Abstract
Spermatogenesis in mammals is achieved by multiple players that pursue a common goal of generating mature spermatozoa. The developmental processes acting on male germ cells that culminate in the production of the functional spermatozoa are regulated at both the transcription and post-transcriptional levels. This review addresses recent progress towards understanding such regulatory mechanisms and identifies future challenges to be addressed in this field. We focus on transcription factors, chromatin-associated factors and RNA-binding proteins necessary for spermatogenesis and/or sperm maturation. Understanding the molecular mechanisms that govern spermatogenesis has enormous implications for new contraceptive approaches and treatments for infertility.
Keywords: male germ cells, Sertoli cells, epididymis, fertility
1. Introduction
Mammalian spermatogenesis is an androgen-dependent developmental process driven by interactions between germ cells and somatic cells. This process generates a continuous supply of functional sperm by using a complex programme of mitotic, meiotic and differentiation events in the seminiferous tubules of the testis. Each of these events depends on the appropriate expression of specific genes.
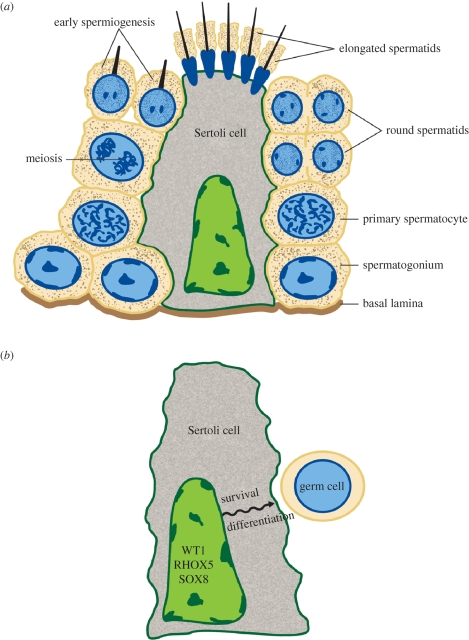
The long journey from a spermatogonial stem cell to a functional sperm involves many steps and the cooperation of many cell types (figure 1a). The most immature germ cells in the testis, spermatogonia, undergo mitotic divisions before converting into spermatocytes, which undergo meiosis to become haploid round spermatids. These latter cells then undergo a series of differentiation events to produce elongated spermatids. All these steps require that the developing germ cells physically interact with Sertoli cells for nourishment and support, as well as instructive guidance (figure 1b). Following completion of cellular differentiation, the immature spermatozoa are released from the seminiferous tubules and traverse to the epididymis. Here, they are exposed to an acidic microenvironment and a variety of secretory proteins that are essential for their complete maturation into spermatozoa (sperm) and their long-term storage in a quiescent state.
Figure 1.
(a) Schematic representation of developing germ cells imbedded in a Sertoli cell in the seminiferous tubule. Spermatogonia undergo mitotic divisions, ultimately giving rise to meiotic spermatocytes. Meiosis causes reductive divisions, leading to the generation of haploid round spermatids. These latter cells undergo a series of differentiation events to become elongated spermatids, which detach from the Sertoli cell and travel down the seminiferous tubule lumen to reach the epididymis (not shown). (b) Recently defined Sertoli-cell-expressed transcription factors that guide the survival and differentiation of the adjacent germ cells.
This review is devoted to recent studies identifying transcriptional and post-transcriptional regulatory proteins that regulate spermatogenesis and sperm maturation. In §2, we cover transcription factors, §3 deals with non-DNA-binding chromatin-associated factors and finally in §4 we cover RNA-binding proteins. We have focused on papers published since our last review on this subject in 2005 (Maclean & Wilkinson 2005). Our review does not attempt to be an exhaustive survey of all recent papers on this subject, but rather it is a selective view that covers papers that we feel particularly advance the field. We apologize if we have missed important papers in this research area and urge readers to look at several other excellent recent review articles on related topics, including chromatin remodelling and post-transcriptional regulation in germ cells (Kimmins & Sassone-Corsi 2005; Kotaja & Sassone-Corsi 2007; Nagamori & Sassone-Corsi 2008).
2. Dna-binding proteins that regulate spermatogenesis
In this section, we review recent studies identifying spermatogenic roles for six transcription factors. Two of them (heat shock factor 2 (HSF2) and OVOL1) are expressed in germ cells and appear to directly act on genes in germ cells to control spermatogenesis. Three of them (WT1, RHOX5 and SOX8) are expressed in Sertoli cells, the post-mitotic nurse cells in the adult testes that physically interact with developing germ cells in the seminiferous tubules and are critical for all stages of male gametogenesis (figure 1b). Finally, one of them (FOXI1) acts in the epididymis to control one or more of the final stages of sperm maturation that occur in this organ.
(a). Heat shock factor 2
HSF2, originally identified based on its sequence similarity to the heat-shock gene activator HSF1, is not induced by heat shock but instead is constitutively expressed during embryogenesis (Sarge et al. 1991; Pirkkala et al. 2001). Consistent with its embryonic expression pattern, HSF2 has several functions during embryogenesis, as revealed by targeted disruption of Hsf2 in mice (Kallio et al. 2002; Wang et al. 2003). However, HSF2's functions are not confined to embryogenesis. In adult mice, HSF2 is abundantly expressed in the nuclei of meiotic and differentiating male germ cells; i.e. early pachytene spermatocytes and round spermatids, respectively (Kallio et al. 2002). Hsf2-mutant male mice are hypofertile, have reduced testes and epididymides size and display elevated apoptosis of pachytene spermatocytes, accompanied by reduced numbers of round and elongated spermatids (Kallio et al. 2002). These data indicates that HSF2 promotes the survival of meiotic germ cells and/or their differentiation into spermatids (figure 2).
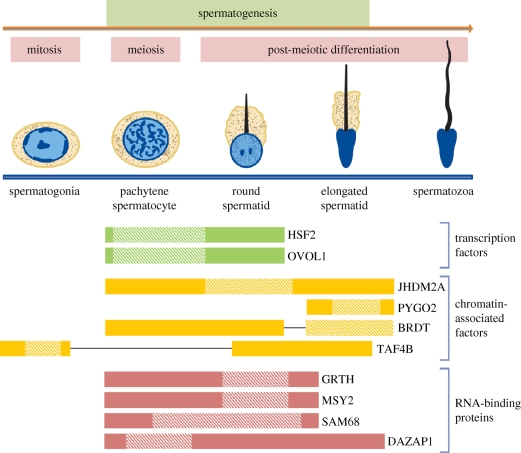
Figure 2.
Germ-cell regulatory factors crucial for spermatogenesis. Shown are germ-cell-expressed transcription factors, chromatin-associated factors, and RNA-binding proteins that have recently been shown to have roles in spermatogenesis. Horizontal bars indicate the approximate germ-cell stage(s) at which these factors are expressed. Shaded areas within the horizontal bars indicate the approximate germ-cell stage(s) at which these factors act. Note that factors that act in the Sertoli cell are not shown here but instead are in figure 1b. Also not shown is FOXI1, which promotes sperm maturation in the epididymis.
Recently, the molecular targets of HSF2 during spermatogenesis were identified at the genome-wide level by Akerfelt et al. (2008). High-resolution chromatin immunoprecipitation (ChIP)-on-chip analysis using promoter microarrays to assay HSF2-binding sites in adult mouse testes revealed that HSF2 primarily interacts with sites on the mouse Y chromosome. Interestingly, some of its target genes (Ssty2 and Sly) are part of multi-copy gene clusters in the male-specific region of the long arm of the mouse Y chromosome (MSYq) that are expressed exclusively during spermatogenesis. Previous studies have shown that deletion of the MSYq region results in sperm head defects and impaired fertility in mice (Toure et al. 2004). Likewise, Hsf2 mutant mice are hypofertile and have a high proportion of sperm with head defects. This phenocopy, along with the fact that many of HSF2's binding sites are in the MSYq region, suggests that the many of the reproductive defects in Hsf2-mutant mice result from misregulation of HSF2 target genes on the Y chromosome. In agreement with this, Hsf2-mutant mice display decreased expression of the MSYq resident genes Ssty2 and Sly.
In addition to altered expression of Y-chromosome genes, Hsf2-mutant mice exhibit pronounced DNA fragmentation and altered levels of chromatin-packing proteins in epididymal spermatozoa: transition protein-2 (TNP2), protamine-1 (PRM1) and protamine-2 (PRM2). This suggests that HSF2 is also essential for directing the unique chromatin reorganization events that occur during late stages of spermatogenesis.
This study by Akerfelt et al. (2008) is a landmark paper, as it is one of very few studies to perform genome-wide analysis of transcription factor-binding sites in vivo rather than in cultured cells. It remains for future studies to determine precisely how HSF2 controls male fertility and whether it directly or indirectly controls chromatin structure.
(b). OVOL1
OVOL1 is a member of the Ovo family of zinc-finger transcription factors. Disruption of Ovol1 in mice leads to abnormalities in several tissues that express OVOL1, including the testis, skin, kidney and urogenital tract (Dai et al. 1998). The defects are most pronounced in the testis, where there is massive germ-cell degeneration, accompanied by dramatically reduced sperm cell count and subfertility. Because the loss of germ cells in the adult testes of Ovol1-mutant mice complicates analysis of Ovol1 function in spermatogenesis, recently Li et al. (2005) examined Ovol1 function during the first wave of spermatogenesis. They found that germ cells lacking OVOL1 from postnatal mice exhibit poor exit from mitotic proliferation in spermatogonia and approximately 50 per cent of the spermatocytes are incapable of proceeding through the late pachytene stage (figure 2). This pachytene arrest is accompanied by dramatically enhanced apoptosis of late pachytene spermatocytes.
Why do Ovol1-mutant germ cells have a propensity to continue proliferating, rather than differentiating? These cells exhibit abnormal precocious nuclear localization of the cyclin B1, suggesting the possibility that this G2-M cell-cycle regulator has a role in the decision to proliferate. Other candidate factors involved were revealed by RNA expression profiling (microarray analysis) of Ovol1-mutant testes. This analysis identified transcripts encoding several other regulators of cell-cycle progression that were upregulated in response to loss of OVOL1 during the first wave of spermatogenesis. Among these was inhibitor of differentiation-2 (ID2), a helix–loop–helix transcription factor that is known to serve as a positive regulator of proliferation and a negative regulator of differentiation in multiple cell lineages. Indeed, loss of Id2 is known to prevent complete differentiation of male germ cells in mice (Yokota 2001). Therefore, the upregulation of ID2 in OVOL1-lacking germ cells is consistent with their propensity to proliferate and their inability to progress through meiosis. Interestingly, biochemical studies and reporter gene assays indicated that OVOL1 directly inhibits Id2 transcription by repressing its promoter (Li et al. 2005). Thus, OVOL1 appears to serve as a direct link to molecules that regulate the decision whether germ cells will undergo proliferation versus differentiation.
(c). WT1
Wilms' tumour (WT1) gene encodes a zinc-finger transcription factor and tumour suppressor that, like OVOL1, is critical for the embryonic development of several organs, including gonads (Park & Jameson 2005). During mouse gonadogenesis, WT1 is first significantly expressed in cells of the genital ridge between day 10.5 and 11.5 post coitum (e10.5–11.5). Targeted disruption of Wt1 causes many cells in the genital ridge to undergo apoptosis, thereby preventing gonad formation. Thus, WT1 is crucial for the development of the early bipotential gonad by virtue of its ability to promote cell survival in this organ (Kreidberg et al. 1993; Brennan & Capel 2004). WT1 expression persists in Sertoli cells after gonad formation, but until recently, its functional role in this context was not known, as Wt1-mutant mice die early during embryogenesis.
The role of WT1 in Sertoli cells after the bipotential gonad stage was recently addressed by two studies (Gao et al. 2006; Rao et al. 2006). Gao et al. (2006) used a conditional knockout approach to disrupt the Wt1 gene in foetal Sertoli cells at approximately e14.5 and found that this severely disrupted testis cord formation in male embryos and subsequently led to the formation of abnormal tubular architecture and reduced testes size in adult males. This result clearly demonstrated that WT1 is crucial for developmental events that convert the bipotential gonad into a male gonad. Rao et al. (2006) used an RNA interference approach to address whether WT1 also has a role in spermatogenesis (Rao et al. 2006). Using a Sertoli cell promoter (from the Rhox5 homeobox gene; see below) to express an artificial microRNA (miRNA) targeting WT1 in postnatal and adult (but not foetal) Sertoli cells, they found that this led to disruption of the apical ectoplasmic specialization (ES), a junctional complex formed between Sertoli cells and elongating/elongated spermatids. The disruption of this adherens junction may be the causal event that led to the other phenotypic alterations in these mice, including increased germ-cell apoptosis, reduced sperm count, reduced sperm motility and subfertility. All of these phenotypic effects were the result of reduced WT1 expression, not ‘off-target’ effects elicited by the miRNA targeting Wt1, as transgenic mice expressing a dominant-negative form of WT1 in postnatal and adult Sertoli cells had essentially the same defects.
RNA expression profiling and follow-up transfection experiments identified several genes misregulated in response to perturbed WT1 in adult Sertoli cells (Rao et al. 2006). Among these were Eps8 and Icap1-α, which encode crucial signalling molecules at the apical ES. Together, these results suggested a model in which WT1 promotes the association of Sertoli cells and germ cells via Rac-mediated signal transduction events that lead to the formation of the apical ES. Future studies will be required to test this model and to elucidate the transcriptional networks downstream of WT1 in Sertoli cells that promote the survival and differentiation of the adjacent germ cells.
(d). RHOX5
Rhox5 is the founding member of the reproductive homeobox on the X chromosome (Rhox) gene cluster (Maclean et al. 2005). This gene cluster contains more than 30 genes in mice, making it the largest homeobox gene cluster known in any species (Jackson et al. 2006; Maclean et al. 2006; Morris et al. 2006; Wang & Zhang 2006; Geyer & Eddy 2008). In adult and postnatal mice, Rhox homeobox genes are preferentially expressed in male and female reproductive tissues, and thus they are likely to encode a large family of homeodomain transcription factors committed to the regulation of reproductive functions. While the roles of most Rhox genes are not yet known, Maclean et al. (2005) recently showed that targeted disruption of Rhox5 causes male germ-cell apoptosis, decreased sperm cell count, impaired sperm motility and subfertility in mice. Because RHOX5 protein is expressed specifically in Sertoli cells within postnatal and adult testis (Lindsey & Wilkinson 1996; Pitman et al. 1998; Sutton et al. 1998), these germ-cell defects are probably caused by a defect in the ability of Sertoli cells to programme or support the germ cells.
To identify downstream genes that mediate Rhox5 function in Sertoli cells, Hu et al. (2010) conducted RNA expression profiling to identify genes differentially expressed in response to forced RHOX5 expression in the 15P-1 Sertoli cell line. A key gene regulated by Rhox5 in 15P-1 cells is Unc5c (Hu et al. 2008). Unc5c is negatively regulated by physiological levels of RHOX5 protein in 15P-1 Sertoli cells and it is also negatively regulated by RHOX5 in vivo, as Rhox5-null testes have elevated levels of Unc5c mRNA. Rhox5 represses Unc5c transcription by acting through a cis element in the Unc5c 5′ untranslated region, but whether RHOX5 acts directly by binding to this element or instead acts through intermediate factors to control Unc5c transcription is not yet known. Unc5c encodes a pro-apoptotic receptor with tumour suppressor activity that was previously best known for its role in neuron development and function in both Caenorhabditis elegans and mice (Leung-Hagesteijn et al. 1992; Ackerman et al. 1997). Hu et al. (2008) established that Unc5c is also crucial for the normal homeostatic control of male germ cells, as loss of Unc5c elicited decreased germ-cell apoptosis in both postnatal and adult mice testes. Together, these data support a model in which RHOX5's pro-germ-cell survival role is mediated in part through its ability to repress the expression of the pro-apoptotic molecule UNC5C.
The Rhox5 gene depends on androgen and androgen receptor (AR) for expression in Sertoli cells (Lindsey & Wilkinson 1996; Maiti et al. 1996; Rao et al. 2003; De Gendt et al. 2004; Bhardwaj et al. 2008). This suggests the possibility that RHOX5 is a transcription factor that mediates some of the actions of testosterone in spermatogenesis. This follows from the fact that Sertoli cells, not germ cells, are responsible for receiving and responding directly to the signals from AR that ultimately drive meiosis and later stages of germ-cell maturation (Chang et al. 2004; De Gendt et al. 2004; Holdcraft & Braun 2004; Maclean & Wilkinson 2005; Tsai et al. 2006). If indeed RHOX5 is responsible for mediating some of the actions of androgen in the testes, it follows that a subset of RHOX5-regulated genes should also be androgen regulated; i.e. they are secondary androgen-response genes (Dean & Sanders 1996). Indeed, Hu et al. (2010) recently identified a set of genes regulated by RHOX5 in Sertoli cell lines and the testes in vivo that are also androgen- and/or AR-regulated in the testes in vivo. Many of these genes, including Unc5c, do not have obvious AR-binding elements in their promoters, suggesting that instead they are indirectly regulated by AR through the ability of AR to induce RHOX5. Future studies will be required to identify the complex network of genes downstream of AR and RHOX5 in Sertoli cells that promote the survival and differentiation of the directly adjacent germ cells.
(e). SOX8
The SRY-related high-mobility group (HMG) box (SOX) transcription factors were discovered by virtue of their having an HMG box, a DNA-binding domain originally identified in SRY, the ‘master’ transcription factor that dictates the formation of the male gonad (Sekido & Lovell-Badge 2009). One of the best-studied SOX proteins, SOX9, acts just downstream of SRY to orchestrate the formation of the testes in mammals. SOX9 and another group-E family member, SOX8, are both expressed in adult Sertoli cells, leading to the possibility that, like WT1 and RHOX5, these SOX transcription factors function in Sertoli cells to drive spermatogenesis. While this has not been addressed in the case of SOX9, recently O'Bryan et al. (2008) examined this issue for SOX8. They found that disruption of the Sox8 gene elicits an age-dependent perturbation of spermatogenesis, characterized by sloughing of spermatocytes and round spermatids, failure in spermiation, disorganization of the spermatogenic cycle and disorientation and inappropriate placement of germ-cell types within the epithelium.
The failure of spermiation and sloughing of germ cells in Sox8-null mice may be the result of defective Sertoli cell–germ cell adhesion in these mutant mice. In normal mice, the apical ES is dissolved between stages VII and VIII of the seminiferous cycle, thereby facilitating the release of the mature spermatids from the tubule. In Sox8-null mice, the breakdown of the apical ES between these stages is impaired, providing a likely explanation for the spermiation failure in these mice. Thus, it appears that, like WT1, SOX8 regulates the expression of genes in Sertoli cells that are crucial for germ cells to adhere to Sertoli cells in a regulated manner. Given that the related factor, SOX9, is also expressed in adult Sertoli cells (Kobayashi et al. 2005), it will be interesting in the future to determine whether SOX9 works together with SOX8 to regulate the association of Sertoli cells with germ cells.
(f). FOXI1
Forkhead box (FOX) proteins are a large family of transcription factors that have diverse roles in many physiological and pathological processes, including ageing and cancer (Fu & Tindall 2008; Partridge & Bruning 2008). While most FOX proteins have been studied in great detail, one that has been largely ignored is FOXI1. To elucidate the role of FOXI1, Blomqvist et al. (2006) generated Foxi1-null mice and found that they have sperm maturation defects in the epididymis such that, following ejaculation, the spermatozoa from these mice fail to migrate through the female genital tract in sufficient numbers to allow fertilization. It is likely that this defect stems from abnormal gene expression in narrow and clear cells, the major proton-secreting cells in the epididymal epithelia necessary for acidification of the luminal environment for sperm activation. Proton secretion in these cells is achieved through a vacuolar H+-ATPase proton pump. Foxi1-null mice lack expression of the B1 subunit of this vacuolar H+-ATPase proton pump, and in vitro transfection experiments strongly suggest that FOXI1 directly regulates the transcription of the B1 subunit gene ATP6V1B1.
This study by Blomqvist et al. (2006) provides strong evidence that FOXI1 is crucial for epididymal epithelial cells to produce the acidic luminal environment required for sperm maturation. By also identifying one of the key downstream targets of FOXI1 responsible for mediating this function, they established a foundation for elucidating the complex transcriptional networks that direct secretory events in the epididymis essential for spermatozoa maturation. It remains for future studies to identify other FOXI1-regulated genes in the epididymis that are important for sperm maturation. One likely candidate is the gene encoding the bicarbonate/chloride transporter pendrin, which, like ATP6V1B1, is positively regulated by FOXI1 in narrow and clear cells in the epididymis (Blomqvist et al. 2006), as well as transfected tissue culture cells (Blomqvist et al. 2004).
3. Chromatin-associated factors that regulate spermatogenesis
In this section, we review recent studies identifying non-DNA-binding transcriptional regulators that control events in spermatogenesis. Rather than binding to DNA directly, this class of factors interacts with DNA-binding factors to gain access to transcriptional control regions. As such, they are part of chromatin. The basic structural unit of chromatin is the nucleosome, which is composed of DNA wrapped around several ubiquitously expressed DNA-binding proteins called histones. The core histones (H2A, H2B, H3 and H4) are small basic proteins that enable compaction of the large eukaryotic genomic DNA to fit into nuclei. Histones undergo a wide variety of post-translational modifications that alter their interaction with DNA as well as with other nuclear proteins, which in turn modulates the transcription rate of the resident genes. Recently, it was discovered that Jumonji-C-domain-containing histone demethylase-2A (JHDM2A), an enzyme that removes methyl groups from specific residues in histone tails, is a key positive regulator of spermatid maturation (figure 2). Here, we discuss this discovery in detail and then go on to discuss two other chromatin-associated factors—PYGO2 and BRDT—that also promote spermatid maturation (figure 2). Interestingly, both of these factors possess domains with the potential to bind to specific histone modifications. PYGO2 has a plant homeodomain (PHD) motif, which typically has the capacity to bind to specific methylated histone residues (Li et al. 2006; Pena et al. 2006; Shi et al. 2006; Matthews et al. 2007), while BRDT has a bromodomain, which in other proteins can bind acetylated histone residues that are known to regulate transcription and chromatin condensation (Matangkasombut & Buratowski 2003; Pivot-Pajot et al. 2003). Finally, we also discuss a chromatin-associated factor that is part of the basal transcription machinery—TAF4B—that was recently shown to have a surprisingly specific role in germ-cell progression and early stages of spermatogenesis.
(a). Jumonji-C-domain-containing histone demethylase-2A
Methylation is a key histone post-translational modification that can either augment or repress transcription, depending on the particular amino acid in the histone tail that undergoes methylation (Martin & Zhang 2005). Different outcomes are achieved by virtue of the ability of different methylated residues to attract different sets of transcriptional effector proteins and chromatin-modifying enzymes. Several years ago, the enzymes responsible for histone methylation—called histone methyltransferases—were discovered (Martin & Zhang 2005). Until recently, it was widely believed that the methyl groups introduced by these enzymes were a stable mark that provided an irreversible signal promoting either repressed or activated transcription (Bannister et al. 2002). This view was erased when enzymes capable of removing the methyl mark—histone demethylases—were discovered (Bannister & Kouzarides 2005; Klose & Zhang 2007). This discovery had two important implications. First, it revealed that regulation of transcription through histone methylation is a reversible process. Second, it indicated that histone methylation had a higher potential for being regulated than previously thought, as not only histone methylation, but also histone demethylation, could be subject to regulation. To begin to understand the in vivo significance of histone demethylation, laboratories have begun to examine the phenotypic consequences of knocking out specific histone demethylase genes. A recent study conducted by Okada et al. (2007) revealed that knockout of one of these genes causes defects in post-meiotic stages of spermatogenesis. They found that targeted mutation of the catalytic domain of JHDM2A (recently renamed lysine demethylase 3A (KDM3A)) led to drastically reduced sperm count and male infertility (Okada et al. 2007). The earliest germ-cell defects occurred in round spermatids, which they found had abnormal morphology and nuclear organization, consistent with the fact that JHDM2A expression normally peaks at this stage. Few round spermatids in Jhdm2a-null mice progressed to the elongated spermatid stage in the testes, and, of the few spermatids that reached the epididymis, most were immotile and had abnormally shaped heads as a result of defective chromatin condensation.
Many post-meiotic events, including chromatin condensation, reorganization of the spermatid nucleus and formation of the acrosome and flagellum, require the synthesis of a number of spermatid-specific histone-like proteins, including transition proteins, protamines and several testis-specific histone variants (Kimmins & Sassone-Corsi 2005). Analysis of the expression of the genes encoding these histone-like and histone variant proteins in Jhdm2a-null mice revealed that JHDM2A specifically regulates the expression of the genes encoding TNP1 and PRM1 (Okada et al. 2007). While it is not certain how JHDM2A accomplishes this, Okada et al. (2007) obtained evidence strongly suggesting that JHDM2A directly regulates these genes by binding to and removing a negative transcriptional mark: the methyl groups from lysine 9 of histone H3 (H3K9). This suggests that JHDM2A is necessary for the proper chromatin reorganization during spermatid maturation by directly promoting the transcription of the Tnp1 and Prm1 genes. Consistent with this model, the phenotype of Jhdm2a-null mice is remarkably similar to the phenotype of mice lacking transition proteins and protamines (Cho et al. 2001; Zhao et al. 2004).
As a caveat, it should be noted that the study conducted by Okada et al. (2007) was limited to only a few genes, and thus it will be important to broaden the scope of the analysis to identify other JHDM2A target genes. It will then be crucial to elucidate which of these target genes control the dramatic alterations in chromatin that are integral to the final stages of male germ-cell maturation. It seems likely that many genes will be involved, as genome-wide studies suggest that H3K9 methylation is widely employed to repress transcription (Barski et al. 2007). Another future challenge will be to identify histone demethylases besides JHDM2A that are important for spermatogenesis. Recently, histone demethylases have been shown to play a role in the germ line and reproductive functions of various model organisms, including fruitflies and C. elegans (Nottke et al. 2009).
(b). PYGO2
Pygopus (Pygo), a member of the PHD-finger protein family, was originally identified on the basis of its being a genetic modifier of Wnt signalling in Drosophila melanogaster (Jessen et al. 2008). The canonical Wnt signalling pathway promotes the stabilization and translocation of β-catenin into the nucleus, where it forms a complex with the LEF/TCF transcription factor to activate the transcription of specific target genes. Pygo serves as a coactivator in this pathway by virtue of its ability to bind and retain β-catenin in the nucleus. In mice, there are two Pygo genes: Pygo1 and Pygo2. While targeted deletion of Pygo1 has no obvious effect on development or fertility (Schwab et al. 2007), targeted deletion of Pygo2 causes severe developmental disorders, including lens agenesis and kidney deformities, which ultimately leads to embryonic mortality (Li et al. 2007; Schwab et al. 2007; Song et al. 2007). While embryonic lethality precluded using Pygo2-null mice to determine whether PYGO2 has a role in spermatogenesis, Nair et al. (2008) recently identified a hypomorphic allele of Pygo2 that proved useful in this regard. They found that mice homozygous for the floxed Pygo2 allele (but that retained the Pygo2 gene because of lack of Cre recombinase) were infertile, had reduced numbers of elongated spermatids and had defects in spermiogenesis, the final stage of spermatid maturation in the testes. Step-9 to -11 elongating spermatids in these Pygo2-mutant mice had incomplete nuclear condensation and low expression of key post-meiotic genes essential for chromatin condensation, including those encoding the histone-replacement proteins PRM1, PRM2 and TNP2, as well as the testis-specific histone H1 family member, H1FNT. Together, these results suggested that that introduction of loxP sites into the Pygo2 locus resulted in a hypomorphic allele that did not impair most of PYGO2's functions but did prevent it from functioning normally in spermiogenesis.
Because the PYGO proteins were originally identified on the basis of their binding to β-catenin and promoting Wnt/β-catenin signalling in flies (Jessen et al. 2008), this leads to the hypothesis that they regulate genes in mammalian germ cells in the same manner. However, Nair et al. (2008) obtained several lines of evidence suggesting that this is probably not the case. First, β-catenin does not copurify with PYGO2 in mice testes. Second, analysis of a Wnt reporter mouse strain indicated that while Wnt signalling occurs in spermatocytes and round spermatids, it does not occur in elongating spermatids, where PYGO2 is expressed and functions. Lastly, round spermatids from Pygo2-hypomorphic mutant testis have normal nuclear levels of β-catenin. The notion that mouse PYGO2 acts independently of Wnt/β-catenin signalling is also supported by an earlier study showing that targeted disruption of Pygo2 causes developmental anomalies in some tissues that do not use the Wnt/β-catenin signalling pathway (Li et al. 2007).
If not through Wnt/β-catenin signalling, how does PYGO2 promote spermiogenesis? Nair et al. (2008) observed that Pygo2-mutant step-9 spermatids had defects in the ability of histone H3 to be acetylated on lysine 9 and 14 (H3K9 and H3K14). This suggested that PYGO2 promotes the global acetylation of these two histone residues, which was further supported by the finding that immunopurified PYGO2 from testes extracts was capable of acetylating both histone H3 and H4 peptide substrates. Given that both H3K9 and H3K14 are associated with transcriptional activation (Shahbazian & Grunstein 2007), this suggests that one mechanism by which PYGO2 activates genes responsible for spermiogenesis is through PYGO2's ability to promote the acetylation of these two particular residues. Whether PYGO2 directly acetylates histones or promotes the action of a histone acetyltransferase (HAT) in germ cells that directly mediates this event is not known.
In addition to eliciting specific histone modifications, PYGO2 may be recruited to particular histone modifications enriched in its target genes. PYGO2 has a PHD motif, a structural domain that typically binds histone H3 tails trimethylated on lysine 4 (H3K4me3), a mark that is strongly associated with transcriptionally active genes (Li et al. 2006; Pena et al. 2006; Shi et al. 2006). Sequence alignment of the PHD motif in the mouse and human PYGO proteins with those in well-studied PHD proteins known to bind to methylated H3 histones indicated that several amino acid residues critical for H3K4me3 binding are conserved in PYGO proteins (Jessen et al. 2008). This indicates that mammalian PYGO proteins may also use their PHD domain to interact with H3K4me3. Together with the evidence that PYGO2 promotes histone acetylation, this suggests a model explaining how PYGO2 transcriptionally activates genes. It binds to genes ‘primed’ for activation by virtue of their being decorated with H3K4me3 and then it promotes histone modifications (H3K9 and H3K4 acetylation) that ultimately lead to transcriptional induction. By analogy, another PHD-finger protein, YNG1, binds to H3K4me3 and promotes histone acetylation by virtue of its associating with HATs (Taverna et al. 2006). Future studies will be required to test this model and to determine whether it explains all of PYGO2's functions in spermatid maturation. PYGO2 may also participate in HAT-independent regulatory mechanisms in germ cells, as PYGO2 not only associates with chromatin, but is in the cytoplasm of elongating and elongated spermatids (Nair et al. 2008).
(c). BRDT
Over 10 years ago, the first enzyme mediating the insertion of acetyl groups into histones was discovered (Brownell et al. 1996; Kuo et al. 1996), which was quickly followed by the discovery of several other HATs (Roth et al. 2001). Acetylation of lysine residues on histone proteins is a major post-translational modification that is important for changes in chromatin organization and for the epigenetic control of gene expression. Acetylated lysine residues are bound by a structural motif called a bromodomain (Yang 2004). Many bromodomain-containing proteins have been identified that are conserved from yeast to humans, including several HATs and transcription coactivators, most of which are involved in transcriptional regulation and/or chromatin remodelling (Zeng & Zhou 2002; Mujtaba et al. 2007). Among these is bromodomain testis-specific protein (BRDT), a member of the bromodomains and extraterminal (BET) subfamily, all of which have two bromodomains and a region of homology in the C-terminus (Florence & Faller 2001; Shang et al. 2007). BRDT is abundantly expressed in the nuclei of primary spermatocytes and round spermatids, and thus it is a good candidate to regulate transcription in germ cells. Indeed, a recent paper by Shang et al. (2007) revealed that loss of BRDT leads to specific defects in male germ cells. These investigators found that Brdt-mutant mice have abnormal elongating spermatids (steps 9–12) that have undergone aberrant morphogenesis. These mice also have aberrant spermatozoa with abnormally shaped heads, low spermatozoa counts and are sterile. While the mechanism responsible for the sperm head abnormalities is not certain, a clue is that the majority of the elongated spermatids in Brdt-mutant mice do not form polarized heterochromatin foci at the nuclear envelope. In normal mice, such heterochromatin foci are believed to facilitate the process of shaping sperm heads (Martianov et al. 2005).
The molecular mechanisms by which BRDT promotes the differentiation of elongated spermatids, including the condensation of their chromatin, is not known. Nonetheless, Shang et al. (2007) identified one downstream gene that might have a role: the testis-specific histone variant gene H1t. They found that Brdt-mutant testes have elevated levels of H1t mRNA and that the H1t promoter is bound by BRDT, which together suggests that the H1t gene is a direct target of BRDT. While H1t is a good candidate to have a role in mediating BRDT's functions in male germ cells, it cannot be sufficient, as H1t-mutant mice have normal spermatogenesis and fertility (Drabent et al. 2000; Lin et al. 2000). Thus, future studies will be required to identify other BRDT target genes and to determine which of these mediates BRDT's ability to promote chromatin remodelling and germ-cell development.
Another issue that has not yet been resolved is the role played by the two bromodomains in BRDT. Human BRDT has been shown to interact with acetylated histone tails in somatic cells, suggesting the possibility that one or both of the bromodomains in BRDT share with classical bromodomains the ability to bind to acetylated histones (Pivot-Pajot et al. 2003). While suggestive, it remains to be definitively shown whether the BRDT bromodomains interact specifically with acetylated histones and, if so, how this promotes the maturation of male germ cells.
(d). TAF4B
A key core component of RNA polymerase II is TFIID, which itself is composed of several subunits, including TATA-binding protein (TBP) and the TBP-associated factors (TAFs). While most TAFs are ubiquitously expressed and are thought to act in all cell types, TAF4B is selectively present in only a few cell types and functions specifically in germ cells. In females, TAF4B is expressed in oocytes, and targeted mutation of Taf4b in mice leads to female infertility and defects in ovarian and oocyte function (Falender et al. 2005b). Likewise, in males, TAF4B is expressed in germ cells and its loss causes infertility. Falender et al. (2005a) found that male infertility is not immediate (it occurs by 11 weeks of age), and it is accompanied by a dramatic depletion of male germ cells and degenerated seminiferous tubules. Infertility appears to be the result of a defect in germ cells, not supporting cells, based on the results of germ-cell transplantation experiments (Falender et al. 2005a). While the full impact of TAF4B loss is not manifest until after the first round of spermatogenesis in adult mice, Falender et al. (2005a) showed that TAF4B initially functions very early in male germ-cell maturation, as there is a striking decrease in spermatogonia in Taf4b-null mice even during the first wave of spermatogenesis. TAF4B may function even earlier—in gonocytes—as it is expressed in these non-dividing cells that serve as the immediate precursors to spermatogonia. Regardless of when precisely it begins to function, TAF4B expression is clearly essential for the survival and/or the proliferation of immature germ cells. It remains for future studies to identify the specific genes regulated by TAF4B and to determine precisely how it controls male germ-cell fate. Given that TAF4B is a core component of Pol II, it will be fascinating to also learn the means by which it directs RNA polymerase II to activate specific genes in germ cells.
4. Rna-binding proteins that regulate spermatogenesis
Protein levels in cells are dictated not only by the rate of transcription but also by the rates of subsequent events, including RNA processing, nuclear RNA export, translation and RNA decay (Bruce & Wilkinson 2005; Gudikote & Wilkinson 2005). These post-transcriptional events are controlled by RNA-binding proteins. In this section, we focus on four RNA-binding proteins that were recently shown to be critical for spermatogenesis (figure 2). Intriguingly, at least three of these proteins promote the translation of subsets of germ-cell mRNAs, reinforcing the long-held view that translational control is an important component of normal spermatogenesis (Kotaja & Sassone-Corsi 2007; Nagamori & Sassone-Corsi 2008). Gonadotrophin-regulated testicular RNA helicase (GRTH) is one of these translation-promoting proteins; it increases the survival of both spermatocytes and round spermatids. Another is Src-associated in mitosis 68 kDa (SAM68); it is required for most germ cells to progress to the spermatid stage; the few germ cells that advance to the point of becoming spermatozoa in the absence of SAM68 have major flagellum defects. The third translation-promoting RNA-binding factor is mouse Y-box protein 2 (MSY2); it controls later events in spermatogenesis, including nuclear condensation in elongating spermatids. The final factor we will discuss in this section is deleted in azoospermia-associated protein-1 (DAZAP1), a nuclear-cytoplasmic shuttling RNA-binding protein that promotes completion of meiosis in the germ cells by an unknown mechanism.
(a). Gonadotrophin-regulated testicular RNA helicase
DEAD-box RNA helicases are multi-functional proteins known to regulate a variety of events, including transcription, RNA unwinding, mRNA nuclear export and translation (Fuller-Pace 2006; Linder 2006, 2008). GRTH is a testis-specific member of the DEAD-box family of RNA helicases that is expressed in meiotic spermatocytes, spermatids and Leydig cells (Dufau & Tsai-Morris 2007). Targeted disruption of Grth in mice causes increased apoptosis of spermatocytes at stage XII of the seminiferous epithelial cycle, arrest of spermatogenesis in late (step 8) round spermatids, azoospermia and male infertility (Tsai-Morris et al. 2004). While it is possible that some or all of these germ-cell defects are a secondary consequence of a defect in Leydig cell function, this seems unlikely because Leydig cells from Grth-null mice have only modest morphological abnormalities, including reduced lipid droplets and swollen mitochondria, and are able to maintain normal circulating levels of testosterone (Tsai-Morris et al. 2004). Thus, GRTH probably promotes the survival of meiotic germ cells in a cell-autonomous manner, allowing them to complete meiosis and undergo differentiation.
Recently, Gutti et al. (2008) obtained evidence how GRTH promotes the survival of spermatocytes. They found that deletion of Grth reduces the levels of anti-apoptotic proteins (e.g. BCL-XL and BCL2) and elevates the levels of pro-apoptotic proteins (e.g. BID, BAD and BAK) in germ cells. Not only were apoptosis proteins increased in level in Grth-lacking germ cells, but events crucial for active apoptosis were triggered: cleavage and/or activation of caspase 9, caspase 3 and poly(ADP-ribose) polymerase.
How does GRTH regulate the expression of apoptotic and anti-apoptotic proteins? While the answer to this question is not yet known, a clue has come from studies demonstrating that GRTH regulates gene expression at the level of translation. The first evidence for this was the discovery that Grth-null mice have reduced levels of several germ-cell proteins but have normal levels of the corresponding mRNAs. For example, the abundance of TNPs and testis-specific angiotensin-converting enzyme is reduced in testes from these null mice without a decrease in their corresponding mRNAs (Tsai-Morris et al. 2004). This suggests that GRTH either stabilizes these proteins or stimulates the translation of their mRNAs. Evidence for the latter is that GRTH associates with polyribosomes in testis extracts (Sheng et al. 2006) and recombinant GRTH stimulates the rate of translation of specific mRNAs in vitro (Tang et al. 1999).
While it appears likely that GRTH selectively associates with a subset of mRNAs in germ cells and stimulates their translation, it remains for future studies to conclusively demonstrate this and to determine how it is achieved. One clue is that GRTH has RNA unwinding activity in the presence of ATP (Tang et al. 1999), and thus GRTH may stimulate translation by removing inhibitory RNA secondary structure at the 5′ end of its target mRNAs. In addition to stimulating translation, GRTH may also promote the export of specific mRNAs from the nucleus. The evidence for this is that the cytoplasmic/total RNA ratios for Pgk2, tAce and Tnp2 mRNA are selectively reduced in Grth-null round spermatids (Sheng et al. 2006). How GRTH promotes the nuclear export of these mRNAs and whether this function requires GRTH's RNA helicase activity remain to be determined. An intriguing finding that may bear on GRTH's roles in both nuclear export and translation is the discovery that Grth-null round spermatids have drastically smaller chromatoid bodies (CBs) (Tsai-Morris et al. 2004). CBs are non-membranous, cytoplasmic foci that contain many factors involved in RNA processing and decay (Nagamori & Sassone-Corsi 2008). While their function is not known, CBs are likely to serve as sites of RNA decay and translation repression in germ cells, just as P-bodies do in somatic cells (Parker & Sheth 2007). By virtue of its ability to dictate the formation of CBs, GRTH is likely to be a major regulatory factor controlling mRNA metabolism and translation in differentiating germ cells.
(b). SAM68
KH-type RNA-binding proteins are intriguing molecules that regulate the differentiation and function of several cell types and influence tumour development (Busa et al. 2007; Lukong et al. 2008). A well-studied member of this family, SAM68, is a ubiquitously expressed factor that regulates many steps of RNA metabolism, including alternative mRNA splicing, nuclear mRNA export and translation. Thus, SAM68 is one of a growing class of RNA-binding proteins that have roles in both the nucleus and the cytoplasm (Shyu & Wilkinson 2000; Wilkinson & Shyu 2001). Given its widespread expression and its multi-functional activities, it was anticipated that loss of SAM68 would lead to embryonic lethality and broad developmental defects. In contrast to this prediction, Sam68-null mice typically survive to adulthood and actually exhibit some advantageous characteristics, including protection from both age-dependent bone loss and mammary gland tumours (Richard et al. 2005, 2008). However, Sam68-null mice do have some defects, including few adipocytes in the bone marrow, suboptimal motor coordination and male sterility (Richard et al. 2005; Lukong & Richard 2008). Recently, Paronetto et al. (2009) investigated the cellular and molecular basis for the sterility of Sam68-null males. They found that adult Sam68-null males have seminiferous tubules largely devoid of round and elongated spermatids. Although they did not determine the precise developmental stage of this defect, they found that the defect occurs between P16 and P25, a developmental window when germ cells complete meiosis and differentiate into round spermatids. Accompanying this loss of spermatids was an increase in postnatal germ-cell apoptosis and various aberrations in elongated spermatids detected by electron microscopy. Few epididymal spermatozoa were present in these null mice, and those that accumulated were mostly abnormally shaped and immotile. Spermatozoa that were initially motile typically lost their flagellum during culture in fertilization medium, suggesting that SAM68 is necessary for the production of components necessary to generate an intact flagellum.
Given that SAM68 is an RNA-binding protein, Paronetto et al. (2009) investigated whether its biological role in germ cells revolved around its ability to bind RNA. Indeed, they obtained evidence that this was the case. Using a ribonucleoprotein particle (RNP) capture assay, they showed that SAM68 associates with bulk poly (A)+ RNA in pachytene spermatocytes. Coimmunoprecipitation experiments coupled with RNA expression profiling identified specific transcripts both bound and regulated in level by SAM68. The authors did not determine how SAM68 regulates the levels of these mRNAs, but instead they obtained several lines of evidence that SAM68 promotes the translation of a subset of germ-cell mRNAs. First, SAM68 cosediments with polysomes. Second, SAM68 interacts with poly(A)+ mRNAs in the polysome fractions. Third, Sam68-null mice exhibit a shift in the polysome profile of SAM68 target mRNAs consistent with their being translated less efficiently. Fourth, Sam68-null mice express reduced levels of proteins encoded by these target mRNAs. Finally, the 3′ untranslated region of one of the SAM68 targets, Spag16, is sufficient to confer SAM68-dependent upregulation on a reporter construct in transfected cells. Interestingly, Spag16 encodes a component of the sperm axoneme whose loss reduces sperm motility and causes infertility (Zhang et al. 2006), and thus it may be a key downstream target of SAM68 responsible for its role in promoting sperm motility.
In the future, it will be important to directly address this possibility, as well as investigate the functional role of the approximately 400 other transcripts regulated in level by SAM68 in mouse testes. It will also be crucial to disentangle which of these are direct versus indirect targets of SAM68. The molecular mechanism of the action of SAM68 in germ cells requires further scrutiny. Clearly, SAM68 does not only serve to promote translation, but also regulates the level of its target transcripts at either the transcriptional or post-transcriptional level. With regard to the latter, SAM68 is known to regulate alternative splicing in somatic cells (Matter et al. 2002; Chawla et al. 2009), and thus it may do the same in germ cells and thereby control the level of specific mRNA isoforms. Alternatively, SAM68 might control the stability of its target mRNAs. Finally, it will be important to know why SAM68 selectively targets only a specific subset of transcripts in germ cells and what dictates the specific effects it has on a given mRNA in a given cell type.
(c). Mouse Y-box protein 2
Y-box proteins were originally identified as DNA-binding proteins that use their conserved cold shock domain to associate with Y-box consensus elements in DNA (Matsumoto & Wolffe 1998). Later, it was discovered that these proteins are also RNA-binding proteins that first interact with mRNAs when they are transcribed in the nucleus. Many Y-box proteins then travel with mRNAs to the cytoplasm, where they regulate their fate (Wilkinson & Shyu 2001). One of the best-studied Y-box proteins, FRGY2, is present at high levels in the cytoplasm of Xenopus laevis oocytes, where it functions to block maternal mRNAs from being prematurely translated (Tafuri & Wolffe 1993; Bouvet & Wolffe 1994). Mouse MSY2, which was originally identified by virtue of its sequence identity with X. laevis FRGY2 (Gu et al. 1998), is specifically expressed in mouse male and female germ cells (Xu & Hecht 2008). To elucidate the functional role of MSY2 in germ cells, Yang et al. (2005b) generated Msy2-null mice. They found that the mutant males had an abnormally high number of apoptotic meiotic spermatocytes, leading to a massive loss of step 8/9 spermatids, and also had misshapen and multi-nucleated spermatids, lacked spermatozoa in the epididymis and were sterile (Yang et al. 2007). Similarly, the mutant females had defects in ovulation, premature loss of oocytes and were infertile (Yang et al. 2005b).
To begin to understand the molecular role of MSY2 in spermatogenesis, Yang et al. (2005a) identified mRNAs specifically bound by MSY2 in male germ cells. They found that the majority of the MSY2-bound mRNAs had a Y-box DNA sequence motif in the promoter region of their respective genes. This suggested the intriguing possibility that MSY2 is initially recruited to specific promoters, and then later is bound to the mRNAs transcribed from these promoters. This provides an efficient and highly selective means to control gene expression, and it is supported by the fact that MSY2, like FRGY2, can bind both DNA and RNA (Matsumoto & Wolffe 1998). Among the genes likely to be controlled by this mechanism are Prm1 and Tnp2, as both have Y-boxes in their promoters and both exhibit drastically reduced expression in Msy2-null testes. Because PRM1 and TNP2 are crucial for the structural reorganization of spermatid chromatin during the final stages of spermatogenesis, their loss of expression is likely to contribute to the nuclear condensation defects that occur in Msy2-null late-stage spermatids.
Because MSY2 interacts with both DNA and RNA, it could potentially act at many different levels to regulate its targets. In follow-up work to their original mouse knockout paper, Yang et al. (2007) investigated this issue. They obtained evidence that loss of Msy2 does not affect transcription, mRNA splicing or mRNA intracellular transport, but instead has a selective effect on the rate of translation. In particular, loss of Msy2 causes a subset of MSY2-bound mRNAs to shift from RNPs to polysomes. This suggests that MSY2 normally serves to repress translation of a subset of germ-cell mRNAs. It will be interesting to identify the signals that release this repression, thereby inducing the translation of MSY2-bound mRNAs at the appropriate time during germ-cell development. Recently, Medvedev et al. (2008) demonstrated that phosphorylation of MSY2 by cyclin dependent kinase-1 triggers the degradation of several oocyte-stored mRNAs. It will be important to determine whether male germ cells employ a similar mechanism to regulate the translation of MSY2-bound paternal mRNAs.
(d). DAZAP1
Heterogeneous nuclear ribonuclear proteins (hnRNPs) are a family of RNA-binding proteins that coat newly synthesized RNAs and participate in a wide variety of events, including mRNA splicing, nuclear mRNA export, and translation (Perrotti & Calabretta 2002; Martinez-Contreras et al. 2007). Most hnRNP family members have two RNP-type RNA-binding domains near the N-terminus and a glycine- or proline-rich domain near the C-terminus. While many members of the hnRNP family were discovered two or more decades ago, a family member relevant to spermatogenesis was isolated more recently. This hnRNP protein—DAZAP1—was isolated on the basis of its interaction with DAZ, a germ-cell-specific RNA-binding protein encoded on the Y chromosome that is deleted in 10 per cent of infertile men with idiopathic azoospermia (Reijo et al. 1995, 1996; Tsui et al. 2000). To determine the in vivo function of DAZAP1, Hsu et al. (2008) generated Dazap1-mutant mice carrying either a floxed neomycin resistant (Fn) hypomorphic allele or a null allele. Both alleles caused widespread developmental defects, including perinatal lethality, retarded growth, obesity and male infertility. To investigate the underlying basis for this male infertility, the investigators used mice with the Fn hypomorphic allele, as the null mice die before two months of age. They found that Fn-mutant mice had testes with significantly increased numbers of apoptotic male germ cells, an accumulation of post-pachytene spermatocytes and a complete absence of post-meiotic germ cells (i.e. spermatids). Similar defects were observed in testes from Dazap1-null mice that survived until four to five weeks of age, providing strong evidence that these defects are indeed caused by loss of DAZAP1 function.
The molecular basis for DAZAP1 action is not known, but like most hnRNP proteins, DAZAP1 has the ability to shuttle between the nucleus and the cytoplasm (Lin & Yen 2006), and thus it may promote the nuclear export of the mRNAs that it binds. It has a strong affinity for RNA in vitro, but whether it has selectivity for binding particular mRNAs in vivo is not known (Hori et al. 2005). Like other hnRNP proteins, DAZAP1 could also regulate many other post-transcriptional events. At steady state, DAZAP1 is predominantly in the nuclei of pachytene spermatocytes and round spermatids, but shifts to the cytoplasm in spermatids when they are undergoing elongation and their transcription is shutting down (Vera et al. 2002). It will be intriguing to determine whether its function concomitantly shifts from regulating nuclear events to cytoplasmic events.
5. Perspective
Spermatogenesis has been studied for decades, but the molecular mechanisms involved in the progression of spermatogonia to mature motile spermatozoa are poorly understood. Such mechanisms are crucial to understand, as they may lead to cures of male infertility as well as the development of new male contraception approaches. In this review, we focused on recent advances in our understanding of molecular pathways that control spermatogenesis. We highlighted transcription factors that act either in germ cells or in somatic cells (such as Sertoli cells) to control spermatogenesis. Analysis of the transcription factors that have been studied to date suggests that those expressed in germ cells are more likely to regulate specific phases of germ-cell progression, while Sertoli cell-expressed transcription factors have roles that are often required through all stages of gametogenesis in seminiferous tubules. Given that there are many stages of germ-cell development, involving mitosis, meiosis and post-meiotic differentiation events, all of which are required to generate mature spermatozoa, it is likely that a vast number of germ-cell-expressed transcription factors are necessary to control and coordinate these events. At present, only a handful of transcription factors have been identified that are involved in a given spermatogenic stage, and thus the field clearly has a long way to go to complete the picture. Furthermore, redundant and regulatory relationships between different transcription factors acting at most stages of spermatogenesis remain largely unknown. A further complicating factor is that most transcription factors with identified roles in the testis also have roles (or are expressed) in other tissues. To address this problem, tissue- and cell-type-specific knockout and knockdown approaches will need to be used with a greater repertoire of cell-type- and stage-specific promoters than are currently available. The identification and study of testes-specific transcription factors is another avenue worth exploring, as in vivo analysis of their function will not be confounded by effects in other tissues.
Regulation of gene expression in post-meiotic male germ cells is controlled by unique mechanisms that are specific to these cells. As spermatids undergo differentiation, their nuclei become largely transcriptionally inactive as a result of replacement of histones with highly basic TNPs and RNPs, extensive nuclear remodelling and compaction of the chromatin (Sassone-Corsi 2002). The recent identification and functional analysis of chromatin regulators in male germ-cell biology has impacted our understanding of these events. As described in this review, these chromatin regulators control key epigenetic events, such as histone modifications, which in turn lead to potent changes in transcription rates in cells. Unfortunately, very few chromatin-associated factors that function in postnatal and adult germs cells have so far been identified, and thus great strides will be necessary to fill this gap so that we can advance our understanding of the epigenetic mechanisms that regulate germ-cell development.
In addition to transcriptional regulation, post-transcriptional regulatory mechanisms are crucial for normal germ-cell development. For example, mRNAs encoding proteins needed for the terminal stages of spermatogenesis must be stabilized because late-stage germ cells undergo chromatin compaction events that halt ongoing transcription. Very little is known about how this mRNA stabilization event is achieved in germ cells. Instead, most progress has been made towards understanding how another post-transcriptional event is regulated in germ cells: translation. Translational regulation is extremely important in late-stage germ cells, as transcription has ceased, and thus it cannot contribute to regulatory control. Finally, the recent identification of non-coding regulatory RNAs in the testes (He et al. 2009) has added yet another level of complexity to spermatogenesis. Clearly, we are a long way from unravelling the complex multi-layered molecular circuits that control germ cell development and function.
Footnotes
One contribution of 17 to a Theme Issue ‘The biology and regulation of spermatogenesis’.
References
- Ackerman S. L., Kozak L. P., Przyborski S. A., Rund L. A., Boyer B. B., Knowles B. B.1997The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature 386, 838–842 (doi:10.1038/386838a0) [DOI] [PubMed] [Google Scholar]
- Akerfelt M., Henriksson E., Laiho A., Vihervaara A., Rautoma K., Kotaja N., Sistonen L.2008Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc. Natl Acad. Sci. USA 105, 11 224–11 229 (doi:10.1073/pnas.0800620105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T.2005Reversing histone methylation. Nature 436, 1103–1106 (doi:10.1038/nature04048) [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Schneider R., Kouzarides T.2002Histone methylation: dynamic or static? Cell 109, 801–806 (doi:10.1016/S0092-8674(02)00798-5) [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K.2007High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (doi:10.1016/j.cell.2007.05.009) [DOI] [PubMed] [Google Scholar]
- Bhardwaj A., Rao M. K., Kaur R., Buttigieg M. R., Wilkinson M. F.2008GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Mol. Cell. Biol. 28, 2138–2153 (doi:10.1128/MCB.01170-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist S. R., Vidarsson H., Fitzgerald S., Johansson B. R., Ollerstam A., Brown R., Persson A. E., Bergstrom G. G., Enerback S.2004Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J. Clin. Invest. 113, 1560–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist S. R., Vidarsson H., Soder O., Enerback S.2006Epididymal expression of the forkhead transcription factor Foxi1 is required for male fertility. EMBO J. 25, 4131–4141 (doi:10.1038/sj.emboj.7601272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Wolffe A. P.1994A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77, 931–941 (doi:10.1016/0092-8674(94)90141-4) [DOI] [PubMed] [Google Scholar]
- Brennan J., Capel B.2004One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 5, 509–521 (doi:10.1038/nrg1381) [DOI] [PubMed] [Google Scholar]
- Brownell J. E., Zhou J., Ranalli T., Kobayashi R., Edmondson D. G., Roth S. Y., Allis C. D.1996Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 (doi:10.1016/S0092-8674(00)81063-6) [DOI] [PubMed] [Google Scholar]
- Bruce S., Wilkinson M. F.2005RNA stability. In Encyclopedic reference of genomics and proteomics in molecular medicine (eds Ganten D., Ruckpaul K.). Berlin, Germany: Springer International [Google Scholar]
- Busa R., Paronetto M. P., Farini D., Pierantozzi E., Botti F., Angelini D. F., Attisani F., Vespasiani G., Sette C.2007The RNA-binding protein SAM68 contributes to proliferation and survival of human prostate cancer cells. Oncogene 26, 4372–4382 (doi:10.1038/sj.onc.1210224) [DOI] [PubMed] [Google Scholar]
- Chang C., Chen Y. T., Yeh S. D., Xu Q., Wang R. S., Guillou F., Lardy H., Yeh S.2004Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc. Natl Acad. Sci. USA 101, 6876–6881 (doi:10.1073/pnas.0307306101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G., Lin C. H., Han A., Shiue L., Ares M., Jr, Black D. L.2009Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol. Cell. Biol. 29, 201–213 (doi:10.1128/MCB.01349-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C., Willis W. D., Goulding E. H., Jung-Ha H., Choi Y. C., Hecht N. B., Eddy E. M.2001Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat. Genet. 28, 82–86 (doi:10.1038/88313) [DOI] [PubMed] [Google Scholar]
- Dai X., Schonbaum C., Degenstein L., Bai W., Mahowald A., Fuchs E.1998The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12, 3452–3463 (doi:10.1101/gad.12.21.3452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. M., Sanders M. M.1996Ten years after: reclassification of steroid-responsive genes. Mol. Endocrinol. 10, 1489–1495 (doi:10.1210/me.10.12.1489) [DOI] [PubMed] [Google Scholar]
- De Gendt K., et al. 2004A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc. Natl Acad. Sci. USA 101, 1327–1332 (doi:10.1073/pnas.0308114100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabent B., Saftig P., Bode C., Doenecke D.2000Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem. Cell. Biol. 113, 433–442 [DOI] [PubMed] [Google Scholar]
- Dufau M. L., Tsai-Morris C. H.2007Gonadotropin-regulated testicular helicase (GRTH/DDX25): an essential regulator of spermatogenesis. Trends Endocrinol. Metab. 18, 314–320 (doi:10.1016/j.tem.2007.09.001) [DOI] [PubMed] [Google Scholar]
- Falender A. E., Freiman R. N., Geles K. G., Lo K. C., Hwang K., Lamb D. J., Morris P. L., Tjian R., Richards J. S.2005aMaintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 19, 794–803 (doi:10.1101/gad.1290105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falender A. E., Shimada M., Lo Y. K., Richards J. S.2005bTAF4b, a TBP associated factor, is required for oocyte development and function. Dev. Biol. 288, 405–419 (doi:10.1016/j.ydbio.2005.09.038) [DOI] [PubMed] [Google Scholar]
- Florence B., Faller D. V.2001You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 6, D1008–D1018 (doi:10.2741/Florence) [DOI] [PubMed] [Google Scholar]
- Fu Z., Tindall D. J.2008FOXOs, cancer and regulation of apoptosis. Oncogene 27, 2312–2319 (doi:10.1038/onc.2008.24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V.2006DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34, 4206–4215 (doi:10.1093/nar/gkl460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Maiti S., Alam N., Zhang Z., Deng J. M., Behringer R. R., Lecureuil C., Guillou F., Huff V.2006The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proc. Natl Acad. Sci. USA 103, 11 987–11 992 (doi:10.1073/pnas.0600994103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer C. B., Eddy E. M.2008Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene 423, 194–200 (doi:10.1016/j.gene.2008.06.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Tekur S., Reinbold R., Eppig J. J., Choi Y. C., Zheng J. Z., Murray M. T., Hecht N. B.1998Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol. Reprod. 59, 1266–1274 (doi:10.1095/biolreprod59.5.1266) [DOI] [PubMed] [Google Scholar]
- Gudikote J., Wilkinson M. F.2005RNA-binding proteins: regulation of mRNA splicing, export and decay. In Encyclopedia of life sciences New York, NY: John Wiley & Sons [Google Scholar]
- Gutti R. K., Tsai-Morris C. H., Dufau M. L.2008Gonadotropin-regulated testicular helicase (DDX25), an essential regulator of spermatogenesis, prevents testicular germ cell apoptosis. J. Biol. Chem. 283, 17 055–17 064 (doi:10.1074/jbc.M708449200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Kokkinaki M., Pant D., Gallicano G. I., Dym M.2009Small RNA molecules in the regulation of spermatogenesis. Reproduction 137, 901–911 (doi:10.1530/REP-08-0494) [DOI] [PubMed] [Google Scholar]
- Holdcraft R. W., Braun R. E.2004Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 131, 459–467 (doi:10.1242/dev.00957) [DOI] [PubMed] [Google Scholar]
- Hori T., Taguchi Y., Uesugi S., Kurihara Y.2005The RNA ligands for mouse proline-rich RNA-binding protein (mouse PRRP) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 33, 190–200 (doi:10.1093/nar/gki153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. C., Chen H. Y., Lin Y. W., Chu W. C., Lin M. J., Yan Y. T., Yen P. H.2008DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA 14, 1814–1822 (doi:10.1261/rna.1152808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Shanker S., Maclean J. A., 2nd, Ackerman S. L., Wilkinson M. F.2008The RHOX5 homeodomain protein mediates transcriptional repression of the netrin-1 receptor gene Unc5c. J. Biol. Chem. 283, 3866–3876 (doi:10.1074/jbc.M706717200) [DOI] [PubMed] [Google Scholar]
- Hu Z., Dandekar D., O'Shaughnessy P. J., De Gendt K., Verhoeven G., Wilkinson M. F.2010Androgen-induced Rhox homeobox genes modulate the expression of AR-regulated genes. Mol. Endocrinol. 24, 60–75 (doi:10.1210/me.2009-0303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M., et al. 2006A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genomics 7, 212 (doi:10.1186/1471-2164-7-212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen S., Gu B., Dai X.2008Pygopus and the Wnt signaling pathway: a diverse set of connections. Bioessays 30, 448–456 (doi:10.1002/bies.20757) [DOI] [PubMed] [Google Scholar]
- Kallio M., et al. 2002Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 21, 2591–2601 (doi:10.1093/emboj/21.11.2591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S., Sassone-Corsi P.2005Chromatin remodelling and epigenetic features of germ cells. Nature 434, 583–589 (doi:10.1038/nature03368) [DOI] [PubMed] [Google Scholar]
- Klose R. J., Zhang Y.2007Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell. Biol. 8, 307–318 (doi:10.1038/nrm2143) [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Chang H., Chaboissier M. C., Schedl A., Behringer R. R.2005Sox9 in testis determination. Ann. N. Y. Acad. Sci. 1061, 9–17 (doi:10.1196/annals.1336.003) [DOI] [PubMed] [Google Scholar]
- Kotaja N., Sassone-Corsi P.2007The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell. Biol. 8, 85–90 (doi:10.1038/nrm2081) [DOI] [PubMed] [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D., Jaenisch R.1993WT-1 is required for early kidney development. Cell 74, 679–691 (doi:10.1016/0092-8674(93)90515-R) [DOI] [PubMed] [Google Scholar]
- Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D.1996Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 (doi:10.1038/383269a0) [DOI] [PubMed] [Google Scholar]
- Leung-Hagesteijn C., Spence A. M., Stern B. D., Zhou Y., Su M. W., Hedgecock E. M., Culotti J. G.1992UNC-5, a transmembrane protein with immunoglobulin and thrombospondin type 1 domains, guides cell and pioneer axon migrations in C. elegans. Cell 71, 289–299 (doi:10.1016/0092-8674(92)90357-I) [DOI] [PubMed] [Google Scholar]
- Li B., et al. 2005Ovol1 regulates meiotic pachytene progression during spermatogenesis by repressing Id2 expression. Development 132, 1463–1473 (doi:10.1242/dev.01658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., Patel D. J.2006Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., et al. 2007Developmental phenotypes and reduced Wnt signaling in mice deficient for pygopus 2. Genesis 45, 318–325 (doi:10.1002/dvg.20299) [DOI] [PubMed] [Google Scholar]
- Lin Y. T., Yen P. H.2006A novel nucleocytoplasmic shuttling sequence of DAZAP1, a testis-abundant RNA-binding protein. RNA 12, 1486–1493 (doi:10.1261/rna.42206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Sirotkin A., Skoultchi A. I.2000Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol. Cell. Biol. 20, 2122–2128 (doi:10.1128/MCB.20.6.2122-2128.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P.2006Dead-box proteins: a family affair-active and passive players in RNP-remodeling. Nucleic Acids Res. 34, 4168–4180 (doi:10.1093/nar/gkl468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P.2008mRNA export: RNP remodeling by DEAD-box proteins. Curr. Biol. 18, R297–R299 (doi:10.1016/j.cub.2008.02.027) [DOI] [PubMed] [Google Scholar]
- Lindsey J. S., Wilkinson M. F.1996Pem: a testosterone- and LH-regulated homeobox gene expressed in mouse Sertoli cells and epididymis. Dev. Biol. 179, 471–484 (doi:10.1006/dbio.1996.0276) [DOI] [PubMed] [Google Scholar]
- Lukong K. E., Richard S.2008Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav. Brain Res. 189, 357–363 (doi:10.1016/j.bbr.2008.01.010) [DOI] [PubMed] [Google Scholar]
- Lukong K. E., Chang K. W., Khandjian E. W., Richard S.2008RNA-binding proteins in human genetic disease. Trends Genet. 24, 416–425 (doi:10.1016/j.tig.2008.05.004) [DOI] [PubMed] [Google Scholar]
- Maclean J. A., 2nd, Wilkinson M. F.2005Gene regulation in spermatogenesis. Curr. Top Dev. Biol. 71, 131–197 [DOI] [PubMed] [Google Scholar]
- Maclean J. A., 2nd, Chen M. A., Wayne C. M., Bruce S. R., Rao M., Meistrich M. L., Macleod C., Wilkinson M. F.2005Rhox: a new homeobox gene cluster. Cell 120, 369–382 [DOI] [PubMed] [Google Scholar]
- Maclean J. A., Lorenzetti D., Hu Z., Salerno W. J., Miller J., Wilkinson M. F.2006Rhox homeobox gene cluster: recent duplication of three family members. Genesis 44, 122–129 (doi:10.1002/gene.20193) [DOI] [PubMed] [Google Scholar]
- Maiti S., Doskow J., Li S., Nhim R. P., Lindsey J. S., Wilkinson M. F.1996The Pem homeobox gene. Androgen-dependent and -independent promoters and tissue-specific alternative RNA splicing. J. Biol. Chem. 271, 17 536–17 546 [DOI] [PubMed] [Google Scholar]
- Martianov I., Brancorsini S., Catena R., Gansmuller A., Kotaja N., Parvinen M., Sassone-Corsi P., Davidson I.2005Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl Acad. Sci. USA 102, 2808–2813 (doi:10.1073/pnas.0406060102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Zhang Y.2005The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell. Biol. 6, 838–849 (doi:10.1038/nrm1761) [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J. F., Revil T., Chabot B.2007hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623, 123–147 (doi:10.1007/978-0-387-77374-2_8) [DOI] [PubMed] [Google Scholar]
- Matangkasombut O., Buratowski S.2003Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11, 353–363 (doi:10.1016/S1097-2765(03)00033-9) [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Wolffe A. P.1998Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol 8, 318–323 (doi:10.1016/S0962-8924(98)01300-2) [DOI] [PubMed] [Google Scholar]
- Matter N., Herrlich P., Konig H.2002Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420, 691–695 (doi:10.1038/nature01153) [DOI] [PubMed] [Google Scholar]
- Matthews A. G., et al. 2007RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 (doi:10.1038/nature06431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev S., Yang J., Hecht N. B., Schultz R. M.2008CDC2A (CDK1)-mediated phosphorylation of MSY2 triggers maternal mRNA degradation during mouse oocyte maturation. Dev. Biol. 321, 205–215 (doi:10.1016/j.ydbio.2008.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L., Gordon J., Blackburn C. C.2006Identification of a tandem duplicated array in the Rhox alpha locus on mouse chromosome X. Mamm. Genome 17, 178–187 (doi:10.1007/s00335-005-0138-4) [DOI] [PubMed] [Google Scholar]
- Mujtaba S., Zeng L., Zhou M. M.2007Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26, 5521–5527 (doi:10.1038/sj.onc.1210618) [DOI] [PubMed] [Google Scholar]
- Nagamori I., Sassone-Corsi P.2008The chromatoid body of male germ cells: epigenetic control and miRNA pathway. Cell Cycle 7, 3503–3508 [DOI] [PubMed] [Google Scholar]
- Nair M., Nagamori I., Sun P., Mishra D. P., Rheaume C., Li B., Sassone-Corsi P., Dai X.2008Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev. Biol. 320, 446–455 (doi:10.1016/j.ydbio.2008.05.553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottke A., Colaiacovo M. P., Shi Y.2009Developmental roles of the histone lysine demethylases. Development 136, 879–889 (doi:10.1242/dev.020966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bryan M. K., et al. 2008Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev. Biol. 316, 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Scott G., Ray M. K., Mishina Y., Zhang Y.2007Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123 (doi:10.1038/nature06236) [DOI] [PubMed] [Google Scholar]
- Park S. Y., Jameson J. L.2005Minireview: transcriptional regulation of gonadal development and differentiation. Endocrinology 146, 1035–1042 (doi:10.1210/en.2004-1454) [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U.2007P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 (doi:10.1016/j.molcel.2007.02.011) [DOI] [PubMed] [Google Scholar]
- Paronetto M. P., et al. 2009Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 185, 235–249 (doi:10.1083/jcb.200811138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Bruning J. C.2008Forkhead transcription factors and ageing. Oncogene 27, 2351–2363 (doi:10.1038/onc.2008.28) [DOI] [PubMed] [Google Scholar]
- Pena P. V., Davrazou F., Shi X., Walter K. L., Verkhusha V. V., Gozani O., Zhao R., Kutateladze T. G.2006Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D., Calabretta B.2002Post-transcriptional mechanisms in BCR/ABL leukemogenesis: role of shuttling RNA-binding proteins. Oncogene 21, 8577–8583 (doi:10.1038/sj.onc.1206085) [DOI] [PubMed] [Google Scholar]
- Pirkkala L., Nykanen P., Sistonen L.2001Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15, 1118–1131 (doi:10.1096/fj00-0294rev) [DOI] [PubMed] [Google Scholar]
- Pitman J. L., Lin T. P., Kleeman J. E., Erickson G. F., MacLeod C. L.1998Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Dev. Biol. 202, 196–214 (doi:10.1006/dbio.1998.8978) [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C., Caron C., Govin J., Vion A., Rousseaux S., Khochbin S.2003Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol. Cell. Biol. 23, 5354–5365 (doi:10.1128/MCB.23.15.5354-5365.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. K., Wayne C. M., Meistrich M. L., Wilkinson M. F.2003Pem homeobox gene promoter sequences that direct transcription in a Sertoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. Mol. Endocrinol. 17, 223–233 (doi:10.1210/me.2002-0232) [DOI] [PubMed] [Google Scholar]
- Rao M. K., Pham J., Imam J. S., Maclean J. A., Murali D., Furuta Y., Sinha-Hikim A. P., Wilkinson M. F.2006Tissue-specific RNAi reveals that WT1 expression in nurse cells controls germ cell survival and spermatogenesis. Genes Dev. 20, 147–152 (doi:10.1101/gad1367806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo R., et al. 1995Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 10, 383–393 (doi:10.1038/ng0895-383) [DOI] [PubMed] [Google Scholar]
- Reijo R., Alagappan R. K., Patrizio P., Page D. C.1996Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet 347, 1290–1293 (doi:10.1016/S0140-6736(96)90938-1) [DOI] [PubMed] [Google Scholar]
- Richard S., et al. 2005Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 1, e74 (doi:10.1371/journal.pgen.0010074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S., Vogel G., Huot M. E., Guo T., Muller W. J., Lukong K. E.2008Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene 27, 548–556 (doi:10.1038/sj.onc.1210652) [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Denu J. M., Allis C. D.2001Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 (doi:10.1146/annurev.biochem.70.1.81) [DOI] [PubMed] [Google Scholar]
- Sarge K. D., Zimarino V., Holm K., Wu C., Morimoto R. I.1991Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 5, 1902–1911 (doi:10.1101/gad.5.10.1902) [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P.2002Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176–2178 (doi:10.1126/science.1070963) [DOI] [PubMed] [Google Scholar]
- Schwab K. R., Patterson L. T., Hartman H. A., Song N., Lang R. A., Lin X., Potter S. S.2007Pygo1 and Pygo2 roles in Wnt signaling in mammalian kidney development. BMC Biol. 5, 15 (doi:10.1186/1741-7007-5-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R., Lovell-Badge R.2009Sex determination and SRY: down to a wink and a nudge? Trends Genet. 25, 19–29 (doi:10.1016/j.tig.2008.10.008) [DOI] [PubMed] [Google Scholar]
- Shahbazian M. D., Grunstein M.2007Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (doi:10.1146/annurev.biochem.76.052705.162114) [DOI] [PubMed] [Google Scholar]
- Shang E., Nickerson H. D., Wen D., Wang X., Wolgemuth D. J.2007The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development 134, 3507–3515 (doi:10.1242/dev.004481) [DOI] [PubMed] [Google Scholar]
- Sheng Y., Tsai-Morris C. H., Gutti R., Maeda Y., Dufau M. L.2006Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J. Biol. Chem. 281, 35 048–35 056 (doi:10.1074/jbc.M605086200) [DOI] [PubMed] [Google Scholar]
- Shi X., et al. 2006ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (doi:10.1038/nature04835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A. B., Wilkinson M. F.2000The double lives of shuttling mRNA binding proteins. Cell 102, 135–138 (doi:10.1016/S0092-8674(00)00018-0) [DOI] [PubMed] [Google Scholar]
- Song N., Schwab K. R., Patterson L. T., Yamaguchi T., Lin X., Potter S. S., Lang R. A.2007Pygopus 2 has a crucial, Wnt pathway-independent function in lens induction. Development 134, 1873–1885 (doi:10.1242/dev.001495) [DOI] [PubMed] [Google Scholar]
- Sutton K. A., et al. 1998Androgen regulation of the Pem homeodomain gene in mice and rat Sertoli and epididymal cells. J. Androl. 19, 21–30 [PubMed] [Google Scholar]
- Tafuri S. R., Wolffe A. P.1993Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4). J. Biol. Chem. 268, 24 255–24 261 [PubMed] [Google Scholar]
- Tang P. Z., Tsai-Morris C. H., Dufau M. L.1999A novel gonadotropin-regulated testicular RNA helicase. A new member of the dead-box family. J. Biol. Chem. 274, 37 932–37 940 (doi:10.1074/jbc.274.53.37932) [DOI] [PubMed] [Google Scholar]
- Taverna S. D., et al. 2006Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 (doi:10.1016/j.molcel.2006.10.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure A., Szot M., Mahadevaiah S. K., Rattigan A., Ojarikre O. A., Burgoyne P. S.2004A new deletion of the mouse Y chromosome long arm associated with the loss of Ssty expression, abnormal sperm development and sterility. Genetics 166, 901–912 (doi:10.1534/genetics.166.2.901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. Y., Yeh S. D., Wang R. S., Yeh S., Zhang C., Lin H. Y., Tzeng C. R., Chang C.2006Differential effects of spermatogenesis and fertility in mice lacking androgen receptor in individual testis cells. Proc. Natl Acad. Sci. USA 103, 18 975–18 980 (doi:10.1073/pnas.0608565103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Morris C. H., Sheng Y., Lee E., Lei K. J., Dufau M. L.2004Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc. Natl Acad. Sci. USA 101, 6373–6378 (doi:10.1073/pnas.0401855101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui S., Dai T., Roettger S., Schempp W., Salido E. C., Yen P. H.2000Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics 65, 266–273 (doi:10.1006/geno.2000.6169) [DOI] [PubMed] [Google Scholar]
- Vera Y., Dai T., Hikim A. P., Lue Y., Salido E. C., Swerdloff R. S., Yen P. H.2002Deleted in azoospermia associated protein 1 shuttles between nucleus and cytoplasm during normal germ cell maturation. J. Androl. 23, 622–628 [PubMed] [Google Scholar]
- Wang X., Zhang J.2006Remarkable expansions of an X-linked reproductive homeobox gene cluster in rodent evolution. Genomics 88, 34–43 (doi:10.1016/j.ygeno.2006.02.007) [DOI] [PubMed] [Google Scholar]
- Wang G., Zhang J., Moskophidis D., Mivechi N. F.2003Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36, 48–61 (doi:10.1002/gene.10200) [DOI] [PubMed] [Google Scholar]
- Wilkinson M. F., Shyu A. B.2001Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23, 775–787 (doi:10.1002/bies.1113) [DOI] [PubMed] [Google Scholar]
- Xu M., Hecht N. B.2008MSY2 and polypyrimidine tract binding protein 2 stabilize mRNAs in the mammalian testis. Int. J. Androl. 31, 457–461 (doi:10.1111/j.1365-2605.2008.00885.x) [DOI] [PubMed] [Google Scholar]
- Yang X. J.2004Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26, 1076–1087 (doi:10.1002/bies.20104) [DOI] [PubMed] [Google Scholar]
- Yang J., Medvedev S., Reddi P. P., Schultz R. M., Hecht N. B.2005aThe DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl Acad. Sci. USA 102, 1513–1518 (doi:10.1073/pnas.0404685102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Medvedev S., Yu J., Tang L. C., Agno J. E., Matzuk M. M., Schultz R. M., Hecht N. B.2005bAbsence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl Acad. Sci. USA 102, 5755–5760 (doi:10.1073/pnas.0408718102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Morales C. R., Medvedev S., Schultz R. M., Hecht N. B.2007In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic arrest. Biol. Reprod 76, 48–54 (doi:10.1095/biolreprod.106.055095) [DOI] [PubMed] [Google Scholar]
- Yokota Y.2001Id and development. Oncogene 20, 8290–8298 (doi:10.1038/sj.onc.1205090) [DOI] [PubMed] [Google Scholar]
- Zeng L., Zhou M. M.2002Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513, 124–128 (doi:10.1016/S0014-5793(01)03309-9) [DOI] [PubMed] [Google Scholar]
- Zhang Z., et al. 2006Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol. Reprod. 74, 751–759 (doi:10.1095/biolreprod.105.049254) [DOI] [PubMed] [Google Scholar]
- Zhao M., et al. 2004Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis 38, 200–213 (doi:10.1002/gene.20019) [DOI] [PubMed] [Google Scholar]