic time scales than other state variables (see electronic supplementary material, equations S10 and S11), balancing internal NADH/NAD and ATP/ADP ratios severely constrains reaction rates and may account for energy spilling observed in bacterial cultures (Dauner et al. 2001).
Figure 3.
Concentration of (a) some state variables and (b) all biological structures in micromoles, except for dC, which is in millimoles.
Figure 4.
Redox ζNADH, and energy ζATP, state variables (µmol mmol−1), and entropy production rate 〈dS/dt〉, averaged over 8-day intervals for the duration of the simulation.
Figure 5.
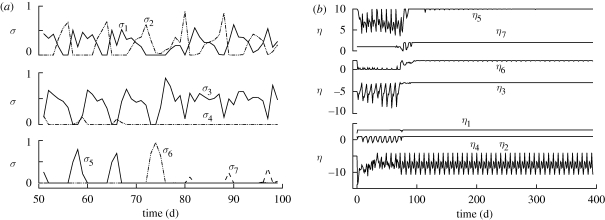
Control variables for (a) partitioning of biological structure synthesis, σi, and (b) ATP coupling to metabolic reactions, ηi. Note that plot (a) shows only control variables about the time of nitrate depletion from 50 to 100 days.
Near the 75-day transition point, control variables change so that σ4 decreases to 0, while σ6 and σ7 turn on for the first time (figure 5a). The latter two control variables allocate biological structure to decomposition of biological structure and detritus, respectively, while σ4 controls 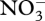 uptake, which is exhausted after 75 days, so is no longer needed. Control variables also alter reaction energy coupling via changes in ηi around the transition point (figure 5b). Prior to
uptake, which is exhausted after 75 days, so is no longer needed. Control variables also alter reaction energy coupling via changes in ηi around the transition point (figure 5b). Prior to 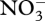 depletion, the system runs at high efficiency, in which ATP synthesis is high (η3 more negative), while ATP requirements for biological structure synthesis, r5, are maintained low (η5 less positive; figure 5b). After
depletion, the system runs at high efficiency, in which ATP synthesis is high (η3 more negative), while ATP requirements for biological structure synthesis, r5, are maintained low (η5 less positive; figure 5b). After 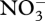 depletion, control variables switch to favour low ATP synthesis (η3 less negative) and high ATP requirements (η5 more positive), a low-efficiency mode which is necessary to maintain energy balance as dictated by ζATP via equation (3.8) (figure 4). Laboratory cultures have also been shown to spill ATP under nutrient-limited growth (Russell & Cook 1995) and hence also exhibit similar low-efficiency modes.
depletion, control variables switch to favour low ATP synthesis (η3 less negative) and high ATP requirements (η5 more positive), a low-efficiency mode which is necessary to maintain energy balance as dictated by ζATP via equation (3.8) (figure 4). Laboratory cultures have also been shown to spill ATP under nutrient-limited growth (Russell & Cook 1995) and hence also exhibit similar low-efficiency modes.
In the latter part of the simulation, detrital C and N (dC, dN) begin to accumulate owing to synthesis of 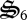 , which degrades biological structure with dC and dN formation (figure 3a). Biological structure is ultimately limited by N availability, so that more entropy can be produced if N is maintained within the biological structure pool,
, which degrades biological structure with dC and dN formation (figure 3a). Biological structure is ultimately limited by N availability, so that more entropy can be produced if N is maintained within the biological structure pool, 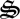 . Consequently, control variables allocate some biological structure to
. Consequently, control variables allocate some biological structure to 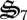 to return N in dN to active catalyst. While
to return N in dN to active catalyst. While 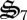 also causes conversion of dC to CH2O, entropy production is not C-limited because of the continuous addition of CH4. While complete oxidation of dC would contribute to entropy production, entropy production per unit of biological structure is higher for CH4 oxidation. Consequently, biological structure is not squandered to completely use dC under the simulated conditions. As a result, dC continuously accumulates (figure 3a). Interestingly, we have also observed long-term accumulation and depletion of dissolved organic C and N in the experimental microcosms (electronic supplementary material, figure S2b) and large amounts of particulate C have visually accumulated in the microcosms (electronic supplementary material, figure S1), but it has not been quantified yet. Synthesis of
also causes conversion of dC to CH2O, entropy production is not C-limited because of the continuous addition of CH4. While complete oxidation of dC would contribute to entropy production, entropy production per unit of biological structure is higher for CH4 oxidation. Consequently, biological structure is not squandered to completely use dC under the simulated conditions. As a result, dC continuously accumulates (figure 3a). Interestingly, we have also observed long-term accumulation and depletion of dissolved organic C and N in the experimental microcosms (electronic supplementary material, figure S2b) and large amounts of particulate C have visually accumulated in the microcosms (electronic supplementary material, figure S1), but it has not been quantified yet. Synthesis of 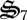 continues until decomposition of dN matches its production, at which point σ7 is downregulated and the system reaches pseudo steady-state at approximately 300 days (figure 3).
continues until decomposition of dN matches its production, at which point σ7 is downregulated and the system reaches pseudo steady-state at approximately 300 days (figure 3).
During the period in which N is bound in dN (75 < t < 300), entropy production is low, but increases to its steady state value once recycling of N from dN is balanced (figure 4). The system reaches pseudo-steady-state near day 300, where entropy production rate is 2.16 J l−1 d−1K−1 and the dominate reaction rates are respiration (r3: 1570 µM d−1), methane oxidation (r1: 815 µM d−1) and CH2O oxidation (r2: 755 µM d−1). Production of biological structure, r5, matches its degradation rate, r6, at 120 µM d−1. The high frequency 8-day oscillations observed throughout the simulation (figures 3–5) obviously result from the 8-day optimization interval used. However, if the sub-interval daily data points are not plotted, then the oscillations disappear. The oscillations are analogous to changes in plant metabolism that occur over the course of the daily photic period. Interestingly, CH2O acts as an ‘internal’ storage mechanism over the sub-interval (figure 3a). The magnitude of CH2O storage increases with longer optimization intervals, or if CH4 is periodically introduced into the system instead of being continuous (data not shown). While no attempt has been made to calibrate the model to experimental data yet, maximizing entropy production over successive intervals does produce results at least qualitatively similar to our observations (see electronic supplementary material).
5. Discussion
The MEP principle states that systems will tend to follow pathways that maximize entropy production at steady state. If internal self-organized structures facilitate entropy production, then those structures are more likely to develop and persist, as occurs with Rayleigh-Benard convection cells and hurricanes (Schneider & Kay 1994). In the MEP perspective, living systems can be viewed as molecular machines which catalyse reactions that both synthesize and degrade molecular machines and dissipate energy via redox reactions in the process, but are otherwise undifferentiated from abiotic systems. Because MEP states that there must be many different ways to produce entropy maximally, the form of the molecular machines is unimportant for biogeochemical MEP. Consequently, we have chosen a metabolic perspective of ecosystem biogeochemistry that only predicts functional characteristics in terms of allocation of molecular machinery. The approach strives for long-term predictability at the expense of short-term dynamic details. However, the metabolic model does predict biological structure allocation (figure 3b), which can be related to observed functional groups and community level gene expression obtained from experiments, and it does produce reasonable results and qualitatively compares with our experimental mesocosm data prior to calibration. Furthermore, the metabolic MEP perspective leads to some interesting extrapolations, which we briefly explore below.
Current MEP theory applies to steady-state systems (Dewar 2003; Niven 2009), but ecosystems seldom operate at steady state and at times can be quite far from it. During major transient events, such as recovery from system collapse, MEP may not be a useful descriptor of system response. Rather, the community composition will likely play the dominant role in system dynamics as it reorganizes and attempts to return to MEP. However, for perturbations that an ecosystem has evolved to cope with, MEP should be an important system attractor, so we expect MEP to be relevant even if a system is not strictly at steady state. In fact, it is the ability of living systems to predict future states in the presence of fluctuations that allows biological systems to produce more entropy than abiotic systems that take the steepest descent approach. To predict future states requires a system to store relevant information. Hence, increases in information (i.e. physical complexity (Adami et al. 2000)) facilitate increases in thermodynamic entropy production—a rather interesting parallel.
Biological systems acquire information through evolution and store it within the metagenome. This information describes how to construct biological structures from the resources available. But maximizing entropy production requires optimal use, and possibly selection of resources at the whole system level (Traulsen & Nowak 2006; Williams & Lenton 2008; Wilson & Wilson 2008), not at the level of the constituent components. Information stored in any genome correlates with the entire system (Adami 2002), including that stored in other genomes, so all ecosystem organisms must be inherently coupled to achieve MEP. Consider a closed ecosystem similar to our methanotrophic system, which contains finite resources and so a limit to the total amount of biological structure that can be synthesized. How should biological structures be allocated to maximize entropy production? While some biological structure must be allocated to oxidizing methane, other structures must be allocated to recycling nutrients, including nutrients contained in biological structures that are no longer useful (Chesson & Kuang 2008). Of course, excessive or insufficient allocation to any one structure will result in sub-optimal entropy production, so system level coordination (e.g. figure 3b) is an emergent property of the MEP principle.
While one can image a single organism that could orchestrate all ecosystem functions, it would be at a competitive disadvantage to any specialist organism that only maintained a fraction of the entire metagenome (Mills et al. 1967). Distributing genomic information allows a system to retain important information not relevant to the current ecosystem state at very low copy number, such as that observed in the rare biosphere (Sogin et al. 2006). Accordingly, a distributed metabolic system can function as a single metabolic entity, but is more efficient with system resources, hence is more likely to be selected for at the system level (Traulsen & Nowak 2006; Williams & Lenton 2008; Wilson & Wilson 2008). Consequently, we view an ecosystem composed of autonomous individuals all attempting to maximize their own fecundity in the Darwinian context, but this leads to an MEP state at the system level. If MEP is a true attractor, then ecosystems that recycle nutrients more effectively should become more spatially dominant. Likewise, systems that produce more entropy over longer time periods should also be selected for. We speculate that systems will evolve towards entropy production that is maximized over infinite time and space. This contrasts abiotic (information lacking) systems that maximize entropy production at a point in space–time (steepest descent).
If a system organizes towards MEP over infinite time and space, then steepest descent routes must be inhibited, but they cannot be prevented from occurring. As already mentioned, a forest fire rapidly increases short-term entropy production, but at the expense of long-term entropy production. Invasive species can produce an identical phenomenon if the invading species propagates via oxidation of biological structure. However, if the invading species causes lower entropy production on longer time scales, then the new state will not be stable. On the contrary, if the invasive species increases averaged entropy production, then it should persist. Entropy production serves as a useful measure of system stability then. While changes in species abundances are often associated with instability, if the change does not alter entropy production, then from the MEP perspective, the system is stable. Changes in biodiversity that lead to lower entropy production would be considered detrimental to system stability.
If a system is subject to external perturbations, then we may expect the system to store energy internally so that entropy production can be maintained unperturbed, such as that observed in CH2O dynamics in our modelled system (figure 3a). One way to store energy is to increase the fraction of large biological structures. From the metabolic theory of ecology, organism respiration increases only as the three-fourth power of its mass (West et al. 1997). Consequently, a unit mass of bacteria has a much higher specific respiration than an equivalent mass of elephant or whale. While our model above can store ‘internal’ energy as CH2O, that cannot occur in the environment unless it is protected from microbial consumption, such as in the form of a large animal. If a perturbation occurs, large animals may perish, but they provide energy to prevent hierarchical collapse of the entire system. This is consistent with the observation that biomass-specific respiration decreases with increase in ecosystem maturity (Odum 1969).
While we have discussed some of the possible implications of the MEP principle in this section, our primary objective of this manuscript has been to demonstrate how MEP can be used quantitatively as a governing principle to understand and model biogeochemical processes. A key aspect of our approach is to replace the organismal focus with a functional one, which should produce more robust predictions, albeit with some loss of details. To further develop this approach, it is necessary to examine ecosystems purely at the functional level. For instance, many higher heterotrophs merely serve to capture and masticate biological structure, while other organisms provide physical structure, such as for trapping sediments on marshes (Mudd et al. 2004), facilitating evapotranspiration (Wang et al. 2007), or other transport-limited phenomena (Meysman et al. 2006). We will need to understand the functions that organisms contribute to ecosystems and to quantify functional rates per unit amount of biological structure allocated. Even though examining ecosystems from an MEP perspective will not always be productive for every interest, it does provide an alternative vantage point that can provide great insight. Based on the MEP principle, living systems exist because they hasten entropy production over greater spatio-temporal scales than abiotic systems.
Acknowledgements
I thank Axel Kleidon for organizing the workshop on MEP in the Earth System, as well as editing this special issue of the Philosophical Transactions of the Royal Society B along with Yadvinder Malhi and Peter Cox. The work presented here was funded by the PIE-LTER programme (NSF OCE-0423565), as well as from NSF CBET-0756562, NSF EF-0928742 and NASA Exobiology and Evolutionary Biology (NNG05GN61G).
Footnotes
One contribution of 17 to a Theme Issue ‘Maximum entropy production in ecological and environmental systems: applications and implications’.
References
- Adami C.2002Sequence complexity in Darwinian evolution. Complexity 8, 49–56 (doi:10.1002/cplx.10071) [Google Scholar]
- Adami C., Ofria C., Collier T. C.2000Evolution of biological complexity. Proc. Natl Acad. Sci. USA 97, 4463–4468 (doi:10.1073/pnas.97.9.4463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberty R. A.2003Thermodynamics of biochemical reactions, 397 pp Hoboken, NJ: Wiley & Sons [Google Scholar]
- Alberty R. A.2006Biochemical thermodynamics: applications of mathematica, 464 pp Hoboken, NJ: Wiley & Sons; [PubMed] [Google Scholar]
- Armstrong R. A., McGehee R.1980Competitive exclusion. Am. Nat. 115, 151–170 (doi:10.1086/283553) [Google Scholar]
- Battley E. H.1999An empirical method for estimating the entropy of formation and the absolute entropy of dried microbial biomass for use in studies on the thermodynamics of microbial growth. Thermochim. Acta 326, 7–15 (doi:10.1016/S0040-6031(98)00584-X) [Google Scholar]
- Battley E.2003Absorbed heat and heat of formation of dried microbial biomass: studies on the thermodynamics of microbial growth. J. Thermal Anal. Calorimetry 74, 709–721 (doi:10.1023/B:JTAN.0000011003.43875.0d) [Google Scholar]
- Bejan A.2007Constructal theory of pattern formation. Hydrology Earth Syst. Sci. 11, 753–768 [Google Scholar]
- Beninca E., Huisman J., Heerkloss R., Johnk K. D., Branco P., Van Nes E. H., Scheffer M., Ellner S. P.2008Chaos in a long-term experiment with a plankton community. Nature 451, 822–825 (doi:10.1038/nature06512) [DOI] [PubMed] [Google Scholar]
- Berman-Frank I., Dubinsky Z.1999Balanced growth in aquatic plants: myth or reality? BioScience 49, 29–37 (doi:10.2307/1313491) [Google Scholar]
- Boudart M.1976Consistency between kinetics and thermodynamics. J. Phys. Chem. 80, 2869–2870 (doi:10.1021/j100567a012) [Google Scholar]
- Brugnano L., Magherini C.2004The BiM code for the numerical solution of ODEs. J. Comput. Appl. Math. 164–165, 145–158 (doi:10.1016/j.cam.2003.09.004) [Google Scholar]
- Carlson D. A., Haurie A. B., Leizarowitz A.1991Infinite horizon optimal control: deterministic and stochastic systems, 332 pp., 2nd edn.Berlin, Germany: Springer-Verlag [Google Scholar]
- Chesson P., Kuang J. J.2008The interaction between predation and competition. Nature 456, 235–238 (doi:10.1038/nature07248) [DOI] [PubMed] [Google Scholar]
- Choi J. S., Mazumder A., Hansell R. I. C.1999Measuring perturbation in a complicated, thermodynamic world. Ecol. Modell. 117, 143–158 (doi:10.1016/S0304-3800(99)00042-3) [Google Scholar]
- Clausius R.1850Ueber die bewegende Kraft der Wärme und die Gesetze, welche sich daraus für die Wärmelehre selbst ableiten lassen. Ann. Physik Chemie 155, 368–397 (doi:10.1002/andp.18501550306) [Google Scholar]
- Crooks G. E.2007Beyond Boltzmann-Gibbs statistics: maximum entropy hyperensembles out of equilibrium. Phys. Rev. E (Statistical, nonlinear, and soft matter physics) 75, 1–5 (doi:10.1103/PhysRevE.75.041119) [DOI] [PubMed] [Google Scholar]
- Dauner M., Storni T., Sauer U.2001Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 183, 7308–7317 (doi:10.1128/JB.183.24.7308-7317.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar R.2003Information theory explanation of the fluctuation theorem, maximum entropy production and self-organized criticality in non-equilibrium stationary states. J. Phys. A: Math. General 36, 631–641 (doi:10.1088/0305-4470/36/3/303) [Google Scholar]
- Dewar R. C.2005Maximum entropy production and the fluctuation theorem. J. Phys. A: Math. General 38, L371–L381 (doi:10.1088/0305-4470/38/21/L01) [Google Scholar]
- Eu B. C.1992Kinetic theory and irreversible thermodynamics, 752 pp Montreal, Canada: John Wiley & Sons [Google Scholar]
- Falkowski P. G., Fenchel T., DeLong E. F.2008The microbial engines that drive earth's biogeochemical cycles. Science 320, 1034–1039 (doi:10.1126/science.1153213) [DOI] [PubMed] [Google Scholar]
- Fath B. D., Patten B. C., Choi J. S.2001Complementarity of ecological goal functions. J. Theor. Biol. 208, 493–506 (doi:10.1006/jtbi.2000.2234) [DOI] [PubMed] [Google Scholar]
- Fernandez A., Huang S., Seston S., Xing J., Hickey R., Criddle C., Tiedje J.1999How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65, 3697–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. S., Hashsham S. A., Dollhopf S. L., Raskin L., Glagoleva O., Dazzo F. B., Hickey R. F., Criddle C. S., Tiedje J. M.2000Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl. Microbiol. Biotechnol. 66, 4058–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans J., Wolinsky M., Dunbar J.2005Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390 (doi:10.1126/science.1112665) [DOI] [PubMed] [Google Scholar]
- Gill P. E., Murray W., Saunders M. A.2005SNOPT: an SQP algorithm for large-scale constrained optimization. SIAM Rev. 47, 99–131 (doi:10.1137/S0036144504446096) [Google Scholar]
- Gnaiger E.1990Concepts on efficiency in biological calorimetry and metabolic flux control. Thermochim. Acta 172, 31–52 (doi:10.1016/0040-6031(90)80557-F) [Google Scholar]
- Graham D. W., Knapp C. W., Van Vleck E. S., Bloor K., Lane T. B., Graham C. E.2007Experimental demonstration of chaotic instability in biological nitrification. ISME J. 1, 385–393 (doi:10.1038/ismej.2007.45) [DOI] [PubMed] [Google Scholar]
- Helling R. B.2002Speed versus efficiency in microbial growth and the role of parallel pathways. J. Bacteriol. 184, 1041–1045 (doi:10.1128/jb.184.4.1041-1045.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra R. U., Edwards J. S., Palsson B. O.2002Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (doi:10.1038/nature01149) [DOI] [PubMed] [Google Scholar]
- Ito K., Kunisch K.2002Receding horizon optimal control for infinite dimensional systems. ESAIM: control, optimisation and calculus of variations 8, 741–760 (doi:10.1051/cocv:2002032) [Google Scholar]
- Jin Q., Bethke C. M.2003A new rate law describing microbial respiration. Appl. Environ. Microbiol. 69, 2340–2348 (doi:10.1128/AEM.69.4.2340-2348.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S. E.1994Review and comparison of goal functions in system ecology. Vie Milieu 44, 11–20 [Google Scholar]
- Jorgensen S. E., Patten B. C., Straskraba M.2000Ecosystems emerging: 4. Growth. Ecol. Modell. 126, 249–284 (doi:10.1016/S0304-3800(00)00268-4) [Google Scholar]
- Khoshmanesh A., Hart B. T., Duncan A., Beckett R.2002Luxury uptake of phosphorus by sediment bacteria. Water Res. 36, 774–778 (doi:10.1016/S0043-1354(01)00272-X) [DOI] [PubMed] [Google Scholar]
- Kirk D. E.1970Optimal control theory: an introduction, 452 pp..1st edn.Englewood Cliffs, NJ: Prentice-Hall, Inc [Google Scholar]
- Lineweaver C. H., Egan C. A.2008Life, gravity and the second law of thermodynamics. Phys. Life Rev. 5, 225–242 (doi:10.1016/j.plrev.2008.08.002) [Google Scholar]
- Lorenz R. D., Lunine J. I., Withers P. G.2001Titan, Mars and Earth: entropy production by latitudinal heat transport. Geophys. Res. Lett. 28, 415–418 (doi:10.1029/2000GL012336) [Google Scholar]
- Lotka A. J.1922Contribution to the energetics of evolution. Proc. Natl Acad. Sci. USA 8, 147–151 (doi:10.1073/pnas.8.6.147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalef R.1968Perspectives in ecological theory, 112 pp Chicago, IL: University of Chicago Press [Google Scholar]
- Mejer H., Jorgensen S. E.1979Exergy and ecological buffer capacity. In State-of-the-art in ecological modelling (ed. Jorgensen S. E.), pp. 829–846 London, UK: Pergamon Press [Google Scholar]
- Meysman F. J. R., Bruers S.2007A thermodynamic perspective on food webs: quantifying entropy production within detrital-based ecosystems. J. Theor. Biol. 249, 124–139 (doi:10.1016/j.jtbi.2007.07.015) [DOI] [PubMed] [Google Scholar]
- Meysman F. J. R., Galaktionov O. S., Gribsholt B., Middelburg J. J.2006Bioirrigation in permeable sediments: advective pore-water transport induced by burrow ventilation. Limnol. Oceanogr. 51, 142–156 [Google Scholar]
- Mills D. R., Peterson R. L., Spiegelman S.1967An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl Acad. Sci. USA 58, 217–224 (doi:10.1073/pnas.58.1.217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowitz H. J.1968Energy flow in biology: biological organization as a problem in thermal physics, 179 pp New York, NY: Academic Press [Google Scholar]
- Moroz A.2008On a variational formulation of the maximum energy dissipation principle for non-equilibrium chemical thermodynamics. Chem. Phys. Lett. 457, 448–452 (doi:10.1016/j.cplett.2008.04.050) [Google Scholar]
- Mudd S. M., Fagherazzi S., Morris J. T., Furbish D. J.2004Flow, sedimentation, and biomass production on a vegetated salt marsh in South Carolina: toward a predictive model of marsh morphologic and ecologic evolution. In The ecogeomorphology of salt marshes (eds Fagherazzi S., Marani M., Blum L. K.), pp. 165–187 Washington, DC: AGU [Google Scholar]
- Murray B. G., Jr2001Are ecological and evolutionary theories scientific? Biol. Rev. 76, 255–289 (doi:10.1017/S146479310100567X) [DOI] [PubMed] [Google Scholar]
- Nielsen S. N., Ulanowicz R. E.2000On the consistency between thermodynamical and network approaches to ecosystems. Ecol. Modell. 132, 23–31 (doi:10.1016/S0304-3800(00)00302-1) [Google Scholar]
- Niven R. K.2009Steady state of a dissipative flow-controlled system and the maximum entropy production principle. Phys. Rev. E (Statistical, nonlinear, and soft matter physics) 80, 021 113–021 115 (doi:10.1103/PhysRevE.80.021113) [DOI] [PubMed] [Google Scholar]
- Odum E. P.1969The strategy of ecosystem development. Science 164, 262–270 (doi:10.1126/science.164.3877.262) [DOI] [PubMed] [Google Scholar]
- Odum H. T.1988Self-organization, transformity, and information. Science 242, 1132–1139 (doi:10.1126/science.242.4882.1132) [DOI] [PubMed] [Google Scholar]
- Odum H. T., Pinkerton R. C.1955Time's speed regulator: the optimum efficiency for maximum power output in physical and biological systems. Am. Sci. 43, 321–343 [Google Scholar]
- O'Neill R. V., DeAngelis D. L., Waide J. B., Allen T. F. H.1986A hierarchical concept of ecosystems. Monographs in population biology, vol. 23, pp. 1–253 Princeton, NJ: Princeton University Press [Google Scholar]
- Paltridge G. W.1975Global dynamics and climate—a system of minimum entropy exchange. Q. J. R. Meteorol. Soc. 104, 927–945 (doi:10.1002/qj.49710444206) [Google Scholar]
- Peters R. H.1976Tautology in evolution and ecology. Am. Nat. 110, 1–12 (doi:10.1086/283045) [Google Scholar]
- Pfeiffer T., Bonhoeffer S.2002Evolutionary consequences of tradeoffs between yield and rate of ATP production. Zeitschrift für Physikal. Chem. 216, 51–63 (doi:10.1524/zpch.2002.216.1.051) [Google Scholar]
- Prigogine I., Nicolis G.1971Biological order, structure and instabilities. Q. Rev. Biophys. 4, 107–148 (doi:10.1017/S0033583500000615) [DOI] [PubMed] [Google Scholar]
- Pueyo S., He F., Zillio T.2007The maximum entropy formalism and the idiosyncratic theory of biodiversity. Ecol. Lett. 10, 1017–1028 (doi:10.1111/j.1461-0248.2007.01096.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Cook G. M.1995Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Mol. Biol. Rev. 59, 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E. D., Kay J. J.1994Complexity and thermodynamics: towards a new ecology. Futures 26, 626–647 (doi:10.1016/0016-3287(94)90034-5) [Google Scholar]
- Schrödinger E.1944What is life? Cambridge, UK: Cambridge University Press [Google Scholar]
- Shapiro J. A.1998Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104 (doi:10.1146/annurev.micro.52.1.81) [DOI] [PubMed] [Google Scholar]
- Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., Arrieta J. M., Herndl G. J.2006Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl Acad. Sci. USA 103, 12 115–12 120 (doi:10.1073/pnas.0605127103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson R.1989Emergent attractors and the law of maximum entropy production: foundations to a theory of general evolution. Syst. Res. 6, 187–197 [Google Scholar]
- Toussaint O., Schneider E. D.1998The thermodynamics and evolution of complexity in biological systems. Comp. Biochem. Physiol. A 120, 3–9 [DOI] [PubMed] [Google Scholar]
- Traulsen A., Nowak M. A.2006Evolution of cooperation by multilevel selection. Proc. Natl Acad. Sci. USA 103, 10952–10955 (doi:10.1073/pnas.0602530103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanowicz R. E.1986Growth and development: ecosystems phenomenology, 203 pp New York, NY: Springer [Google Scholar]
- Ulanowicz R. E., Hannon B. M.1987Life and the production of entropy. Proc. R. Soc. Lond. B 232, 181–192 (doi:10.1098/rspb.1987.0067) [Google Scholar]
- Ulanowicz R. E., Platt T.1985Ecosystem theory for biological oceanography. Can. Bull. Fish. Aquat. Sci. 213, 260 pp [Google Scholar]
- Vallino J. J.2003Modeling microbial consortiums as distributed metabolic networks. Biol. Bull. 204, 174–179 (doi:10.2307/1543554) [DOI] [PubMed] [Google Scholar]
- Vallino J. J., Hopkinson C. S., Hobbie J. E.1996Modeling bacterial utilization of dissolved organic matter: optimization replaces Monod growth kinetics. Limnol. Oceanogr. 41, 1591–1609 [Google Scholar]
- Vuddagiri S. R., Hall K. R., Eubank P. T.2000Dynamic modeling of reaction pathways on the Gibbs energy surface. Ind. Eng. Chem. Res. 39, 508–517 (doi:10.1021/ie9904091S0888-5885(99)00409-1) [Google Scholar]
- Wang J., Bras R. L., Lerdau M., Salvucci G. D.2007A maximum hypothesis of transpiration. JGR Biogeosci. 112, 1–11 (doi:10.1029/2006JG000255) [Google Scholar]
- Washida T.1995Ecosystem configurations consequent on the maximum respiration hypothesis. Ecol. Modell. 78, 173–193 (doi:10.1016/0304-3800(93)E0102-9) [Google Scholar]
- Weber B. H., Depew D. J., Smith J. D.1988Entropy, information, and evolution: new perspective on physical and biological evolution, 390 pp Cambridge, MA: MIT Press [Google Scholar]
- West G., Brown J. H., Enquist B. J.1997A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 (doi:10.1126/science.276.5309.122) [DOI] [PubMed] [Google Scholar]
- Williams H. T. P., Lenton T. M.2008Environmental regulation in a network of simulated microbial ecosystems. Proc. Natl Acad. Sci. USA 105, 10 432–10 437 (doi:10.1073/pnas.0800244105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. S., Sober E.1989Reviving the superorganism. J. Theor. Biol. 136, 337–356 (doi:10.1016/S0022-5193(89)80169-9) [DOI] [PubMed] [Google Scholar]
- Wilson D. S., Wilson E. O.2008Evolution ‘for the Good of the Group’. Am. Sci. 96, 380–389 (doi:10.1511/2008.74.1) [Google Scholar]
- Wittebolle L., Vervaeren H., Verstraete W., Boon N.2008Quantifying community dynamics of nitrifiers in functionally stable reactors. Appl. Environ. Microbiol. 74, 286–293 (doi:10.1128/AEM.01006-07) [DOI] [PMC free article] [PubMed] [Google Scholar]