Abstract
Sexual reproduction depends on the production of haploid gametes, and their fusion to form diploid zygotes. Here, we discuss sperm production and function in a molecular and functional evolutionary context, drawing predominantly from studies in model organisms (mice, Drosophila, Caenorhabditis elegans). We consider the mechanisms involved in establishing and maintaining a germline stem cell population in testes, as well as the factors that regulate their contribution to the pool of differentiating cells. These processes involve considerable interaction between the germline and the soma, and we focus on regulatory signalling events in a variety of organisms. The male germline has a unique transcriptional profile, including expression of many testis-specific genes. The evolutionary pressures associated with gene duplication and acquisition of testis function are discussed in the context of genome organization and transcriptional regulation. Post-meiotic differentiation of spermatids involves very dramatic changes in cell shape and acquisition of highly specialized features. We discuss the variety of sperm motility mechanisms and how various reproductive strategies are associated with the diversity of sperm forms found in animals.
Keywords: spermatogenesis, evolution, gene expression
1. Introduction
Sexual reproduction in diploid organisms requires the formation of two haploid gametes, which have to find each other, recognize that they are from the same species and fuse to create a new diploid organism. In isogamous species, for example, most yeasts, the gametes are very similar, differing only in mating type. Isogamy is probably the ancestral state for sexual reproduction; however, most sexual eukaryotes have evolved to give two sexes, with the process of gametogenesis being the key sexually dimorphic feature—females make eggs, males make sperm. Sperm differs from eggs significantly in morphology, thus these species are anisogamous. While eggs are typically immotile, large and endowed with a large cytoplasmic maternal contribution to embryonic development, sperm are typically motile, small and carrying only a relatively small cytoplasmic contribution to embryonic development. With the evolution of dimorphism of gamete morphology comes significant differences in the process of gametogenesis in the two sexes. The theoretical considerations underlying the evolution of anisogamy, and selective pressures acting on gamete size and number have been reviewed by Lessells et al. (2009). Typically, many more sperm than eggs are produced, and each egg requires much more investment. In simple terms, one can think of the egg production process as being optimized for quality, with quantity being compromised, while in the much higher throughput process of sperm production quality might have to be compromised for quantity. Here, we consider only spermatogenesis, particularly concentrating on the conservation and divergence of the spermatogenic process at a genetic and cellular level.
The basic processes of spermatogenesis are astonishingly similar in even very different animals, and the genes responsible are highly conserved (Bonilla & Xu 2008). Sperm production typically continues through adult life, maintained by a stem cell population, and we will discuss the evolutionary similarities in molecular mechanisms defining spermatogenic stem cells and ensuring their maintenance and protection. Spermatogonia arising from these male germline stem cells act as transit amplifying populations, undergoing limited proliferation in a semi-committed state, before differentiating into spermatocytes. Sister spermatocytes are linked by persistent cytoplasmic bridges that are generated owing to incomplete cytokinesis in each of the mitotic amplification divisions (Gou & Zheng 2004). Thus, sister cells form ‘cysts’, whose members proceed through spermatogenesis in synchrony. The transition from spermatogonial cell to spermatocyte involves exit from the mitotic cell cycle, and commitment to the spermatogenic differentiation programme, including activation of the meiotic process. DNA replication typically occurs early in the primary spermatocyte stage, after which the cells enter an extended meiotic prophase. In addition to carrying out the meiosis-specific chromosome processes of pairing and recombination, the chromosomes in male germline cells activate transcription of a diverse set of spermatogenesis-specific genes. These genes are responsible for endowing the spermatozoa with their unique features, for example, their free-living habit and motility. After the meiotic divisions, the spermatids progress through dramatic morphological events, to result in mature sperm. This review concentrates on conserved aspects of spermatogenesis, and for many of the statements here there will be exceptions. We have tried to focus on the coherent underlying similarities between systems, rather than the outliers, fascinating though these are. We will discuss the nature of these testis-specific genes, how they are expressed, how they evolved their testis functions and how these functions are implemented in the formation of the mature male gamete.
2. Germline stem cells—an evolutionary perspective
The spermatogonial stem cells (SSCs) carry out one of the most important functions in the male organism. They ensure consistent supplies of germ cells, which differentiate into mature sperm, ready to fertilize and thus pass on the individual's genetic content to the next generation. Humans, for example, produce 100 million spermatozoa every day, all derived from a relatively small population of stem cells (Oatley & Brinster 2008). The feature that makes adult stem cells stand out among other cells in the organism is their capacity for production of daughter cells with different fates; on average, a stem cell division gives rise to a daughter cell, which then further divides and differentiates, and also to a new ‘mother cell’ with stem cell identity to repeat this process indefinitely. In the case of male germline stem cells, only one type of differentiated cell is produced in the process, i.e. the sperm. In addition to this asymmetric division property, adult stem cells have also to be able to undergo symmetric division to compensate for the stochastic loss of stem cells over time. Stem cell specification, maintenance and self-renewal therefore are key factors for spermatogenesis. The micro-environment, or stem cell niche, plays an essential role in all these processes (Yamashita et al. 2005). Understanding the interaction between stem cells and their niche is of wider clinical importance—cancer stem cells are also intimately associated with their niches, and further insight into the nature of this relationship could potentially lead to clinical applications (reviewed in Li & Neaves 2006). Primordial germ cells (PGCs), the embryonic precursors of SSCs, contain many features which are conserved throughout the metazoan kingdom. PGCs in both male and female embryos have conspicuous electron dense aggregates, which have been termed basophilic cytoplasmic bodies, dense bodies, nuage, mitochondrial clouds, chromatoid bodies, yolk nuclei or Balbiani bodies, depending on the species (Kloc & Etkin 2005). They are generally composed of endoplasmic reticulum (ER), mitochondria, proteins and RNAs. Interestingly, two of the earliest and highly conserved PGC markers, nanos and vasa, are also associated with this organelle in metazoans as diverse as teleost fishes and aphids (Chang et al. 2007; Li et al. 2009). Despite the commonalities, there are significant dissimilarities concerning mode and timing of germ cell specification in different species (reviewed in Wylie 1999).
(a). The embryonic origin of male germline stem cells
In the majority of animals, germline precursors are determined early in development, when they become distinct from somatic cells. Exceptionally, for example, in hydra, the germline cells can arise late in development (Kusnetsov et al. 2001). There are two distinct modes of germline establishment in metazoa; preformation and epigenesis (induction) (Extavour & Akam 2003). Preformation involves the asymmetric localization of germline determinants in eggs, this is incorporated into a subset of cells during cleavage, and they are then destined to become germline. In epigenesis, the germline precursors are induced during embryogenesis (or at later stages in some cases) by signalling pathways. In Drosophila, the oocyte is connected to sister nutritive cells (nurse cells) (King 1970). The Drosophila oocyte is transcriptionally quiescent, while the nurse cells are highly transcriptionally active, transferring gene products to the growing oocyte during oogenesis (Zhou & King 2004; Caceres & Nilson 2005). The components of germ cell plasm are therefore produced by nurse cells, and specifically localized to the posterior pole of the oocyte. It is thus ‘preformed’ and assigned to impose germline fate on embryonic cells inheriting it, even before fertilization. During gastrulation, these pole cells, which are initially on the surface of the embryos, are swept into the developing gut, as PGCs, and migrate through the gut, and then anteriorly. When they meet somatic gonad precursors, they accumulate, coalesce and start gonadogenesis (reviewed in Santos & Lehmann 2004). Pole cells that fail to interact with the presumptive gonadal mesoderm die, rather than contributing to any somatic lineage, implying that they need continued inputs from gonadal somatic niche cells for survival and proliferation (Coffman et al. 2002). After coalescing into a gonad, the developing testis is organized so that a population of PGCs surround a developing signalling centre, the hub (Le Bras & van Doren 2006), and these PGCs mature into germline stem cells (GSCs). Some PGCs apparently are unable, probably by virtue of their position in the developing gonad, to interact with the niche. These PGCs apparently progress directly into spermatogenesis, contributing to the first spermatogenic cysts in testes (Sheng et al. 2009). In a similar scenario in C. elegans, P-granules, containing factors that determine germ cell fate, are present in the oocyte before fertilization; however, they are uniformly distributed. Just after fertilization, they are re-localized to the posterior pole of the embryo. At each early embryonic cleavage division, the P-granules localize to a single blastomere, and the capacity to contribute to the germline is limited to the P-granule containing blastomere. Finally, symmetric division of the P4 blastomere distributes the P-granules to both daughter cells, and these generate the germline precursors that populate the gonad arms (Strome 2005). Other, although still controversial, examples of preformation include some amphibian, fish and avian germline development (for review, see Extavour & Akam 2003).
Epigenetic determination of PGCs, on the other hand, has been reported for the most intensively studied mammalian model organisms. In the mouse, PGCs cannot be identified until 6.5 days after fertilization, when they are detected in extraembryonic tissue. From there, they migrate to the developing urogenital ridges (Anderson et al. 2000). Transplantation experiments have shown that the proximal epiblast, the region from which PGCs derive, epigenetically induces germ cell fate (Tam & Zhou 1996). The other well-studied example of definitive epigenetic germline induction comes from urodele amphibians, in which similar transplantation experiments have shown that the ventral endoderm is sufficient to induce PGC formation in the lateral plate mesoderm (for review, see Extavour & Akam 2003). Testes formation in mammals starts with the separation of seminiferous cord bundles from the surface epithelium, which are then divided into individual cords by mesenchyme. The PGCs at this stage are called gonocytes, and they transform into SSCs a few days after birth. Interestingly, as in Drosophila, a sub-population of gonocytes can proceed directly into spermatogonia and initiate the first spermatogenic wave, bypassing a self-renewing stem cell stage (Yoshida et al. 2006). The remaining subset would develop into the stem cell population, which will be maintained throughout adulthood. In adult mouse testis, SSCs are referred to as As spermatogonia, s standing for single, as they are undivided and do not contain intercellular bridges (Huckins 1978). Whether this dual pathway from gonocytes to spermatogonia is mouse-specific or a general mammalian theme is not clear, although the parallel with Drosophila is very intriguing, and could indicate that direct development from PGCs to spermatogonia is a relatively broadly used strategy.
The question of preformation versus epigenesis in embryology has caused debates since Aristotle's De Generatione Animalium (van Speybroeck et al. 2002), and even though epigenesis has emerged as the general pattern of the development of an organism, there is ongoing discussion concerning the origin of germ cells. It is clear now that both modes are present across metazoans, but which is ancestral? The main impediment of drawing a conclusion is the paucity of available data. Although there are a few very well-studied model organisms, including Drosophila melanogaster, C. elegans and Mus musculus, they do not satisfactorily represent the diversity of metazoa. Model organisms with good descriptive data only represent two of the three major branches of animals (deutrosomes and ecdysozoans), and there is very little data from any species in the Lophotrocozoa (Aguinaldo et al. 1997; Takahashi et al. 2009). In addition, the method of germline formation seems to differ even within a class—whereas PGCs arise via epigenesis in urodele amphibians, preformation is found in anuran amphibians. In summary, including data from less investigated organisms, the current hypothesis favours epigenesis as the more ancestral mechanism (Extavour & Akam 2003). However, it seems that in some cases lack of evidence of preformation (e.g. if no localized early germline determinants are detected) leads to the assumption of epigenesis. The discussion of the evolutionary origin of germline formation will therefore continue.
(b). Male germline stem cell maintenance versus differentiation
Once the PGCs associate with somatic gonadal precursors, gonadogenesis can commence, and, in males, the PGCs transform into male germline stem cells. Depending on the organism, spermatogenesis is initiated either immediately or delayed until a later stage during development. However, the stem cell population ordained at that early stage has to be maintained and protected until adulthood, and throughout the reproductive span of the animal. It is therefore of vital importance to ensure stem cell preservation and self-renewal. The stem cell niche is the specialized micro-environment in which stem cells reside, and which is uniquely conducive to normal stem cell behaviour. The concept of a niche is common to all stem cells, including tumour stem cells (Li & Xie 2005). However, despite this general theoretical framework, experimental evidence for the role of the niche for the early germline is sparse. In most organisms, it has proved very difficult so far to pinpoint the exact dimensions of the niche, although good progress is being made in locating the stem cell support cues in the mouse (Ogawa et al. 2005). Most of our understanding of the mechanism of stem cell maintenance is derived from the two organisms with a well-defined and readily accessible cellular organization within the testes—D. melanogaster and C. elegans. The understanding of stem cell behaviour in mouse testes has lagged behind these invertebrate systems predominantly because it has been extremely difficult until recently to distinguish between a true stem cell and an early spermatogonial cell (Oatley & Brinster 2006, 2008).
The mammalian Sertoli cells are well established as major contributors to the stem cell niche, but tubular myoid and Leydig cells may additionally contribute to the niche. Besides that, vascularization has also been shown to be important for defining the niche (Yoshida et al. 2007). The release of cytokine colony-stimulating factor-1 as well as follicle-stimulating hormone (FSH) and luteinizing hormone (LH) influence self-renewal and stem cell proliferation (Oatley & Brinster 2008). Sertoli cells in mice express the Ets-related transcription factor ERM, which is necessary for stem cell maintenance (Chen et al. 2005), and in vitro experiments demonstrated that fibroblast growth factor (bFGF), and possibly EGF, in combination with glial cell line-derived neurotrophic growth factor (GDNF) regulate stem cell self-renewal (Oatley & Brinster 2008), but the underlying mechanisms and interactions are still unclear. The conserved RNA-binding protein NANOS2 has also recently been shown to be required to maintain murine SSCs; disruption of this protein depleted stem cell reserves, while over-expression results in accumulation of additional stem cells (Sada et al. 2009).
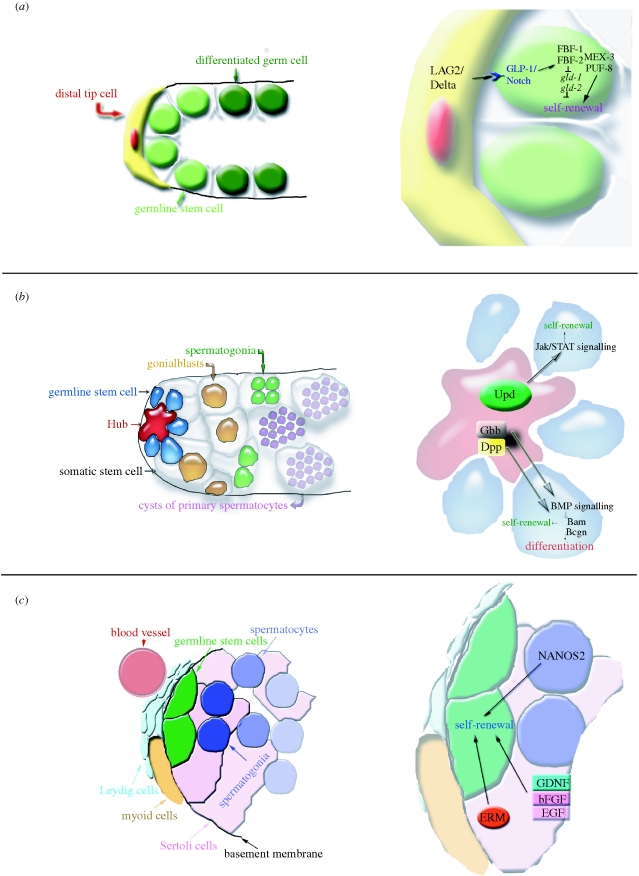
Although the architecture of testes is very different between D. melanogaster and C. elegans, the regulatory mechanisms within the stem cell zone are similar (figure 1). Both have tubular testes, and differentiation progresses from the blind end. Caenorhabditis elegans has males and hermaphrodites; the hermaphrodite ovo-testis initially produces sperm, switching on the last moult to producing oocytes. In this nematode model, a single (or pair, in the testis) somatic support cell, termed distal tip cell (DTC), orchestrates stem cell maintenance, early mitotic and late meiotic divisions in the germline (Kimble & White 1981). Even though the identification of GSCs in C. elegans is not undisputed (Cinquin 2009), it has clearly been shown that ablation of the DTC leads to loss of the germline owing to differentiation, demonstrating its necessity for stem cell maintenance. Notch signalling from the DTC to the GSCs in C. elegans requires direct contact (Henderson et al. 1994; Hansen et al. 2004), whereas Wnt signalling diffuses and regulates cell fate of more proximal cells (Lam et al. 2006). Similarly, a globular cluster of somatic support cells (called the hub) at the apical tip of the D. melanogaster testis is crucial to prevent stem cell loss by differentiation. Like the DTC in nematodes, it regulates stem cell and gonialblast fate by short- and long-range signalling. In the Drosophila testis, the ligand of the Jak/STAT pathway, Unpaired, is secreted from the hub and received only by the directly attached GSCs, where it promotes self-renewal (Kiger et al. 2001; Tulina & Matunis 2001). Transforming growth factor (TGF-beta) signalling has also emerged as a promoter of stem cell identity, but derives from the somatic cyst stem cells (CySC), which are also physically attached to the hub cells, intermingled with the GSCs. Signalling via the morphogenetic protein (BMP) family provides a long-range gradient, acting on more distant cells, spermatogonia and cyst cells (the differentiated daughters of the CySCs) (Bunt & Hime 2004).
Figure 1.
Stem cell niche organization and signalling. (a) In the proximal arm of the male gonad of C. elegans, the DTC provides the necessary cues for stem cell maintenance to the closest germline stem cells. The outline on the right indicates the signalling molecules involved. (b) In the Drosophila testis, a somatic cluster of cells called the hub acts as stem cell niche for the closely associated germline and somatic stem cells. The signalling pathways involved in the decision between self-renewal and differentiation are outlined on the right. (c) In the mouse testis, the stem cell niche comprises different factors; Sertoli cells, Leydig cells and vascularization seem to play a role. The outline on the right depicts the factors promoting self-renewal of the germline stem cells.
Asymmetric stem cell division produces a daughter cell, which is destined to differentiate, and a stem cell resuming its pluripotency. The daughter cell, or gonialblast, undergoes a limited—and species-specific—number of transient mitotic amplifications before entering meiosis and emerging as spermatocyte. During the period of mitotic divisions, the germ cells appear to be in an intriguing in-between state. They are not stem cells anymore, as they have lost their ability to divide asymmetrically and regenerate stem cells, but they are not irreversibly determined to become spermatozoa. In flies, gonialblasts have the ability to revert to a stem cell should the opportunity arise (Brawley & Matunis 2004). Similarly, under particular conditions, mouse differentiating spermatogonia can de-differentiate to become germline stem cells (Barroca et al. 2009). Equivalent experiments for C. elegans are still awaiting the appropriate experimental tools. Meanwhile, DTC ablation experiments mentioned above indicated that the first nine rows of early germ cells at the distal end of the gonad are not committed to differentiation without enforcing signals, and thus might still be able to revert to stem cell identity.
It has been speculated that the switch to differentiation resembles the tipping of a balance between antagonistic factors—those in favour of differentiation and a combination of cues that prevent differentiation and/or maintain the undifferentiated state. In Drosophila, two distinct signalling pathways seem to be in contest. As described above, the hub cells act directly on the GSC via the Jak/STAT pathway to ensure stem cell maintenance. A gradient of Gbb/Dpp signalling, members of the BMP family, decreases with distance from the hub to inhibit differentiation (Shivdasani & Ingham 2003). Intracellularly, the concentration of Bam (bag of marbles, Kawase et al. 2004), on the other hand, increases towards gonialblast and spermatocyte stages, thus opposing the BMP signalling and promoting differentiation. Both factors, Dpp and Bam, also repress each other. Expression of bam in combination with bgcn (benign gonial cell neoplasm) limits the number of transit-amplifying mitotic transitions in Drosophila to four, before they enter meiosis. Bam has only been found in Drosophila species so far, and seems to be absent even from other dipterans such as mosquitoes, although recently a protein very distantly related to Bam has been described in mouse. RNA interference has been shown to be another important level of regulation to maintain GSCs. Members of the Musashi and the Argonaute families of RNA-binding proteins (Musashi and Piwi, respectively) are necessary to preserve the stem cell population (Cox et al. 1998, 2000; Siddall et al. 2006). A very similar scenario is found in C. elegans (reviewed by Kimble & Crittenden 2007), where the DTC secretes the ligand LAG-2, which activates the Notch pathway in the adjacent stem cells by binding the GLP-1 receptor. This appears to downregulate gld-1 (a translational repressor) and gld-2 (cytoplasmic poly-A polymerase), via FBF-2, which also acts as a regulator of translation. FBF-2 is a member of the conserved PUF family, which includes pumilio, a conserved translational regulator preserving GSC maintenance. Recently, two novel PUF family members have been identified in C. elegans, PUF-8 and MEX-3, which act redundantly to promote stem cell self-renewal (Ariz et al. 2009).
An imbalance between factors endorsing an undifferentiated state and regulators in favour of differentiation will finally build up and push the cells out of the spermatogonial phase and into the spermatocyte phase; the transient mitotic amplifications might resemble the state before the decision is finalized (Cinquin 2009). The evolutionary history of this specific relationship between somatic and germline cells—the stem cell niche—is impossible to explain at this time, owing to the lack of data. Even in the well-studied model organisms, conclusive data are accumulating very slowly, and far too few taxa have been investigated to allow us to infer the evolutionary steps that have occurred. The regulatory pathways involved, Notch, BMP and Jak/STAT signalling, are conserved throughout the metazoan kingdom. Notch is an ancient and conserved pathway, which probably evolved as a regulator of segmentation in the common ancestors of the metazoans, and has since been integrated into different functions, for example, in the specification of the germ layers (Shi & Stanley 2006). The Jak/STAT pathway is another well-conserved pathway, which is involved in a wide variety of cellular processes, for many of which it might actually play a supervisory role, conducting other signalling cascades (Hou et al. 2002; Arbouzova & Zeidler 2006). BMPs are a family of proteins at least as old as body patterning, and might have played a very active role in evolution towards Chordata (Brown et al. 2008). More recently, in evolutionary terms, they have adopted more specialized functions, including inducing PGCs in mammals. It is possible that the BMPs involved in primary PGC induction have later been recruited to support germline proliferation (White-Cooper et al. 2009). The importance of RNA interference for stem cell self-renewal is a widespread and highly conserved phenomenon found in all types of stem cells—embryonic, somatic tissue and germline stem cells (Gangaraju & Lin 2009). In particular, members of the Argonaute RNA-binding protein family are found in yeast and plants, although their preference to bind to testes-enriched RNAs is a feature of the piwi subclade of the Argonaute family, which is conserved throughout ciliates, slime moulds and animals (Seto et al. 2007). The target sequences piwi proteins can bind to are extensive and include transposons and repetitive sequences, both remnants of the RNA world, providing further support of the evolutionary conservation of this mechanism.
3. Testis-specific genes, their evolutionary origin and their expression
As the male germline cells transit out of the spermatogonial amplification divisions, they undergo a dramatic change in their potential. They lose the ability to revert to a stem cell character, and even lose the ability to amplify via mitotic division. Instead, they enter a commitment phase, where they initiate expression of genes required for their final destiny, i.e. they begin to express the genes they need to become mature sperm. At a cellular and structural level, spermatozoa are highly specialized entities, distinct from every other cell in the organism. The mechanisms driving very rapid adaptive evolution of sperm include sexual selection, sexual conflict and sperm competition (Swanson & Vacquier 2002). Sequence analyses of a range of insect, mammalian and marine invertebrate species confirmed that on average reproductive genes and proteins evolve faster than their non-reproductive counterparts (Swanson & Vacquier 2002; Swanson et al. 2003). A comparison of genes from 12 Drosophila species provided evidence that sex- and reproduction-related genes underwent lineage-specific accelerated evolution, thus driving species divergence (Haerty et al. 2007). Evolution at high speed might also be the reason for the variety observed in metazoan spermatozoa in respect to their specialized cellular architecture, and the sperm-specific functions of cell components. However, it is important to note that many spermatogenesis proteins are under significant functional constraints, and evolve very slowly. Included in this category are several structural proteins, for example, the testis-specific beta-tubulin isoform, a major component of the spermatid axoneme, has barely changed in 60 million years (Nielsen et al. 2006).
In very general terms, it is obvious that genes expressed in testis can be ubiquitously expressed, or can have testis-specific expression. Testis-specific or testis-enriched genes for an individual species can be broadly grouped into two categories: those genes with obvious paralogues expressed in other tissues and those without such paralogues. The second category can further be subdivided into those present as a single copy in the genome and those present in multiple copies, all of which are testis-specific or biased in their expression. More than 10 per cent of all D. melanogaster predicted protein-coding genes are expressed specifically in testis or are highly testis-enriched for their expression (Chintapalli et al. 2007). This represents a significant proportion of all testis-expressed genes. Similarly, in mouse, 11 per cent of genes expressed in spermatocytes are found to be testis-specific in their expression (Choi et al. 2007). This raises a variety of interesting questions regarding the evolution of testis-specific genes. What functions do they have, and are these functions conserved? Why are so many genes testis-specific in their expression? How have these testis-specific genes arisen?
Let us first consider testis-specific genes with paralogues in expressed in other tissues. By definition, these gene pairs arise by a gene duplication event. Immediately after a gene duplication event, a second, redundant, copy of a functional gene exists. If the duplication is generated by duplicating a genomic region, this second copy is likely to contain the parent gene's transcriptional regulatory regions. In contrast, a retroposed duplicate will rely on the insertion site, and regulatory elements carried within the transcript itself, for its transcriptional control elements. After duplication, the gene pair can remain as functionally redundant duplicates, and this situation can be selected for if it is beneficial, e.g. if high expression of the gene product is advantageous. If the duplicates diverge in function, one copy of the gene can maintain the original function while the other adopts no function (i.e. it degenerates), or the pair can each take on a subset of the original function (subfunctionalization), or one copy can evolve a new function never carried out by the pre-duplication gene, while the other retains the original function (neofunctionalization). Parsimoniously, one would expect a paralogous pair of genes, where one copy is testis-specific and the other is ubiquitous to originate from a ubiquitously expressed parent gene. Intuitively, it seems inefficient for the male germ cells to express testis-specific copies of ubiquitously expressed genes, when they could equally express the ubiquitous copy—why have more genes than is necessary? One advantage for duplicating genes in this way is to overcome intrinsic problems with expressing the ubiquitous copy, for example, if the parent gene is on the X chromosome, an autosomal copy might be strongly selected as it will have escaped the X chromosome transcriptional inactivation that occurs in many primary spermatocytes (Turner 2007) (see below). As an example in mouse, the ubiquitously expressed gene for the glycolytic pathway enzyme phosphoglycerate kinase (Pgk1) is on the X chromosome. Its expression declines as spermatocytes enter the meiotic phase. The functional retroposed copy of this gene Pgk2, located autosomally, is expressed in spermatocytes, and thus allows these cells to continue glycolysis (reviewed in Eddy 2002). Duplication of genes also allows copies to functionally specialize, for example, the spermatogenesis-specific paraologues of glycolytic enzymes in mouse have acquired modifications that promote their localization the sperm midpiece (reviewed by Eddy 2002).
The evolutionarily more intriguing class of genes is that expressed exclusively in spermatogenesis. Some of these gene products will be components of mature sperm, while many others will function during the spermatogenic process, but will not be incorporated in the final product. These genes are likely to function to give sperm its unique characteristics, and indeed approximately half of the proteins detected in mature sperm are testis-enriched in their expression (Dorus et al. 2006). Single-copy testis-specific genes could have evolved from ancestral genes that were ubiquitous; if the sequence has diverged enough, it will not be possible to infer this relationship. Newly evolved testis-specific genes are often under positive selection, decreasing our chances of understanding their origin (Dorus et al. 2008). For example, in all somatic cells, DNA is neatly packaged into the 10 nm chromatin fibre by nucleosomes. Packaging these units to give the 30 nm solenoid are linker histones, which are less well conserved than the core nucleosomal histones, and can vary in number and assembly. In contrast to somatic cells, the DNA in sperm nuclei is usually found compacted into a distinct, non-nucleosomal toroid configuration, associated with small basic protamine or protamine-like proteins. In both humans and mouse, some sperm chromatin remains nucleosomal, although it is not clear if this retention of a histone-bound chromatin pool is universal (Pittoggi et al. 1999). During spermatogenesis in many species histones are replaced first by transition proteins, which are then substituted with protamines (Braun 2001). In other species, the transition from histone to protamines may be direct. This repackaging of the chromatin leads to the most condensed form of DNA, taking up 95 per cent less volume than in a somatic cell. Somatic histones contain lysine-rich C- and N-terminal ends, but very little arginine. On the other hand, more than half of the amino acids constituting protamines are arginine. An evolutionary link between these two proteins has therefore been dismissed for many years, despite their similar roles in packaging chromatin. It has recently been shown that a frameshift mutation in the tail of the sperm-specific histone H1 provided the key step in the evolution of protamines, and that these are true descendant of histones (Lewis et al. 2003, 2004a,b).
Most exciting among the reports of evolution of new testis-specific gene function has been the finding that genes encoding functional RNAs or proteins can evolve from previously non-transcribed genomic sequence. The detection of such events has been heavily reliant on the availability of genome sequences of several closely related species. The evolution of new testis-specific genes has been documented for several Drosophila species (Levine et al. 2006; Begun et al. 2007). Such new genes are not necessarily testis-specific, as seen in the report of de novo gene origins in humans (Knowles & McLysaght 2009). Testis-specific genes typically have short promoters, e.g. transcription of the mouse spermatid-specific gene SP-10 is driven by a <300 bp promoter region, and it is tempting to speculate that the use of such short promoters facilitates the de novo acquisition of transcriptional activity in any given genomic region. The use of genomic tiling arrays, along with next generation sequencing technology, has already revealed that much more of the genome is transcribed than was previously suspected (Johnson et al. 2004). This transcription of non-coding genomic regions, in spermatogenic cells, increases the chances of evolution of new protein-coding genes, as fortuitous mutation in spermatocyte or spermatid DNA, allowing production of a new protein can readily be selected for (or against) in the individual sperm directly derived from the cell in which the mutation occurred.
(a). Genomic organization of testis-specific genes
How is the transcription of testis-specific genes controlled? There are two features to testis specificity: the expression in testis and the lack of expression in other cell types. These features could be linked, e.g. by the action of repressors and activators working at the same promoter elements, or they could be independent, e.g. activation could depend on specific transcription factors while repression could depend on genes residing in a ‘repressive’ chromosomal region. In both D. melanogaster and vertebrates, it is remarkable that the promoter regions that confer testis-specific expression are relatively small, compared, for example, with genes activated during embryonic development. For example, in Drosophila, individual cis-regulatory modules for expression of genes in specific mesodermal lineages are typically several hundred base pairs in length, and many genes have more than one such module (Zinzen et al. 2009). In contrast, a fragment of just 76 bp is sufficient to direct testis-specific expression of the Drosophila beta-2 tubulin gene (Michiels et al. 1989). When a promoter fragment from a testis-specific gene has been used to drive expression from a reporter construct, it is typically capable of conferring both testis-specific expression and somatic non-expression to the reporter. In D. melanogaster, this is true even when the transgene is inserted at random positions in the genome. Thus, the pattern of gene expression is conferred by a short DNA sequence. However, genome scale analysis has revealed non-random distributions of testis-specific genes (Andrews et al. 2000; Parisi et al. 2003, 2004). For organisms with a heterogametic male genotype (XY, e.g. Drosophila, humans), it is proposed that sexually antagonistic selection pressures favour the localization of genes with female-specific functions on the X chromosome, while the X chromosome is not a favourable location for genes with male-specific functions. This is because the X chromosome spends more evolutionary time in females (where it is present in two copies) than males (which have a single copy). The bias against the X chromosome for genes required in spermatocytes is exacerbated in many organisms by the phenomenon of sex chromosome inactivation, by which transcription of the X chromosome is dramatically reduced in primary spermatocytes (Handel et al. 1994). These ideas are brought together by the sexual antagonism and X inactivation (SAXI) hypothesis (Wu & Xu 2003). In mouse, the X chromosome is enriched for genes expressed in spermatogonia, and depleted for genes expressed in spermatocytes (Wang et al. 2001). Also consistent with theory, the density of testis-specifically expressed genes is much lower on the X chromosome than the autosomes in Drosophila, and there is a strong bias for retroposition of testis-expressed genes off the X chromosome (Betran et al. 2002). However, while global gene expression studies have found this bias, several multi-copy testis-specific gene families are highly expressed off the X chromsome in both mouse and Drosophila (Ranz et al. 2003; Mueller et al. 2008).
The bias against an X chromosome location for testis-specific genes has even been found for the entire X chromosome in the species Drosophila pseudoobscura, whose X chromosome is actually a fusion of an ancestral X and an ancestral autosome. The neo-X (i.e. the region recently derived from an autosome) shows the same paucity of testis-specific genes as the ancestral X (Sturgill et al. 2007). The loss of testis-specific genes from the neo-X has been caused both by loss of individual genes, by translocation (or retroposition) of genes from the neo-X to the autosomes and by preferential gain of new testis-specific genes on autosomes. Intriguingly, testis-specific genes still residing on the D. pseudoobscura neo-X have not significantly reduced their expression in response to their new chromosomal context (Sturgill et al. 2007).
In addition to chromosome-scale gene localization bias, there is also significant clustering of testis-specific genes within the genome (Miller et al. 2004), indeed in D. melanogaster one-third of the testis-specific genes may reside in clusters (Boutanaev et al. 2002). Many clusters can trivially be attributed to tandem gene duplications, where the paralogues are adjacent genes. Their testis coexpression probably derives from the testis expression of the parental gene; however, even after exclusion of this class, the clustering is still apparent. The remaining, and more interesting from a mechanistic point of view, are clusters of testis coexpressed genes which do not obviously originate from a testis-expressed parental gene. Given the constant genomic shuffling (e.g. inversion, translocations, etc.) occurring over evolutionary time, the modern situation of clusters can only be explained by postulating evolutionary selection pressure for coexpressed genes to become or remain clustered. Higher order chromatin structures have been postulated to be important for allowing coexpression of clustered genes, e.g. spreading of a transcriptionally permissive chromatin structure activating gene expression in spermatocytes or spermatids (Kramer et al. 1998; Boutanaev et al. 2002; Spellman & Rubin 2002; Kalmykova et al. 2005). Equally all the genes in a particular expressed cluster could interact with the same local transcription factory in the nucleus (Osborne et al. 2004). In this scenario, the activation of transcription of one testis-specific gene, and its association with a transcription factory could potentiate activation of nearby genes, by increasing the probability of them contacting the same transcription factory. The arrangement of genes in clusters does not appear to be essential for their transcription, but might make the transcription pattern more robust. This model is consistent with the finding that genes normally resident in clusters retain their normal regulation (spacio-temporal) even when moved to other chromosomal locations, although the absolute expression levels vary depending on chromosomal location (Hense et al. 2007) (H. White-Cooper & C. Morris 2009, unpublished data). Transcription factories have been proposed to facilitate pairing of homologous chromosomes in meiotic prophase (Xu & Cook 2008). Clustering of testis-specific genes into chromosomal domains is also potentially important for their transcriptional silence in the soma. Recent experiments in Drosophila have indicated that the testis-specific gene clusters are associated with repressive sub-nuclear regions, particularly with the nuclear lamina in the soma (Shevelyov et al. 2009). Physical tethering of the testis genes to the nuclear periphery potentially keeps them transcriptionally silent. It is notable that testis-specific genes are less frequently tagged in transposon-mediated mutagenesis screens than other genes. New insertions are usually selected on the basis of expression of a marker gene in the eye; the transcriptional silence imposed on the testis-specific genes is also imposed on the transgene, thus the marker gene is not expressed. This finding is consistent with repression of testis gene expression in the soma depending on chromatin territories, rather than on specific sequence.
4. Differentiation of spermatozoa and acquisition of motility
The meiotic divisions result in four haploid spermatids for every diploid spermatocyte. These spermatids will undergo substantial morphological changes as they transform themselves into mature sperm. Here, we will discuss just a few of these changes at a cell biological level, considering the evolutionary forces that have driven different taxa to adopt a variety of strategies for making sperm.
(a). Sperm diversity and motility
The origin of sex, as opposed to mating type, is defined as the onset of anisogamy—gametes specialize in different directions. Oocytes are by definition the larger gamete. As oocyte production is costly, in terms of resources, ooctyes are typically produced in small(ish) numbers. It is therefore in the interest of the oocyte-bearing gender, the female, to be as selective as possible to ensure the highest quality of fertilization. Sperm, on the other hand, evolved into being the small, usually motile gamete, which is produced in large numbers. Consequently, the male strategy to maximize reproductive success is to aim for high quantity of fertilization rates. Each sperm is made with as little investment as possible, only consisting of the bare minimum—its payload of a nucleus and centriole packaged within the structures needed for it to find and fertilize the egg. Thus, the sperm will typically comprise a motility apparatus (e.g. flagellum), source of energy production (mitochondria) and egg entry apparatus (acrosome), as well as the nucleus and centriole. Some sperm also supply proteins or RNAs required for early embryonic development, e.g. PLCzeta from mammalian sperm activates Ca2+ oscillations in the egg on fertilization (Saunders et al. 2002), while the Spe-11 protein in C. elegans sperm is critical for embryogenesis (Hill et al. 1989). It is not surprising that the different approaches used by males and females lead to sexual conflict and selection, the underlying mechanisms of the rapid evolution of sexual traits. Sperm morphology and its immense diversity is one of these traits. The evolutionary driving force behind the visible variety in shape, size and motility seems to be sperm competition (Roldan et al. 1992). There are relatively few species which have a bona fide monandrous mating system; in most species, polyandry means that any sperm deposited could potentially be out-competed by a sperm of a rival male. It would be tempting to speculate that selection in those circumstances would simply favour the fastest sperm, which holds true in many cases, but the evolution of motility provides a more complex picture. In a simplified model, for external fertilizers such as fishes and amphibians, one would expect sperm motility to increase with sperm competition. This has been confirmed in frogs (Byrne et al. 2003), but not in fishes (Stockley et al. 1997). For internal fertilizers, sperm size is predicted to remain small despite competition (Parker & Begon 1993). Hence, it is surprising to find such an enormous variety of sperm lengths just within insects—ranging from 12 µm in some Hymenoptera to almost 6 cm in Drosophila bifurca (Werner & Simmons 2008). A positive correlation between sperm length and sperm competition has been shown in Lepidoptera and birds, but not in mammals (Gage et al. 2002; Gage & Morrow 2003), whereas selection has favoured short sperm in beetles (Garcia-Gonzalez & Simmons 2007) and crickets (Gage & Morrow 2003). Concluding from conflicting evidence, a better understanding of sperm function will hopefully lead to a more conclusive picture. For primates, for example, it has been shown that the volume of the midpiece is significantly correlated to sperm competition. This region contains the mitochondria, which are essential for motility. Selection might therefore favour the fastest sperm, which might (Gomendio & Roldan 2008), or might not be the longest (Anderson & Dixson 2002).
(b). Flagellate sperm
The basic plan for flagellate sperm consists of head, midpiece and tail, or flagellum, which provides motility. Within this framework, however, the structure of the flagellum has diverged remarkably over time. Insect sperm follows the fundamental layout for spermatozoa—comprising a head with an acrosome, a middle section called transitional centriole adjunct and a flagellum. Interestingly, the insect flagellum contains two long mitochondrial derivatives flanking the axoneme, whereas mitochondria are usually located in the midpiece of other flagellate sperm, indicating that the insect flagellum might actually be homologous to the midpiece of more typical sperm (Werner & Simmons 2008). The axoneme, or axial filament, provides the source of sperm motility. The basic axonemal design has been evolutionarily conserved in cilia and flagella throughout eukaryotes, with a 9 + 2 structure—a circle of nine pairs of microtublues surrounds two single central microtubules (Manton 1953). The insect sperm flagellum, however, shows a variation of this basic plan—most insects possess a 9 + 9 + 2 pattern, with an additional ring of doublet microtubules outside the canonical set (Phillips 1970). This deviation can be further modified, for example, 9 + 9 + 0 in the mayfly Chloeon dipteron, 14 + 0 in Acerentomon majus, a proturan insect, and even tight swirls of 75–91 microtubules in iceryine scale insects (Normark 2009). The various combinations of microtubules result in slight alterations in the precise pattern of sperm movement. Comparisons between the American and the Asian horseshoe crab, for example, showed that the 9 + 2 structure of the former led to a planar wave formation, whereas the 9 + 0 version of its Asian counterpart exhibited helical movement (Inaba 2007). Therefore, losing the central microtubule complex does not necessarily impair motility, as has been suggested for other species—mutants of Chlamydomonas or Drosophila with a 9 + 0 instead of their normal 9 + 2 structure are immotile (Mottier-Pavie & Megraw 2009). Surprisingly, some insect spermatozoa have the peculiar ability to swim backwards as well as forwards. This movement is generated by reversing the propagation of the wave, and has been reported, for example, in the louse Pediculus humanus, and in tephritid flies, Megaselia scalaris (Baccetti et al. 1989). A similar phenomenon has also been described for the spermaotoza of some freshwater and marine invertebrates, which reverse their swimming direction in response to a drop in Ca2+ levels (Ishijima et al. 1994). The biological function of bidirectional sperm movement remains unclear, but could be advantageous to manoeuvre within the narrow female tract (Werner & Simmons 2008). Another rare, yet fascinating, evolutionary invention observed in insects is multiflagellate sperm, which has otherwise only been observed in gastropod paraspermatozoa and in annelids (Riparbelli & Callaini 2007). In the termite Mastotermes darwiniensis, about 100 flagella are attached to a conical head. The axonemes are void of central microtubules, displaying a 9 + 0 structure, and the sperm has been described as ‘feebly motile’ (Bacetti & Dallai 1977). Mammalian sperm also deviates from the basic 9 + 2 pattern, as it contains a set of outer dense fibres (ODFs) around the microtubule annulus, giving it an alternative 9 + 9 + 2 configuration. Although ODFs are restricted to vertebrates, genes homologous to some ODF components have been identified in urochordates (Ciona intestinalis), echinoderms (S. purpuratus), insects and platyhelminths (Schistosoma japonicum), which indicates an ancient origin for at least some of the molecular components of this structure (White-Cooper et al. 2009). This is further supported by ODF homologues found in protists, which are asexually reproducing, but flagellated, indicating that ancestral flagellum proteins were recruited to form the first sperm flagellum (Dawe et al. 2005).
Although clearly the flagellum is homologous to the cilium, its origin has been controversial. At the base of the developing cilia or flagellum lies another feature with a specialized function—the centriole. A pair of centrioles is part of the centrosome, or microtubule organizing centre (MTOC) although experiments in Drosophila indicate that the centriole is not essential for cell division, while it is essential for normal cilia and flagella function (Basto et al. 2006). During spermatogenesis, the developing centrioles transform into basal bodies, providing a template for the growing axoneme. Centrioles normally duplicate during S-phase, but at least in multiflagellate M. darwiniensis, centrioles can be synthesized de novo (Riparbelli et al. 2009). Proper formation of the basal body is essential for sperm motility, as has been shown by a mutation in the Drosophila homologue of the evolutionary conserved centriole protein Bld10, which is part of the somatic centriole as well as of the basal body. Mutations in Bld10 result in flies which are viable but male sterile, owing to loss of the central region of the axoneme and associated sperm immotility (Mottier-Pavie & Megraw 2009), indicating that its centriolar function is not essential for viability, but it is essential for fertility. The discovery of a new division of bacteria called Verrucomicrobia has recently led to a novel symbiotic theory for the origin of the flagellum (Li & Wu 2005). In contrast to any other bacteria, Verrucomicrobia contain tubulins, an important prerequisite for flagellum formation. One Verrucomicrobia group, the epixenosomes, live as ectosymbionts on marine ciliates. Their tubulins share more homology with eukaryotic tubulins than archeal FtsZ (the bacterial gene assumed to be the tubulin ancestral gene) does, and are probably able to form dimers and thus construct microtubules. It is hypothesized that epixenosomes lived in increasingly dependent symbiosis with their host, before becoming endosymbionts and developing interactions with host-specific primitive actin filaments and myosin (Li & Wu 2005). This hypothesis is an intriguing idea, and would suggest that cilia which are found on epithelial cells in metazoa as well as in protozoa, such as flagellates and ciliates, evolved from flagellae, and not vice versa. The evidence would also suggest that centrioles are derived from basal bodies.
5. Aflagellate and amoeboid sperm
Assuming that sperm competition for the fertilization of a limited number of eggs will produce fast swimming sperm, the opposite should hold true—if the selective pressure of competition is removed, motility could be lost without reducing reproductive success. Loosening of the selective pressure by sperm competition does not necessarily equal monogamy (Morrow 2004). Other mechanisms of reducing or removing sperm competition could include self-fertilization, as has been shown for the hermaphroditic form of Dicemydae; or population dynamics—in the sea spiders Pycnogonum littorale, the male to female ratio has shifted, thus limiting the number of available males. Leptophlebiidae are the only members of the monogamous mayfly family with immotile sperm (Gaino 1991; Jamieson et al. 1999). As adults only live for one day, there might simply not be enough time for female to remate, and thus reduced opportunity for sperm competition. Based on currently collected data, it appears that aflagellate spermatozoa have evolved independently at least 36 times across a wide range of taxa (Morrow 2004). In 19 of these cases, loss of flagellae correlates with loss of motility; however, in at least 10 of these examples an alternative mechanism has been employed and the sperm retains motility. Surprisingly, one of the most primitive organisms that indulges in sexual reproduction, the red algae (Rhodophyta), have immotile aflagellate sperm, which are released directly into the water for fertilization (Manton 1970). It could be assumed that aflagellate sperm are ancestral; however, this is unlikely as most of the closely related species have flagellate sperm, thus the flagellum was probably lost in Rhodophyta. This conclusion that aflagellate sperm evolve via loss of a flagellum, rather than being ancestral holds true in other taxa. However, owing to the lack of behavioural studies supporting the morphological data, it is still not clear whether the loss of flagellum correlates with weak or no sperm competition (Morrow 2004).
A lack of flagellum does not necessarily suggest sperm immotility. Locomotion by other means has been observed in a variety of species, although the molecular mechanisms are seldom described (Morrow 2004). Several species within the Macrostomorpha (flatworms) have aflagellate but motile sperm, which produce wiggling movements via the action of a singlet microtubule (Newton 1980). Evolutionary loss of flagella has also been observed among chordates—the fish family of Gymnarchidae produce aflagellate motile sperm, although the underlying machinery is unclear (Mattei 1972; Mattei 1991). Interestingly, their reproductive behaviour suggests low sperm competition again—the mating partners become ventrally physically attached from courtship to spawning, and the males then carry the fertilized eggs in their mouth, making cross-fertilization unlikely. Nematodes and their sister phylum Nematomorpha have developed a specialized form of aflagellar spermatozoa—they produce amoeboid motile sperm (Schmidt-Rhaesa 1997/98). This is the best described atypical motility system, as the molecular mechanism has been extensively studied in C. elegans. Surprisingly, despite the morphological similarity between nematode sperm and amoeboid organisms (such as slime moulds), the underlying mechanism of motility is not conserved. In most amoeboid cells, movement is based on the actin cytoskeleton, in contrast C. elegans spermatozoal pseudopodal motiliy depends not on actin, but on assembly and disassembly of major sperm protein (MSP) filaments (Smith 2006).
In summary, flagellate sperm is believed to be the ancestral form of male gametes. However, the flagellum has been lost independently during evolution in a wide variety of species. Many aflagellate spermatozoa are motile, having employed or evolved alternative mechanisms of movement. A link between low sperm competition and a connected decrease in selective pressure on sperm motility and loss of flagellum has been suggested.
(a). Acquisition of sperm motility
For species generating motile sperm, once spermatocytes have segregated their genomes to become haploid, and undergone cellular rearrangements and morphological modifications to transform into spermatozoa, they only lack motility in order to perform their ultimate task of fertilization. In many organisms, including Drosophila, motility is acquired during the latter stages of spermatogenesis, and mature sperm is the direct end product, requiring no further activation (Lefevre & Jonsson 1962; Tokuyasu 1974). In a wide range of other species, however, spermatids have to undergo a maturation process called capacitation in mammals, or spermiogenesis in C. elegans. Even within insects, some species produce immotile sperm that need to be activated on mating to achieve full motility (Cooper 1950; Werner & Simmons 2008). Spermatozoa change their behaviour, morphology and even their display of surface molecules at this stage. The initial step of sperm activation can take place in the epididymis or spermiduct, followed by final adjustments—depending on the species—within the female genital tract or in water after spawning (Werner & Simmons 2008). Many female-derived or seminal fluid factors are responsible for these events. Examples include motility-activating factors from the egg jelly/egg coat (Hansbrough & Garbers 1981; Suzuki 1990; Suzuki & Yoshino 1992), sperm attractants from the egg (Spehr et al. 2003; Fukuda et al. 2004), de-capacitation factors (DFs) from the seminal plasma (Kawano & Yoshida 2007), hormones present in the genital tract, etc. (Correia et al. 2007). The concept of sperm activating factors released by the egg seems to be evolutionarily conserved; examples are found in mammals, sea urchin, teleost fishes, corals, starfishes and ascidians (for review Yoshida et al. 2008).
A very detailed analysis of the process of sperm activation in a simple organism comes once again from C. elegans. The nematode provides a special feature, which makes it a very popular model for reproduction biologists. It comes in only two sexes—males and hermaphrodites, and in the latter case practises self-fertilization. Hermaphrodites produce a few hundred sperm during their larval stages, then switch to oocyte production. Fertilization takes place in the spermatheca at an extremely efficient rate—almost every gamete is used (Singson 2001). Hermaphrodites can, however, mate with male worms, in which case the non-self sperm outcompetes the hermaphrodite sperm to promote outbreeding (LaMunyon & Ward 1995). The hermaphrodite sperm is stored in the spermatheca, and activated on ovulation of the first oocyte (Ward & Carrel 1979). When a male mates with a hermaphrodite, he transfers immotile sperm, and these are activated in the female tract as soon as possible after ejaculation. Premature maturation in the male genital tract reduces the efficiency of sperm transfer on mating. Swm-1, a predicted secreted serine protease inhibitor, has proved to be essential for regulation of sperm activation. Males mutant for Swm-1 transfer very few sperm into the female reproductive tract, owing to premature activation within the male seminal duct, making the sperm ‘sticky’ (Stanfield & Villeneuve 2006). Thus, Swm-1 must be important for restricting acquisition of sperm motility. Although direct targets of Swm-1 are unconfirmed, Spe-19 has been put forward as a candidate protease, and genes of the spe-8 group are possible targets. Spermatids of nematode species with conserved spe-8 group genes can be activated by a protease mix (for review, Singson 2006). In summary, studies on C. elegans emphasize the importance of the right timing for sperm activation.
Sperm activation or capacitation in chordates often relies on egg-derived factors, as mentioned above, and these factors can also sometimes act as chemoattractants. Even though the compounds found vary and seem to have evolved very rapidly and species-specifically, the concept has not changed throughout evolution. Sperm chemoattractants are even found in bracken fern and algae (for review, Yoshida et al. 2008). Interestingly, sperm activation in the female genital tract requires protease activity provided in seminal fluid (Friedlander et al. 2001), while the mammalian-sperm-associated fertilin metalloproteases are themselves proteolytically processed, and thus activated, during sperm transit in the epididymis (Blobel 2000). Thus, the use of proteases in sperm activation has broad phylogenetic use, and there is potential conservation of the mechanism described for C. elegans. Immediate changes in response to sperm activation are usually triggered by modifications of ionic concentrations, for example, Ca2+ influx, which induces motility (Ren et al. 2001). Mammalian spermatozoa appear to have evolved an additional gear—they have to reach hypermotility upon capacitation in order to be able to penetrate the zona pellucida in preparation for fertilization (Suarez 2008). The driving force to send mammalian sperm into hyperspeed is still not entirely clear, but seems to reside in the seminal fluid. CatSper2, a voltage-gated cation channel, has been shown to be essential for hyperactivation in mice (Quill et al. 2003). Confusingly, de-capacitating activities are also found in the seminal fluid, although their functions are unclear (Maxwell et al. 2007). Candidate DFs include DF, which binds to a GPI-anchored membrane receptor on the sperm surface and regulates Ca2+-ATPase activity (Fraser et al. 1990). Recently, Raf kinase inhibitor protein-1 (RKIP-1) and platelet-activating factor (PAF) have been identified as modulators of capacitation (Nixon et al. 2006; Zhu et al. 2006). The triggers of motility, if they are separated from spermatogenesis, seem to have become more elaborate in higher taxa, resulting in the complex multi-step process of motility, capacitation inducing hypermotility and acrosome reaction to inhibit non-species-specific fertilization.
6. Concluding remarks
In this review, we have considered the diversity of mechanisms employed by animals to generate sperm capable of fertilizing the eggs produced by females of their own species. This process is under strong evolutionary selective forces and we have considered how these forces are manifest in the final process. Much of this discussion is limited to comparison of morphological features because of the limited data on molecular mechanisms that are available for the majority of species, the model systems of mouse, Drosophila and C. elegans being notable exceptions. In considering evolution of spermatogenic processes, it is essential to examine changes in gene sequence, expression and function. It is also intriguing how new genes can be generated, by duplication of existing genes or even de novo, and to consider how these new gene products can integrate themselves into the spermatogenic process, to become essential for efficient production of normal sperm.
Footnotes
One contribution of 17 to a Theme Issue ‘The biology and regulation of spermatogenesis’.
References
- Aguinaldo A., Turbeville J., Linford L., Rivera M., Garey J., Raff R., Lake J.1997Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387, 489–493 (doi:10.1038/387489a0) [DOI] [PubMed] [Google Scholar]
- Anderson M., Dixson A.2002Sperm competition: motility and the midpiece in primates. Nature 2002, 496 (doi:10.1038/416496a) [DOI] [PubMed] [Google Scholar]
- Anderson R., Copeland T. K., Scholer H., Heasman J., Wylie C.2000The onset of germ cell migration in the mouse embryo. Mech. Dev. 91, 61–68 (doi:10.1016/S0925-4773(99)00271-3) [DOI] [PubMed] [Google Scholar]
- Andrews J., Bouffard G. G., Cheadle C., Lu J. N., Becker K. G., Oliver B.2000Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Gen. Res. 10, 2030–2043 (doi:10.1101/gr.10.12.2030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbouzova N. I., Zeidler M. P.2006JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development 133, 2605–2616 (doi:10.1242/dev.02411) [DOI] [PubMed] [Google Scholar]
- Ariz M., Mainpal R., Subramaniam K.2009C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 326, 295–304 (doi:10.1016/j.ydbio.2008.11.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacetti B., Dallai R.1977The first multi-flagellate spermatozoa in the animal kingdom, discovered in Mastotermes darwiniensis. C. R. Acad. Sci. Hebd. Seances Acad. Sci. D 285, 785–788 [PubMed] [Google Scholar]
- Baccetti B., Gibbons B. H., Gibbons I. R.1989Bidirectional swimming in spermatozoa of tephritid flies. J. Submicrosc. Cytol. Pathol. 21, 619–625 [PubMed] [Google Scholar]
- Barroca V., Lassalle B., Coureuil M., Louis J., Le Page F., Testart J., Allemand I., Riou L., Fouchet P.2009Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat. Cell Bio. 11, 190–196 (doi:10.1038/ncb1826) [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C., Khodjakov A., Raff J.2006Flies without centrioles. Cell 125, 1375–1386 (doi:10.1016/j.cell.2006.05.025) [DOI] [PubMed] [Google Scholar]
- Begun D., Lindfors H., Kern A., Jones C.2007Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176, 1131–1137 (doi:10.1534/genetics.106.069245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E., Thornton K., Long M.2002Retroposed new genes out of the X in Drosophila. Genome Res. 12, 1854–1859 (doi:10.1101/gr.6049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel C.2000Functional processing of fertilin: evidence for a critical role of proteolysis in sperm maturation and activation. Rev. Reprod. 5, 75–83 (doi:10.1530/ror.0.0050075) [DOI] [PubMed] [Google Scholar]
- Bonilla E., Xu E. Y.2008Identification and characterization of novel mammalian spermatogenic genes conserved from fly to human. Mol. Hum. Reprod. 14, 137–142 (doi:10.1093/molehr/gan002) [DOI] [PubMed] [Google Scholar]
- Boutanaev A. M., Kalmykova A. I., Shevelyov Y. Y., Nurminsky D. I.2002Large clusters of co-expressed genes in the Drosophila genome. Nature 420, 666–669 (doi:10.1038/nature01216) [DOI] [PubMed] [Google Scholar]
- Braun R.2001Packaging paternal chromosomes with protamine. Nat. Genet. 28, 10–12 (doi:10.1038/88194) [DOI] [PubMed] [Google Scholar]
- Brawley C., Matunis E.2004Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331–1334 (doi:10.1126/science.1097676) [DOI] [PubMed] [Google Scholar]
- Brown F. D., Prendergast A., Swalla B. J.2008Man is but a worm: chordate origins. Genesis 46, 605–613 (doi:10.1002/dvg.20471) [DOI] [PubMed] [Google Scholar]
- Bunt S., Hime G. R.2004Ectopic activation of Dpp signalling in the male Drosophila germline inhibits germ cell differentiation. Genesis 39, 84–93 (doi:10.1002/gene.20030) [DOI] [PubMed] [Google Scholar]
- Byrne P. G., Simmons L. W., Roberts J. D.2003Sperm competition and the evolution of gamete morphology in frogs. Proc. R. Soc. Lond. B 270, 2079–2086 (doi:10.1098/rspb.2003.2433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L., Nilson L.2005Production of gurken in the nurse cells is sufficient for axis determination in the Drosophila oocyte. Development 132, 2345–2353 (doi:10.1242/dev.01820) [DOI] [PubMed] [Google Scholar]
- Chang C. C., Lin G. W., Cook C. E., Horng S. B., Lee H. J., Huang T. Y.2007Apvasa marks germ-cell migration in the parthenogenetic pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Dev. Genes Evol. 217, 275–287 (doi:10.1007/s00427-007-0142-7) [DOI] [PubMed] [Google Scholar]
- Chen C., et al. 2005ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436, 1030–1034 (doi:10.1038/nature03894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V., Wang J., Dow J.2007Using FlyAtlas to identify better Drosophila models of human disease. Nat. Genet. 39, 715–720 (doi:10.1038/ng2049) [DOI] [PubMed] [Google Scholar]
- Choi E., et al. 2007Integrative characterization of germ cell-specific genes from mouse spermatocyte unigene library. BMC Genomics 8, 256 (doi:10.1186/1471-2164-8-256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O.2009Purpose and regulation of stem cells: a systems-biology view from the Caenorhabditis elegans germ line. J. Pathol. 217, 186–198 (doi:10.1002/path.2481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman C. R., Strohm R. C., Oakley F. D., Yamada Y., Przychodzin D., Boswell R. E.2002Identification of X-linked genes required for migration and programmed cell death of Drosophila melanogaster germ cells. Genetics 162, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. W.1950Normal spermatogenesis in Drosophila. In Biology of Drosophila (ed. Demerec M.), pp. 1–61 London, UK: Chapman and Hall [Google Scholar]
- Correia J. N., Conner S. J., Kirkman-Brown J. C.2007Non-genomic steroid actions in human spermatozoa. Persistent tickling from a laden environment. Semin. Reprod. Med. 25, 208–219 (doi:10.1055/s-2007-973433) [DOI] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Baker J., Chang L., Qiao D., Lin H.1998A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (doi:10.1101/gad.12.23.3715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Lin H.2000piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 [DOI] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Portman N., Shaw M. K., Gull K.2005The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J. Cell Sci. 118, 5421–5430 (doi:10.1242/jcs.02659) [DOI] [PubMed] [Google Scholar]
- Dorus S., Busby S. A., Gerike U., Shabanowitz J., Hunt D. F., Karr T. L.2006Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genetics 38, 1440–1445 (doi:10.1038/ng1915) [DOI] [PubMed] [Google Scholar]
- Dorus S., Freeman Z. N., Parker E. R., Heath B. D., Karr T. L.2008Recent origins of sperm genes in Drosophila. Mol. Biol. Evol. 25, 2157–2166 (doi:10.1093/molbev/msn162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy E. M.2002Male germ cell gene expression. Recent Prog. Horm. Res. 57, 103–128 (doi:10.1210/rp.57.1.103) [DOI] [PubMed] [Google Scholar]
- Extavour C. G., Akam M.2003Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884 (doi:10.1242/dev.00804) [DOI] [PubMed] [Google Scholar]
- Fraser L. R., Harrison R. A., Herod J. E.1990Characterization of a decapacitation factor associated with epididymal mouse spermatozoa. J. Reprod. Fertil. 89, 135–148 [DOI] [PubMed] [Google Scholar]
- Friedlander M., Jeshtadi A., Reynolds S.2001The structural mechanism of trypsin-induced intrinsic motility in Manduca sexta spermatozoa in vitro. J. Insect Physiol. 47, 245–255 (doi:10.1016/S0022-1910(00)00109-8) [DOI] [PubMed] [Google Scholar]
- Fukuda N., Yomogida K., Okabe M., Touhara K.2004Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J. Cell Sci. 117, 5835–5845 (doi:10.1242/jcs.01507) [DOI] [PubMed] [Google Scholar]
- Gage M. J., Morrow E. H.2003Experimental evidence for the evolution of numerous, tiny sperm via sperm competition. Curr. Biol. 13, 754–757 (doi:10.1016/S0960-9822(03)00282-3) [DOI] [PubMed] [Google Scholar]
- Gage M. J., Parker G. A., Nylin S., Wiklund C.2002Sexual selection and speciation in mammals, butterflies and spiders. Proc. R. Soc. Lond. B 269, 2309–2316 (doi:10.1098/rspb.2002.2154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaino E., Mazzini M.1991Aflagellate sperm in three species of Leptophlebiidae (Ephemeroptera). Int. J. Insect Morphol. Embryol. 20, 119–125 [Google Scholar]
- Gangaraju V. K., Lin H.2009MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 10, 116–125 (doi:10.1038/nrm2621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez F., Simmons L. W.2007Shorter sperm confer higher competitive fertilization success. Evolution 61, 816–824 (doi:10.1111/j.1558-5646.2007.00084.x) [DOI] [PubMed] [Google Scholar]
- Gomendio M., Roldan E. R.2008Implications of diversity in sperm size and function for sperm competition and fertility. Int. J. Dev. Biol. 52, 439–447 (doi:10.1387/ijdb.082595mg) [DOI] [PubMed] [Google Scholar]
- Gou G., Zheng G.2004Hypotheses for the functions of intercellular bridges in male germ cell development and its cellular mechanisms. J. Theor. Biol. 229, 139–146 [DOI] [PubMed] [Google Scholar]
- Haerty W., et al. 2007Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177, 1321–1335 (doi:10.1534/genetics.107.078865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel M., Park C., Kot M.1994Genetic control of sex-chromosome inactivation during male meiosis. Cytogen. Genome Res. 66, 83–88 (doi:10.1159/000133672) [DOI] [PubMed] [Google Scholar]
- Hansbrough J. R., Garbers D. L.1981Speract. Purification and characterization of a peptide associated with eggs that activates spermatozoa. J. Biol. Chem. 256, 1447–1452 [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T.2004Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131, 93–104 (doi:10.1242/dev.00916) [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J.1994lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913–2924 [DOI] [PubMed] [Google Scholar]
- Hense W., Baines J. F., Parsch J.2007X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 5, e273 (doi:10.1371/journal.pbio.0050273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D., Shakes D., Ward S., Strome S.1989A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11. Dev. Biol. 136, 154–166 (doi:10.1016/0012-1606(89)90138-3) [DOI] [PubMed] [Google Scholar]
- Hou S. X., Zheng Z., Chen X., Perrimon N.2002The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev. Cell 3, 765–778 (doi:10.1016/S1534-5807(02)00376-3) [DOI] [PubMed] [Google Scholar]
- Huckins C.1978Spermatogonial intercellular bridges in whole-mounted seminiferous tubules from normal and irradiated rodent testes. Am. J. Anat. 153, 97–121 (doi:10.1002/aja.1001530107) [DOI] [PubMed] [Google Scholar]
- Inaba K.2007Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann. N. Y. Acad. Sci. 1101, 506–526 (doi:10.1196/annals.1389.017) [DOI] [PubMed] [Google Scholar]
- Ishijima S., Ishijima S. A., Afzelius B. A.1994Movement of Myzostomum spermatozoa: calcium ion regulation of swimming direction. Cell Motil. Cytoskel. 28, 135–142 (doi:10.1002/cm.970280205) [DOI] [PubMed] [Google Scholar]
- Jamieson B. G. M., Dallai R., Afzelius B. A.1999Insects their spermatozoa and phylogeny Enfield, NH: Science Publishers Inc [Google Scholar]
- Johnson J. M., Edwards S., Shoemaker D., Schadt E. E.2004Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21, 93–102 (doi:10.1016/j.tig.2004.12.009) [DOI] [PubMed] [Google Scholar]
- Kalmykova A. I., Nurminsky D. I., Ryzhov D., Shevelyov Y. Y.2005Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 33, 1435–1444 (doi:10.1093/nar/gki281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano N., Yoshida M.2007Semen-coagulating protein, SVS2, in mouse seminal plasma controls sperm fertility. Biol. Reprod. 76, 353–361 (doi:10.1095/biolreprod.106.056887) [DOI] [PubMed] [Google Scholar]
- Kawase E., Wong M. D., Ding B. C., Xie T.2004Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131, 1365–1375 (doi:10.1242/dev.01025) [DOI] [PubMed] [Google Scholar]
- Kiger A. A., Jones D. L., Schulz C., Rogers M. B., Fuller M. T.2001Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542–2545 (doi:10.1126/science.1066707) [DOI] [PubMed] [Google Scholar]
- Kimble J., Crittenden S.2007Control of germline stem cells, entry into meiosis, and the sperm/oocyte decision in C. elegans. Annu. Rev. Cell Dev. Biol. 23, 405–433 (doi:10.1146/annurev.cellbio.23.090506.123326) [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G.1981On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81, 208–219 (doi:10.1016/0012-1606(81)90284-0) [DOI] [PubMed] [Google Scholar]
- King R. C.1970Ovarian development in Drosophila melanogaster New York, NY: Academic Press [Google Scholar]
- Kloc M., Etkin L.2005RNA localization mechanisms in oocytes. J. Cell Sci. 118, 269–282 (doi:10.1242/jcs.01637) [DOI] [PubMed] [Google Scholar]
- Knowles D., McLysaght A.2009Recent de novo origin of human protein-coding genes. Genome Res. 19, 1752–1759 (doi:10.1101/gr.095026.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J., McCarrey J., Djakiew D., Krawetz S.1998Differentiation: the selective potentiation of chromatin domains. Development 125, 4749–4755 [DOI] [PubMed] [Google Scholar]
- Kusnetsov S., Lyanguzowa M., Bosch T.2001Role of epithelial cells and programmed cell death in Hydra spermatogenesis. Zoology 104, 25–31 [DOI] [PubMed] [Google Scholar]
- Lam N., Chesney M. A., Kimble J.2006Wnt signaling and CEH-22/tinman/Nkx2.5 specify a stem cell niche in C. elegans. Curr. Biol. 16, 287–295 (doi:10.1016/j.cub.2005.12.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon C. W., Ward S.1995Sperm precedence in a hermaphroditic nematode (Caenorhabditis elegans) is due to competitive superiority of male sperm. Experientia 51, 817–823 (doi:10.1007/BF01922436) [DOI] [PubMed] [Google Scholar]
- Le Bras S., Van Doren M.2006Development of the male germline stem cell niche in Drosophila. Dev. Biol. 294, 92–103 [DOI] [PubMed] [Google Scholar]
- Lefevre G., Jr, Jonsson U. B.1962Sperm transfer, storage, displacement, and utilization in Drosophila melanogaster. Genetics 47, 1719–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessells C. M., Snook R. R., Hosken D. J.2009The evolutionary origin and maintenance of sperm: selection for a small, motile gamete mating type. In Sperm biology: an evolutionary perspective (eds Birkhead T., Hosken D., Pitnick S.), pp. 43–67 New York, NY: Academic Press [Google Scholar]
- Levine M., Jones C., Kern A., Lindfors H., Begun D.2006Novel genes derived from non-coding DNA in Drosophila melanogaster are frequently X-linked and show testis-biased expression. Proc. Natl Acad. Sci. USA 103, 9935–9939 (doi:10.1073/pnas.0509809103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J., Song Y., de Jong M., Bagha S., Ausio J.2003A walk through vertebrate and invertebrate protamines. Chromosoma 111, 473–482 (doi:10.1007/s00412-002-0226-0) [DOI] [PubMed] [Google Scholar]
- Lewis J. D., de Jong M. E., Bagha S. M., Tang A., Gilly W. F., Ausio J.2004aAll roads lead to arginine: the squid protamine gene. J. Mol. Evol. 58, 673–680 (doi:10.1007/s00239-004-2589-8) [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Saperas N., Song Y., Zamora M. J., Chiva M., Ausio J.2004bHistone H1 and the origin of protamines. Proc. Natl Acad. Sci. USA 101, 4148–4152 (doi:10.1073/pnas.0308721101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Neaves W. B.2006Normal stem cells and cancer stem cells: the niche matters. Cancer Res. 66, 4553–4557 (doi:10.1158/0008-5472.CAN-05-3986) [DOI] [PubMed] [Google Scholar]
- Li J., Wu C.2005New symbiotic hypothesis on the origin of eukaryotic flagella. Naturwissenschaften 92, 305–309 (doi:10.1007/s00114-005-0635-z) [DOI] [PubMed] [Google Scholar]
- Li L., Xie T.2005Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 21, 605–631 (doi:10.1146/annurev.cellbio.21.012704.131525) [DOI] [PubMed] [Google Scholar]
- Li M., Hong N., Xu H., Yi M., Li C., Gui J., Hong Y.2009Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. [DOI] [PubMed] [Google Scholar]
- Manton I.1953Number of fibrils in the cilia of green algae. Nature 171, 485–486 (doi:10.1038/171485b0) [DOI] [PubMed] [Google Scholar]
- Manton I.1970Plant spermatozoids. In Comparative spermatology (ed. Baccetti B.), pp. 143–158 Rome, Italy: Academia Nazionale Dei Lincei [Google Scholar]
- Mattei X.1991Spermatozoon ultrastructure and its systematic implictions in fishes. Can. J. Zool. 69, 3038–3055 (doi:10.1139/z91-428) [Google Scholar]
- Mattei X., Mattei C., Reizer C., Chevalier J.-L.1972Ultrastructure des spermatozoids aflagelles des Mormyres (poisson teleosteens). J. Microsc. 15, 67–78 [Google Scholar]
- Maxwell W. M., de Graaf S. P., Ghaoui Rel H., Evans G.2007Seminal plasma effects on sperm handling and female fertility. Soc. Reprod. Fertil. Suppl. 64, 13–38 [DOI] [PubMed] [Google Scholar]
- Michiels F., Gasch A., Kaltschmidt B., Renkawitz-Pohl R.1989A 14 bp promoter element directs the testis specificity of the Drosophila b2-tubulin gene. EMBO J. 8, 1559–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Cutter A., Yamamoto I., Ward S., Greenstein D.2004Clustered organization of reproductive genes in the C. elegans genome. Curr. Biol. 14, 1284–1290 (doi:10.1016/j.cub.2004.07.025) [DOI] [PubMed] [Google Scholar]
- Morrow E. H.2004How the sperm lost its tail: the evolution of aflagellate sperm. Biol. Rev. Camb. Phil. Soc. 79, 795–814 [DOI] [PubMed] [Google Scholar]
- Mottier-Pavie V., Megraw T. L.2009Drosophila Bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J., Mahadevaiah S., Park P., Warburton P., Page D., Turner J.2008The mouse X chromsome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40, 794–799 (doi:10.1038/ng.126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton W. D.1980Ultrastructural analysis of the motile apparatus of the aflagellate spermatozoon of macrostomum tubum. J. Ultrastruct. Res. 73, 318–330 (doi:10.1016/S0022-5320(80)90091-X) [DOI] [PubMed] [Google Scholar]
- Nielsen M., Caserta J., Kidd S., Phillips C.2006Functional constraint underlies 60 million year stasis of dipteran testis-specific beta-tubulin. Evol. Dev. 8, 23–29 [DOI] [PubMed] [Google Scholar]
- Nixon B., MacIntyre D. A., Mitchell L. A., Gibbs G. M., O'Bryan M., Aitken R. J.2006The identification of mouse sperm-surface-associated proteins and characterization of their ability to act as decapacitation factors. Biol. Reprod. 74, 275–287 (doi:10.1095/biolreprod.105.044644) [DOI] [PubMed] [Google Scholar]
- Normark B. B.2009Unusual gametic and genetic systems. In Sperm biology: an evolutionary perspective (eds Birkhead T. R., Hosken D. J., Pitnick S.), pp. 507–538 New York, NY: Academic Press [Google Scholar]
- Oatley J. M., Brinster R. L.2006Spermatogonial stem cells. Meth. Enzymol. 419, 259–282 [DOI] [PubMed] [Google Scholar]
- Oatley J. M., Brinster R. L.2008Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24, 263–286 (doi:10.1146/annurev.cellbio.24.110707.175355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Ohmura M., Ohbo K.2005The niche for spermatogonial stem cells in the mammalian testis. Int. J. Hematol. 82, 381–388 (doi:10.1532/IJH97.05088) [DOI] [PubMed] [Google Scholar]
- Osborne C. S., et al. 2004Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36, 1065–1071 (doi:10.1038/ng1423) [DOI] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G. G., Malley J., Andrews J., Eastman S., Oliver B.2003Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299, 697–700 (doi:10.1126/science.1079190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., et al. 2004A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5, R40 (doi:10.1186/gb-2004-5-6-r40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G. A., Begon M. E.1993Sperm competition games: sperm size and number under gametic control. Proc. R. Soc. Lond. B 253, 255–262 (doi:10.1098/rspb.1993.0111) [DOI] [PubMed] [Google Scholar]
- Phillips D. M.1970Insect sperm: their structure and morphogenesis. J. Cell Biol. 44, 243–277 (doi:10.1083/jcb.44.2.243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittoggi C., et al. 1999A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J. Cell Sci. 112, 3537–3548 [DOI] [PubMed] [Google Scholar]
- Quill T. A., Sugden S. A., Rossi K. L., Doolittle L. K., Hammer R. E., Garbers D. L.2003Hyperactivated sperm motility driven by CatSper2 is required for fertilization. Proc. Natl Acad. Sci. USA 100, 14 869–14 874 (doi:10.1073/pnas.2136654100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz J., Ponce A., Hartl D. L., Nurminsky D.2003Origin and evolution of a new gene expressed in the Drosophila sperm axoneme. Genetica 118, 233–244 (doi:10.1023/A:1024186516554) [PubMed] [Google Scholar]
- Ren D., Navarro B., Perez G., Jackson A., Hsu S., Shi Q., Tilly J., Clapham D.2001A sperm ion channel required for sperm motility and male fertility. Nature 413, 603–609 (doi:10.1038/35098027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M. G., Callaini G.2007The Drosophila parkin homologue is required for normal mitochondrial dynamics during spermiogenesis. Dev. Biol. 303, 108–120 (doi:10.1016/j.ydbio.2006.10.038) [DOI] [PubMed] [Google Scholar]
- Riparbelli M. G., Callaini G., Mercati D., Hertel H., Dallai R.2009Centrioles to basal bodies in the spermiogenesis of Mastotermes darwiniensis (Insecta, Isoptera). Cell Motil. Cytoskel. [DOI] [PubMed] [Google Scholar]
- Roldan E., Gomendio M., Vitullo A.1992The evolution of eutherian spermatozoa and underlying selective forces: female selection and sperm competition. Biol. Rev. Camb. Phil. Soc. 67, 551–593 [DOI] [PubMed] [Google Scholar]
- Sada A., Suzuki A., Suzuki H., Saga Y.2009The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325, 1394–1398 (doi:10.1126/science.1172645) [DOI] [PubMed] [Google Scholar]
- Santos A. C., Lehmann R.2004Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 14, R578–R589 (doi:10.1016/j.cub.2004.07.018) [DOI] [PubMed] [Google Scholar]
- Saunders C., Larman M., Parrington J., Cox L., Royse J., Blayney L., Swann K., Lai F.2002PLCzeta: a sperm-specific trigger of Ca2+ oscillations in eggs and embryo development. Development 129, 3533–3544 [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A.1997/98Phylogenetic relationships of the Nematomorpha—a discussion of current hypotheses. Zool. Anzeiger 236, 203–216 [Google Scholar]
- Seto A. G., Kingston R. E., Lau N. C.2007The coming of age for Piwi proteins. Mol. Cell 26, 603–609 (doi:10.1016/j.molcel.2007.05.021) [DOI] [PubMed] [Google Scholar]
- Sheng X., Posenau T., Gumulak-Smith J., Matunis E., Van Doren M., Wawersik M.2009Jak-STAT regulation of male germline stem cell establishment during Drosophila embryogensis. Dev. Biol. 334, 335–344 (doi:10.1016/j.ydbio.2009.07.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov Y. Y., Lavrov S., Mikhaylova L., Nurminsky I., Kulathainal R., Egorova K., Rozovsky Y., Nurminsky D. I.2009The B-type lamin is required for somatic repression of testis-specifc gene clusters. Proc. Natl Acad. Sci. USA 106, 3282–3287 (doi:10.1073/pnas.0811933106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Stanley P.2006Evolutionary origins of Notch signaling in early development. Cell Cycle 5, 274–278 [DOI] [PubMed] [Google Scholar]
- Shivdasani A. A., Ingham P. W.2003Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr. Biol. 13, 2065–2072 (doi:10.1016/j.cub.2003.10.063) [DOI] [PubMed] [Google Scholar]
- Siddall N. A., McLaughlin E. A., Marriner N. L., Hime G. R.2006The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc. Natl Acad. Sci. USA 103, 8402–8407 (doi:10.1073/pnas.0600906103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A.2001Every sperm is sacred: fertilization in Caenorhabditis elegans. Dev. Biol. 230, 101–109 (doi:10.1006/dbio.2000.0118) [DOI] [PubMed] [Google Scholar]
- Singson A.2006Sperm activation: time and tide wait for no sperm. Curr. Biol. 16, R160–R162 (doi:10.1016/j.cub.2006.02.039) [DOI] [PubMed] [Google Scholar]
- Smith H.2006Sperm motility and MSP. WormBook (ed. The C. elegans research community), 1–8 See http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M., Gisselmann G., Poplawski A., Riffell J. A., Wetzel C. H., Zimmer R. K., Hatt H.2003Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299, 2054–2058 (doi:10.1126/science.1080376) [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Rubin G. M.2002Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1, 5 (doi:10.1186/1475-4924-1-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield G. M., Villeneuve A. M.2006Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr. Biol. 16, 252–263 (doi:10.1016/j.cub.2005.12.041) [DOI] [PubMed] [Google Scholar]
- Stockley P., Gage M. J., Parker G. A., Møller A. P.1997Sperm competition in fishes: the evolution of testis size and ejaculate characteristics. Am. Nat. 149, 933–954 (doi:10.1086/286031) [DOI] [PubMed] [Google Scholar]
- Strome S.2005Specification of the germ line. In WormBook (eds The C. elegans Reasearch Community), pp. 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Zhang Y., Parisi M., Oliver B.2007Demasculinization of X chromosomes in the Drosophila genus. Nature 4450, 238–241 (doi:10.1038/nature06330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez S.2008Control of hyperactivation in sperm. Hum. Rep. Update 14, 647–657 (doi:10.1093/humupd/dmn029) [DOI] [PubMed] [Google Scholar]
- Suzuki N.1990Structure and function of sea urchin egg jelly molecules. Zool. Sci. 7, 355–370 [Google Scholar]
- Suzuki N., Yoshino K.1992The relationship between amino acid sequences of sperm-activating peptides and the taxonomy of echinoids. Comp. Biochem. Physiol. B 102, 679–690 (doi:10.1016/0305-0491(92)90064-X) [DOI] [PubMed] [Google Scholar]
- Swanson W. J., Vacquier V. D.2002The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 (doi:10.1038/nrg733) [DOI] [PubMed] [Google Scholar]
- Swanson W. J., Nielsen R., Yang Q.2003Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20, 18–20 [DOI] [PubMed] [Google Scholar]
- Takahashi T., McDougall C., Troscianko J., Chen W., Jayaraman-Nagarajan A., Shimeld S., Ferrier D.2009An EST screen from the annelid Pomatoceros lamarckii reveals patterns of gene loss and gain in animals. BMC Evol. Biol. 9, 240 (doi:10.1186/1471-2148-9-240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. P., Zhou S. X.1996The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 178, 124–132 (doi:10.1006/dbio.1996.0203) [DOI] [PubMed] [Google Scholar]
- Tokuyasu K. T.1974Dynamics of spermiogenesis in Drosophila melanogaster. 3. Relation between axoneme and mitochondrial derivatives. Exp. Cell Res. 84, 239–250 (doi:10.1016/0014-4827(74)90402-9) [DOI] [PubMed] [Google Scholar]
- Tulina N., Matunis E.2001Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294, 2546–2549 (doi:10.1126/science.1066700) [DOI] [PubMed] [Google Scholar]
- Turner J.2007Meiotic sex chromsome inactivation. Development 134, 1823–1831 (doi:10.1242/dev.000018) [DOI] [PubMed] [Google Scholar]
- Van Speybroeck L., De Waele D., Van de Vijver G.2002Theories in early embryology: close connections between epigenesis, preformationism, and self-organization. Ann. N. Y. Acad. Sci. 981, 7–49 [PubMed] [Google Scholar]
- Wang P., McCarrey J., Yang F., Page D.2001An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27, 422–426 (doi:10.1038/86927) [DOI] [PubMed] [Google Scholar]
- Ward S., Carrel J. S.1979Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73, 304–321 (doi:10.1016/0012-1606(79)90069-1) [DOI] [PubMed] [Google Scholar]
- Werner M., Simmons L. W.2008Insect sperm motility. Biol. Rev. Camb. Phil. Soc. 83, 191–208 [DOI] [PubMed] [Google Scholar]
- White-Cooper H., Doggett K., Ellis R. E.2009The evolution of spermatogenesis. In Sperm biology: an evolutionary perspective (eds Birkhead T. R., Hosken D. J., Pitnick S. S.), pp. 151–183 New York, NY: Academic Press [Google Scholar]
- Wu C.-I., Xu E.2003Sexual antagonism and X inactivation—the SAXI hypothesis. Trends Genet. 19, 243–247 (doi:10.1016/S0168-9525(03)00058-1) [DOI] [PubMed] [Google Scholar]
- Wylie C.1999Germ cells. Cell 96, 165–174 (doi:10.1016/S0092-8674(00)80557-7) [DOI] [PubMed] [Google Scholar]
- Xu M., Cook P.2008The role of specialized transcription factories in chromsome pairing. Biochim. Biophys. Acta (BBA) 1783, 2155–2160 (doi:10.1016/j.bbamcr.2008.07.013) [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Fuller M. T., Jones D. L.2005Signalling in stem cell niches: lessons from the Drosophila germline. J. Cell Sci. 118, 665–672 (doi:10.1242/jcs.01680) [DOI] [PubMed] [Google Scholar]
- Yoshida S., Sukeno M., Nakagawa T., Ohbo K., Nagamatsu G., Suda T., Nabeshima I.2006The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 133, 1495–1505 (doi:10.1242/dev.02316) [DOI] [PubMed] [Google Scholar]
- Yoshida S., Sukeno M., Nabeshima Y.2007A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science 317, 1722–1726 (doi:10.1126/science.1144885) [DOI] [PubMed] [Google Scholar]
- Yoshida M., Kawano N., Yoshida K.2008Control of sperm motility and fertility: diverse factors and common mechanisms. Cell Mol. Life Sci. 65, 3446–3457 (doi:10.1007/s00018-008-8230-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., King M. L.2004Sending RNAs into the future: RNA localisation and germ cell fate. IUBMB Life 56, 19–27 [DOI] [PubMed] [Google Scholar]
- Zhu J., Massey J. B., Mitchell-Leef D., Elsner C. W., Kort H. I., Roudebush W. E.2006Platelet-activating factor acetylhydrolase activity affects sperm motility and serves as a decapacitation factor. Fertil. Steril. 85, 391–394 (doi:10.1016/j.fertnstert.2005.07.1303) [DOI] [PubMed] [Google Scholar]
- Zinzen R., Gairardot C., Gagneur J., Braus M., Furlong E. E. M.2009Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (doi:10.1038/nature08531) [DOI] [PubMed] [Google Scholar]