Abstract
Large-scale systematic resequencing has been proposed as the key future strategy for the discovery of rare, disease-causing sequence variants across the spectrum of human complex disease. We have sequenced the coding exons of the X chromosome in 208 families with X-linked mental retardation (XLMR), the largest direct screen for constitutional disease-causing mutations thus far reported. The screen has discovered nine genes implicated in XLMR, including SYP, ZNF711 and CASK reported here, confirming the power of this strategy. The study has, however, also highlighted issues confronting whole-genome sequencing screens, including the observation that loss of function of 1% or more of X-chromosome genes is compatible with apparently normal existence.
Mental retardation is defined as a disability characterized by “significant limitations both in intellectual functioning and in adaptive behaviour as expressed in conceptual, social and practical adaptive skills” with onset before the age of 18 years1,2. Mental retardation is one of the main reasons for referral to clinical pediatric, neurological and genetics services and is responsible for 5–10% of health care expenditure in some developed countries3-6.
Mental retardation may be caused by constitutional genetic abnormalities7,8. A significant proportion of these are large deletions, duplications or aneuploidies that affect multiple genes9-11. Mental retardation may also be due to mutations of individual genes and is a feature of autosomal dominant, autosomal recessive and X-linked genetic diseases (Supplementary Table 1 online). The most common cause of X-linked mental retardation (XLMR) is expansion of a trinucleotide repeat in the 5′ untranslated region of the FMR1 gene in fragile-X syndrome. Approximately 80 additional genes involved in XLMR have now been identified through genetic linkage analysis and positional cloning, candidate gene analysis or cytogenetic studies12,13. Each of these accounts for only a small number of families with XLMR and, despite this success, the mutated genes responsible for XLMR in most affected families have not been identified14. Thus, mental retardation generally and XLMR in particular are notable models of genetic heterogeneity in common disease susceptibility.
The identification of further genes involved in XLMR using conventional approaches has become problematic. For many XLMR-associated genes, mental retardation is the only clinical feature or is associated with relatively nonspecific and inconsistent accessory phenotypic characteristics, a clinical picture termed nonsyndromic XLMR. This similarity of phenotype precludes pooling of linkage information from different families. Moreover, mental retardation is common and phenocopies within XLMR families can further compromise mapping of the underlying genes. Although candidate gene studies have occasionally proved fruitful, genes involved in XLMR encode proteins with diverse or unknown biological roles. Mental retardation therefore exemplifies the problems associated with identification of rare disease-causing variants in many complex diseases.
The X chromosome is 155 Mb long and comprises approximately 5% of a haploid human genome15. There are short pseudoautosomal regions at both telomeric ends, which recombine with cognate regions on the Y chromosome. The annotated X-chromosome sequence permits an alternative strategy for the identification of genes involved in XLMR. Systematic sequencing of all coding exons in individuals with XLMR will provide a catalog of X-chromosome coding sequence variants in each case. In some individuals one of these variants will be causative of mental retardation, with the remainder being background genetic variation. This approach is empowered by the characteristic pattern of transmission that allows preselection of families with mental retardation likely due to an abnormality on the X chromosome. Furthermore, it does not depend on genetic linkage information or patterns of accessory phenotypic features and requires a sample from only a single individual in each family. In this study we have implemented this strategy by sequencing most coding exons of X-chromosome genes in families with clinical features compatible with XLMR.
RESULTS
Sequence variants in families with XLMR
Genomic DNA from a male proband, or in five instances a female obligate carrier, from 208 families with multiple individuals with mental retardation and a pattern of transmission compatible with X linkage (see Table 1 for clinical summary) was sequenced through the coding exons of 718 X-chromosome genes (Supplementary Table 1). This set was composed of 699 out of 829 genes from the Vega database and 19 X-chromosome genes not included in Vega but present in Ensembl/NCBI (Supplementary Table 1). The average coverage of the 718 genes screened was 75%; therefore, the coverage of the full protein-coding sequences of the Vega X chromosome was 65%. Sixteen of the genes screened are in the pseudoautosomal regions common to the X and Y chromosomes and 702 are in the X-specific part. The screened DNA corresponds to ~1 Mb of coding sequence per sample and >200 Mb in total. The 208 families were prescreened and found negative for cytogenetic abnormalities at 500G banding resolution, for expansion of the FMR1 trinucleotide repeat and for unambiguous disease-causing sequence variants in the XLMR-causing genes published when the study was initiated (Supplementary Table 1).
Table 1.
Summary of the clinical features of the mental retardation probands studied
| No. families (n = 208) | |
|---|---|
| No. affected males | |
| 2 (sib pair) | 45 (21.6%) |
| 2 (other) | 14 (6.7%) |
| 3 | 52 (25.0%) |
| >3 | 97 (46.6%) |
| Ancestry | |
| Asian | 3 (1.4%) |
| Black African | 1 (0.5%) |
| European | 197 (94.7%) |
| European/Aboriginal | 1 (0.5%) |
| European/Asian | 1 (0.5%) |
| No data | 5 (2.4%) |
| Severity of mental retardation | |
| Severe (IQ 20–34) | 55 (26.4%) |
| Moderate (IQ 35–49) | 96 (46.2%) |
| Mild (IQ 50–69) | 51 (24.5%) |
| No data | 6 (2.9%) |
| Head circumference | |
| Macrocephaly | 24 (11.5%) |
| Microcephaly | 25 (12.0%) |
| Normal | 147 (70.7%) |
| No data | 12 (5.8%) |
| Epilepsy | |
| Yes | 46 (22.1%) |
| No | 148 (71.2%) |
| No data | 14 (6.7%) |
| Speech and language | |
| Absent | 26 (12.5%) |
| Delayed | 177 (85.1%) |
| Normal | 2 (1.0%) |
| No data | 3 (1.4%) |
| Dysmorphic features | |
| Yes | 66 (31.7%) |
| No | 140 (67.3%) |
| No data | 2 (1.0%) |
| Neurological features | |
| Yes | 47 (22.6%) |
| No | 157 (75.5%) |
| No data | 4 (1.9%) |
The slash indicates mixed ancestry.
We detected 1,858 different coding sequence variants, 1,769 from the X-specific and 89 from the pseudoautosomal X-chromosome regions (Table 2). We found that 1,814 were single-nucleotide changes: of these, 980 caused missense amino-acid substitutions, 22 caused nonsense (termination) codons, 13 were abnormalities at highly conserved bases at splice acceptor and donor sites and 799 were synonymous (silent) changes. Three variants were missense double-nucleotide substitutions, and 41 variants were small insertions and deletions, of which 26 were in-frame and 15 caused translational frameshifts.
Table 2.
Summary of all variants
| X specific |
Pseudoautosomal |
||||||
|---|---|---|---|---|---|---|---|
| Nonrecurrent | Recurrent | Total | Nonrecurrent | Recurrent | Total | Grand total | |
| Nonsense substitutions | 18 | 2 | 20 | 1 | 1 | 2 | 22 |
| Missense substitutions | 531 | 409 | 940 | 22 | 21 | 43 | 983 |
| Silent substitutions | 328 | 428 | 756 | 21 | 22 | 43 | 799 |
| In-frame ins/dels | 12 | 14 | 26 | 26 | |||
| Out-of-frame ins/dels | 13 | 1 | 14 | 1 | 1 | 15 | |
| Splice | 12 | 1 | 13 | 13 | |||
| Total | 914 | 855 | 1,769 | 44 | 45 | 89 | 1,858 |
Read-through variants have been included with frameshifting insertions/deletions.
The dataset allows direct characterization of the pattern of haplotypic coding sequence variation of individual X chromosomes. Although ascertained from individuals with XLMR, only a small fraction of the observed variants is likely to cause mental retardation and, therefore, the set predominantly represents background population variation. Of the 1,769 coding sequence variants from the X-specific part of the X chromosome, 914 were nonrecurrent (that is, observed in only one XLMR-affected family) and 855 were recurrent (observed in multiple XLMR-affected families, Table 2). We identified 63% of the recurrent and 16% of the nonrecurrent variants in the dbSNP database. The sequences of any two individuals differed on average by 109 variants. Of these, six were nonrecurrent, including four missense and two synonymous variants, and 103 were recurrent, including 40 missense, 60 synonymous and two in-frame insertions/deletions. The results illustrate that most coding sequence differences between individuals are recurrent (‘common’) variants despite the existence of a larger number of different nonrecurrent (‘rare’) variants.
Sequence variants that truncate proteins
A subset of the sequence variants is predicted to introduce a premature termination codon and hence truncate the wild-type protein sequence. Truncating variants are usually highly deleterious to protein function: they constitute a substantial proportion of monogenic (mendelian) disease-causing mutations but a relatively small proportion of polymorphisms. Therefore, as the first analytic step to identify new genes involved in mental retardation, we considered the set of truncating variants detected in the screen.
We observed 42 different truncating variants in 30 genes (Table 3); 40 were in 28 genes from the X-specific region of the X chromosome and 2 were in 2 genes from the pseudoautosomal region. In addition, we found four ‘read-through’ variants that cause a translational frameshift close to the wild-type termination codon and extend the open reading frame into previously untranslated 3′ DNA (described further in Supplementary Note online).
Table 3.
Truncating and read-through variants identified in the screen
| Region of X chrom |
Gene | Family number |
LOD | Truncating variants in cases |
Stop position |
Protein size (aa) |
Truncating variants in controls |
Frequency in controls |
Mutations in known MR genes |
Abnormal transcript identified |
Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Truncating | ||||||||||||
| Recurrent | X specific | MAGEE2 | – | – | 358G>T, E120* | 120 | 524 | 358G>T, E120* | 4/244 | |||
| X specific | UBE2NL | – | – | 266T>G, L89* | 89 | 154 | 266T>G, L89* | 100/283 | ||||
| PAR | GTPBP6 | – | – | 118C>T, Q40* | 40 | 287 | 118C>T, Q40* | 117/243 | ||||
| Nonrecurrent (>1 variant/gene) | X specific | AP1S2 | 445 | 3.85 | 106C>T, Q36* | 36 | 157 | 0/726 | 16 | |||
| 502 | 2.36 | 154C>T, R52* | 52 | 157 | 0/726 | 16 | ||||||
| 63 | 2.2 | IVS 3–2 delTACA | 64 | 157 | 0/726 | 16 | ||||||
| X specific | BRWD3 | 322 | 0.03 | IVS 29 +1 G>T | 1,093 | 1,802 | 0/520 | Yes | 20 | |||
| 336 | 0.26 | 946_947insA, R316fs*22 | 337 | 1,802 | 0/520 | 20 | ||||||
| X specific | CUL4B | 42 | 0.58 | 1007_1011delTTATA, I336fs*2 | 338 | 913 | 0/637 | 17 | ||||
| 307 | 0.88 | 1162C>T, R388* | 388 | 913 | 0/637 | 17 | ||||||
| 43 | 2.93 | 2566C>T, R856* | 856 | 913 | 0/637 | 17 | ||||||
| 329 | 0.59 | IVS 7–2 A>G | 308 | 913 | 0/637 | Yes | 17 | |||||
| 363 | 0.42 | 2493G>A, T831Ta | 806 | 913 | 0/637 | Yes | 17 | |||||
| X specific | ITIH5L | 32 | −1.38 | 670C>T, R224* | 224 | 1,313 | 0/489 | IL1RAPL1 | ||||
| 74 | −0.63 | IVS 7 +1 ins T | 1,313 | IVS 7 +1 ins T | 1/528 | |||||||
| X specific | SLC9A6 | 197 | 1.79 | 1402C>T, R468* | 468 | 669 | 0/282 | 21 | ||||
| 227 | 0.87 | 51 1_512delAT | 230 | 669 | 0/282 | 21 | ||||||
| X specific | UPF3B | 407 | −0.55 | 674_677delGAAA, R225fs*22 | 275 | 483 | 0/730 | 18 | ||||
| 309 | 0.56 | 867_868delAG, G290fs*2 | 292 | 483 | 0/730 | 18 | ||||||
| 62 | 2.66 | 1288C>T, R430* | 430 | 483 | 0/730 | 18 | ||||||
| X specific | ZDHHC9 | 152 | 0.23 | 175_176insCGCT, Y59fs*33 | 92 | 364 | 0/445 | 19 | ||||
| 602 | 0.49 | IVS 3 +5 G>C | 43 | 364 | 0/445 | Yes | 19 | |||||
| Nonrecurrent (1 variant/gene) | X specific | ARSF | 263 | 0.26 | IVS 8 +5 G>A | 591 | 0/217 | |||||
| X specific | ATXN3L | 324 | −1.47 | 76G>T, 26 * | 26 | 355 | 76G>T, E26* | 1/336 | ||||
| X specific | BEX4 | 232 | 1.03 | IVS 1 +1 G>A | 447 | IVS 1 +1 G>A | 2/530 | |||||
| X specific | DRP2 | 306 | −3.76 | IVS 10 +1 G>C | 372 | 957 | 0/247 | Yes | ||||
| X specific | FAM47B | 241 | 0.66 | 331C>T, Q111* | 111 | 645 | 0/233 | MECP2 | ||||
| X specific | MAP3K15 | 430 | −1.11 | 1069C>T, R357* | 357 | 788 | 1831C>T, R611* | 2/272 | ||||
| 1789C>T, R597* | 1/243 | |||||||||||
| X specific | MAP7D3 | 141 | −1.05 | IVS 9 +1 G>C | 567 | 877 | 0/460 | Yes | ||||
| X specific | P2RY4 | 400 | 0.26 | 1043G>A, W348* | 348 | 366 | 303G>A, W101* | 3/249 | ||||
| X specific | RPL9P7 | 116 | −0.71 | 75delT, I26fs*2 | 27 | 192 | 0/266 | |||||
| X specific | SAGE1 | 32 | 0.88 | 621T>A, C207* | 207 | 904 | 0/273 | IL1RAPL1 | ||||
| X specific | SATL1 | 197 | −6.46 | 1325G>A, W442* | 442 | 632 | 0/284 | SLC9A6 | ||||
| X specific | SSX6 | 81 | −0.03 | 111G>A, W37* | 37 | 188 | 0/274 | |||||
| X specific | SYP | 346 | 0.49 | 274_275insA, T92fs*45 | 137 | 313 | 0/272 | |||||
| X specific | SYTL5 | 438 | −0.51 | 1633G>T, G545* | 545 | 730 | 0/464 | |||||
| X specific | USP9X | 383 | 1.18 | 7505delA, Q2502fs*18 | 2,520 | 2,547 | 0/282 | |||||
| X specific | VSIG4 | 468 | 0.00 | 973A>T, R325* | 325 | 399 | 0/524 | |||||
| X specific | ZCCHC13 | 267 | −1.37 | 45G>A, W15* | 15 | 168 | 0/263 | |||||
| X specific | ZNF183 | 581 | 1.20 | 901C>T, Q301* | 301 | 343 | 0/252 | |||||
| X specific | ZNF711 | 208 | 1.31 | 2157_2158delTG, 719fs*1 | 720 | 771 | 0/252 | |||||
| PAR | IL9R | 432 | – | 58C>T, R20* | 20 | 522 | 0/248 | |||||
| Read-through | ||||||||||||
| Recurrent | PAR | ASMTL | – | – | 1864delT, *622fs*8 | 630 | 622 | 1864delT, *622fs*8 | 71/223 | |||
| X specific | H2BFWT | – | – | 510delA, Q171fs*30b | 201 | 175 | 0/260 | |||||
| Nonrecurrent | X specific | NSDHL | 577 | 1.06 | 1098_1099insT, R367fs*31 | 398 | 374 | 0/224 | ||||
| X specific | CXorf12 | 340 | 0.26 | 782delG, R261fs*11 | 272 | 262 | 0/293 | MECP2 |
This variant introduces a cryptic splice site that causes a truncation.
This variant was seen in only two families and generated a lod score of 0.83 in each.
Three truncating variants were recurrent (UBE2NL:266T>G, L89*; MAGEE2: 358G>T, E120* and GTPBP6: 118C>T, Q40*) (Table 3). These were found in controls at a similar prevalence to XLMR-affected families and so are unlikely to be responsible for mental retardation in the families in which they were identified. They each, however, are predicted to cause substantial truncation of the encoded proteins. Therefore, loss of some, or all, functions of UBE2NL, MAGEE2 and GTPBP6 seems compatible with normal development and intellectual function.
Thirty-eight truncating variants observed in the 702 genes from the X-specific part of the X chromosome were each found in only a single XLMR-affected family (that is, they were nonrecurrent variants). One gene (CUL4B) had five different nonrecurrent truncating variants, two genes (AP1S2 and UPF3B) had three, four genes (BRWD3, ZDHHC9, ITIH5L, SLC9A6) had two, and 19 genes had a single nonrecurrent truncating variant (Table 3 and Supplementary Fig. 1 online). Simulating a random distribution of truncating variants through the 702 genes and comparing it to the distribution observed provided strong evidence for clustering of these nonrecurrent truncating variants (P < 0.001) in a subset of genes. The clustering is consistent with this subset of genes being involved in XLMR, but other explanations cannot be excluded at this stage of analysis.
To evaluate further the genes with multiple truncating variants, we examined segregation of the variants in the families in which they were observed. Some of these results have been previously published16-21. In brief, truncating variants in AP1S2, CUL4B, BRWD3, UPF3B, ZDHHC9 and SLC9A6 segregated completely with mental retardation in the families in which they were identified; that is, each truncating variant was present in all genotyped subjects with mental retardation and absent in unaffected males (Supplementary Fig. 1). We sequenced all the coding exons of these six genes in control X chromosomes and did not find the truncating variants detected in XLMR-affected subjects or any other truncating variants (Table 3). The clustering of multiple different truncating variants in these genes, the evidence for segregation with mental retardation and the absence of truncating variants in controls indicate strongly that AP1S2, CUL4B, BRWD3, UPF3B, ZDHHC9 and SLC9A6 are XLMR genes. Five missense or in-frame variants in CUL4B, ZDHHC9 and SLC9A6 also showed evidence of involvement in mental retardation17,19,21. Mental retardation–causing variants in AP1S2, CUL4B, BRWD3, UPF3B, ZDHHC9 and SLC9A6 together account for the disease in 22 (10.6%) families out of the 208 screened.
By contrast, neither of the two truncating variants in ITIH5L segregated completely with mental retardation (Table 3 and Supplementary Fig. 1). We analyzed the complete coding sequence of ITIH5L in controls and found one of the truncating variants previously observed in a subject with mental retardation. The lack of segregation with mental retardation, the presence of a truncating variant in normal controls and the recent finding of a likely mental retardation–causing IL1RAPL1 deletion in one family with an ITIH5L truncating variant (unpublished data, Table 3) suggests that truncating variants in ITIH5L are not the cause of mental retardation in the families in which they were identified. Nevertheless, the strong evidence overall for the role in mental retardation of genes with more than one nonrecurrent truncating variant is reflected in the heterogeneity lod score of 18.3, with an estimated 92% families in this subset due to the truncating variant.
A single nonrecurrent truncating variant was found in 19 genes from the X-specific part of the X chromosome. Analysis of segregation in each family revealed that the truncating variant in nine genes (ATXN3L, DRP2, MAP3K15, MAP7D3, RPL9P7, SATL1, SSX6, SYTL5 and ZCCHC13) did not segregate with mental retardation (Table 3 and Supplementary Fig. 1). In nine of the remaining ten genes there was full segregation with the disease and in one, VSIG4, additional DNA samples were unavailable for testing. A heterogeneity lod score of 2.4 was obtained for the truncating variants in these 19 families, with mental retardation in 43% attributable to the truncating variant. Sequencing of the complete coding sequences of the 19 genes in male controls revealed one or more truncating variants in ATXN3L, BEX4, MAP3K15 and P2RY4 (Table 3). Furthermore, likely MR-causing abnormalities in MECP2, SLC9A6 and IL1RAPL1 have recently been found in affected individuals with single non-recurrent truncating variants in FAM47B, SATL1 and SAGE1, respectively (Table 3). Taken together, the results from 6 of the 19 genes with a single nonrecurrent truncating variant remain compatible with involvement in the causation of mental retardation (SYP, ZNF711, ARSF, ZNF183, VSIG4, and USP9X), whereas 13 others have one or more inconsistencies. To evaluate these six genes further, we sequenced their complete coding sequences in a further 914 male index subjects from XLMR-affected families and 1,129 male controls (Supplementary Table 2 online).
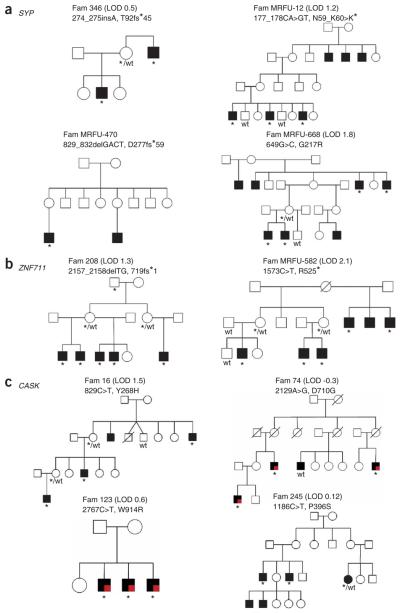
In SYP (also known as synaptophysin or p38) an additional nonsense variant was found in one of the 914 additional XLMR-affected subjects and an additional 4-bp deletion was identified in a second XLMR-affected subject (Fig. 1a and Table 4). The nonsense mutation showed evidence of segregation with mental retardation (lod score 1.2). Samples were not available for evaluation of the 4-bp deletion. No SYP truncating variants were found in the additional 1,129 controls. Together with the data from the primary screen, three SYP truncating variants were found in 1,122 XLMR-affected subjects examined, two of which have been examined and segregate with the disease (combined lod score 1.7), and there were no truncating variants in 1,401 controls. A missense variant found in a single subject with mental retardation at an amino acid residue that is highly conserved and which segregated with mental retardation (lod score 1.8) is also likely to be implicated in disease causation (Table 4). These data implicate SYP in XLMR. In the three families with truncating variants, mental retardation was mild to moderate and there were no consistent additional features, although epilepsy was noted in some individuals. SYP encodes an integral membrane protein of small synaptic vesicles.
Figure 1.
Pedigrees of families with likely deleterious variants in the SYP, ZNF711 and CASK genes. Shaded symbols indicate individuals with mental retardation and open symbols indicate individuals who are unaffected. Symbols containing a red square indicate individuals with both mental retardation and nystagmus. An asterisk indicates the presence of the variant allele and ‘wt’ indicates the presence of the wild-type allele. lod scores are shown in parentheses. (a) SYP. In family MRFU-470, additional individuals were unavailable to genotype. (b) ZNF711. In family 208 the maternal grandfather of the proband (I.1) is thought likely to have mild mental retardation, though this individual was unavailable to enable a detailed clinical assessment. (c) CASK. In family 74, individual III.4 was considered to have milder mental retardation compared to the other affected males and did not present with nystagmus. A lod score of −0.3 was generated when individual III.4 was considered as affected with mental retardation, and a lod score of 1.1 was obtained when his disease status was considered unknown.
Table 4.
Likely deleterious variants in SYP and ZNF711
| Gene | Family number |
lod | Mutation class | Mutation |
|---|---|---|---|---|
| SYP | 346 | 0.5 | Truncatinga | 274_275insA, T92fs*45 |
| SYP | MRFU-12 | 1.2 | Truncating | 177_178CA>GT, N59_K604K* |
| SYP | MRFU-470 | ND | Truncating | 829_832delGACT, D277fs*59 |
| SYP | MRFU-668 | 1.8 | Nonsynonymous | 649G>C, G217R |
| ZNF711 | 208 | 1.3 | Truncatinga | 2157_2158delTG, 719fs*1 |
| ZNF711 | MRFU-582 | 2.1 | Truncating | 1573C>T, R525* |
ND, not determined.
Found in original screen.
In ZNF711, an additional truncating variant was found in one subject (Fig. 1b and Table 4) and showed strong evidence of segregation with mental retardation (lod score 2.1). No ZNF711 variants were found in controls. Together with the results from the primary screen, two truncating variants were found in 1,122 XLMR-affected individuals, both of which segregate with the disease (combined lod score 3.4), and no truncating variants were found in 41,200 controls. These results indicate that ZNF711 is also an XLMR-associated gene. The two families with truncating ZNF711 mutations had moderate mental retardation without consistent additional distinctive features. ZNF711 encodes a zinc-finger protein of unknown function.
In ARSF three additional truncating variants were found among the 914 subjects with XLMR; only one could be evaluated and did not segregate with mental retardation. At least one additional truncating variant was found in each of ARSF, VSIG4 and ZNF183 in controls. In total, we found four truncating variants in ARSF in 1,122 XLMR-affected subjects and five in 1,346 controls. In both VSIG4 and ZNF183 one truncating variant was found in 1,122 cases and one in 1,653 and 1,391 controls, respectively. The results therefore suggest that none of these three genes is likely to be involved in mental retardation. No further USP9X truncating variants were found in cases or controls; consequently, the role of USP9X in XLMR remains uncertain.
Nonsynonymous and synonymous variants
We identified 983 different single-base substitution missense variants (Table 2 and Supplementary Table 3 online). The 26 in-frame deletions/insertions found are listed in Supplementary Table 4 online and described further in the Supplementary Note. As appears to be the case for truncating variants, missense variants may include a subset that causes mental retardation, with the remainder representing background population variation. However, the prevalence of missense variants in normal individuals is much higher than that of truncating variants, and the disruption of protein function they entail is generally more modest. Therefore, disease-causing missense variants are likely to represent a relatively small fraction of the total and distinguishing them from rare polymorphisms is problematic. We applied two analytic approaches to identify potential mental retardation–causing missense variants.
Disease-causing missense variants generally alter amino-acid residues that are more highly conserved during evolution than polymorphisms. Thus, we ranked the 983 missense variants according to a score that reflects the conservation of each amino acid (see Methods). Scrutiny of the top ranking variants from this analysis highlighted CASK. Only two missense variants in CASK were identified in the primary sequencing screen, and these are positioned second and third in the ranking (Fig. 1c and Supplementary Table 5 online). In silico and RT-PCR analyses indicate that one of these, 2129A>G (D710G), introduces a splice site that removes 27 bp of the coding sequence and thus nine amino acids of the CASK protein. Two further missense variants in CASK were found in a screen of 150 additional families with XLMR, both of which are at highly conserved amino acids and would score second and sixth in the ranking of missense variants. We did not find any missense variants in the complete coding region of CASK in 390 control X chromosomes. Mental retardation was mild to moderate in the four families with missense variants. In two, it was accompanied by nystagmus, a highly unusual accessory feature of XLMR, in multiple affected individuals. Three of the four variants segregate completely with mental retardation (Fig. 1c). The fourth variant, in family 74, is present in the three individuals with both mental retardation and nystagmus, but is absent from an individual with mental retardation without nystagmus (III-4), who may be a phenocopy. While this manuscript was under review, heterozygous inactivating mutations of CASK were reported to cause severe cerebral malformation in females22 and, in a male, a hemizygous truncation caused early neonatal lethality. These results are consistent with the discovery here of multiple different missense variants, which are likely to be less deleterious than truncating variants, in viable males with XLMR. CASK encodes a calcium/calmodulin-dependent serine protein kinase that is a member of the membrane-associated guanylate kinase (MAGUK) family and is located at the postsynaptic membrane of central nervous synapses23.
As a further strategy to identify missense variants causing mental retardation, we investigated the number of variants in each gene. Genes that show more amino acid variation in a human population than expected from their rate of evolution are identifiable by the McDonald-Kreitman test24. Application of this test to a random population sample identifies positively selected genes. Application to X-chromosome genes in a sample ascertained for XLMR, however, would be expected to identify both positively selected genes and those with excess missense variants that cause mental retardation. We restricted application of the McDonald-Kreitman test to nonrecurrent variants, as recurrent variants are less likely to be implicated in mental retardation. The results highlighted ZFX (P = 0.0014) and G6PD (P = 0.008), both of which have previously been identified as strong candidates for recent positive selection25,26. It also highlighted, however, four known genes involved in XLMR at similar levels of significance: HUWE1 (P = 0.007), OPHN1 (P = 0.001), MED12 (0.004) and PGK1 (P = 0.002). As these genes do not show evidence of recent positive selection25, their excess variation may be due to mental retardation–causing missense variants. By contrast, zero genes known to cause diseases other than mental retardation (excluding G6PD) were highlighted. Of genes not yet implicated in a monogenic disease, only one was highlighted: SMARCA1 (P = 0.009). These results suggest that missense variants in known and possibly additional XLMR-associated genes account for the disease in a further subset of families.
We observed 428 recurrent and 328 nonrecurrent synonymous variants in the X-specific part of the X chromosome. Although most synonymous variants are biologically silent, a small subset may exert cryptic biological effects through alterations in transcript processing or splicing. To search for additional cryptic splice variants, we applied the program NNsplice27 to all synonymous and missense base substitutions. Three synonymous and seven missense variants were predicted with a high score (>0.9) to introduce a new splice site (Supplementary Table 6 online). One of these is the missense variant in CASK (described above) that causes an abnormality of splicing. Of the remainder, five were recurrent and four were nonrecurrent variants. As all the likely mental retardation–causing variants thus far discovered in this study are nonrecurrent, these results suggest that many of the variants predicted to alter splicing are not implicated in mental retardation.
Characteristics of XLMR genes identified in this screen
Detailed clinical descriptions of the families with mental retardation–causing variants in six of the nine genes implicated in XLMR identified in this screen have been published (AP1S2, BRWD3, CUL4B, SLC9A6, UPF3B, ZDHHC9)16-21 and therefore their features are only reviewed briefly here. Mental retardation ranged from mild to severe and most families were previously classified as having nonsyndromic mental retardation. Following the identification of the genes involved in mental retardation, however, phenotypic characteristics common to some affected males were identified: for example, epilepsy and ataxia (SLC9A6), macrocephaly (BRWD3), relative macrocephaly, hypogonadism, central obesity and tremor (CUL4B), Marfanoid habitus (ZDHHC9), elements of the FG and Lujan-Fryns syndromes (UPF3B), epilepsy (SYP) and nystagmus (CASK). The encoded proteins have roles in vesicle trafficking (AP1S2), chromatin structure (BRWD3), nonsense-mediated RNA decay (UPF3B), ubiquitination (CUL4B), post-translational modification by palmitoylation (ZDHHC9), NA+/H+ exchange (SLC9A6), synaptic function (SYP) and synaptic signal transduction (CASK).
Structural and evolutionary characteristics of XLMR genes
The known biological functions of proteins encoded by genes involved in XLMR are diverse. To explore further the attributes of these genes and their encoded proteins, we compared features of currently identified genes associated with XLMR (80 genes) to X-chromosome genes associated with disease phenotypes that do not include cognitive impairment (61 genes) and X-chromosome genes that have not yet been associated with a mendelian disease (608 genes).
Genes involved in XLMR are more constrained in their evolution between human and macaque as measured by the dN/dS ratio (nonsynonymous changes per nonsynonymous site/synonymous changes per synonymous site) of 0.17 than genes associated with a disease other than mental retardation (0.31, P = 0.009) or genes not associated with a genetic disease (0.32, P = 0.003). Similarly, their amino acid compositions are more highly conserved between human and mouse compared to the other two classes (P < 0.01 for both comparisons). Previous studies have reported that genes implicated in nervous system diseases, genes associated with neurological functions and genes expressed in the brain undergo more purifying selection than genes in other functional classes28-30. The source of this evolutionary constraint is unclear.
Both genes involved in XLMR and those associated with other diseases are characterized by longer protein-coding sequences (P < 0.01), larger genomic footprints (P < 0.02) and greater numbers of exons (P < 0.01) than X-chromosome genes not associated with a disease. In part, this may be attributable to ascertainment bias. Larger genes are likely to have a higher mutation rate, because they constitute a bigger target for mutational processes, and therefore may have a greater prevalence of disease-causing mutations in the population. The higher the prevalence of disease-causing mutations, the more likely a gene is to have been identified as a disease gene. The comparatively large size of disease genes generally has previously been reported31. Significant differences between the three groups of genes were not found in levels of brain expression, tolerance of common sequence variation or the presence of paralogs in the human genome.
DISCUSSION
This direct, systematic sequencing screen of the coding exons of the X chromosome has identified nine XLMR-associated genes, which can now be used in the provision of genetic diagnostics to families with mental retardation. Mental retardation–causing variants in these genes are predominantly in families previously classified as having nonsyndromic mental retardation, emphasizing the utility of a systematic sequencing strategy in resolving phenotypically similar sets of disease cases that are heterogeneous with respect to their underlying genetic causation.
The study highlights, however, some analytical challenges of large-scale, systematic sequencing screens to detect rare disease-causing variants. The extent of genetic heterogeneity can be substantial. Inactivating variants in ~10% of genes on the X chromosome cause XLMR, of which approximately one-third are nonsyndromic. Moreover, individual genes may account for a very small fraction of cases. Mental retardation–causing mutations in SYP and ZNF711 each accounted for only ~0.3% cases, even in a set highly enriched by selection of families compatible with X linkage. Furthermore, disease-causing variants in many XLMR-associated genes constitute only a subset of the rare, nonsynonymous variants that are present. Thus, both SYP and ZNF711 would have been very difficult to detect if most of their disease-causing variants had not been truncating and highly penetrant. Indeed, that we detected them at all in the primary screen of 208 XLMR cases suggests that there are several more XLMR genes that contribute as infrequently.
Identification of protein-truncating variants has been a mainstay of mendelian disease gene discovery in the past, and has been similarly fruitful here. However, the evidence suggests that more than half of the 30 genes in which one or more truncating variants was found are not implicated in the causation of mental retardation. Therefore, a small number of truncating variants in a gene found through a genome-scale screen of a large number of disease cases requires careful evaluation and cannot be regarded as strong evidence of disease causation on its own.
Of the 19 genes with truncating variants in controls and/or in which the truncating variant does not segregate with mental retardation, eight (40%) have only a single exon, compared to 10% (73/718) of X-chromosome genes screened overall (P = 0.004). The enrichment in single-exon structures suggests that some may be retrotransposed copies that are being progressively excluded from the human genome by acquisition of truncating variants32,33. For UBE2NL and RPL9P7 the presence of a multiexon copy with very high sequence identity elsewhere in the genome provides evidence in favor of this hypothesis. However, other genes (ARSF, BEX4, DRP2, MAP3K15, MAP7D3, SAGE1, SATL1, SSX6, SYTL5, USP9X, VSIG4 and ITIH5L) have multiple exons. In the case of ARSF, there may be functional redundancy through closely related arylsulfatase genes elsewhere in the genome. In principle, it is possible that the deleterious effects of some of these truncating variants may be averted by transcript processing. The results overall, however, indicate that >1% of the currently defined protein-coding gene set on the X chromosome are not required for normal existence and cognitive function in humans.
Although we have sequenced most of the protein-coding exons of the X chromosome, the abnormal genes responsible for disease in most of the XLMR-affected families examined have not been identified. Of the subjects with XLMR screened, the likely genetic basis of 53 (25%) has now been established by detection of truncating, missense, in-frame or copy number variants found in this screen or by others analyzing the same cases in parallel (Supplementary Table 7 online). There are several plausible reasons for this. A proportion of X-chromosome genes were intractable to PCR primer design and, even when screened, coverage may not be complete. Moreover, a subset of disease-causing variants usually are in promoter regions, introns and other noncoding sequences, which we have not examined. Other classes of abnormality, for example copy number changes, inversions and other chromosomal rearrangements, are not detectable by a PCR-based resequencing approach. Indeed, some protein-coding genes may not yet have been annotated and it is conceivable that some XLMR-associated genes are not protein coding. It is also possible that mental retardation in a proportion of the families in the screen may not be due to an underlying abnormality on the X chromosome. In particular, some sibling pairs could be chance clusters of sporadic mental retardation cases or be due to autosomal recessive mutations.
It is also plausible that mental retardation in some of the families screened is due to missense variants. Several of the known mental retardation–associated genes initially identified through the presence of truncating changes have disease-causing missense variants reported in other families. Moreover, null alleles due to truncating variants of some X chromosome genes may be lethal in males, whereas missense changes may entail lesser developmental and functional consequences that manifest as mental retardation. However, identification of XLMR genes in which rare, exclusively missense variants are responsible for the disease is challenging. Only a small fraction of missense variants identified in a systematic screen are likely to cause mental retardation. Moreover, judging from the pattern of, mental retardation–causing truncating variants, we predict that they will be distributed over several different genes and that the number in each gene will be small. We adopted strategies based on the conservation of the altered amino acid and the number of missense variants in each gene, to highlight missense mental retardation–causing variants. Both of these were productive. However, the discriminative power of such approaches may be limited and the proportion of mental retardation–causing missense variants they have highlighted is unknown. Further study of the remaining missense variants identified here is now indicated. A critical test is whether there is a difference in the prevalence of a putative disease-causing missense variant, or group of variants, between disease cases and controls. However, the size of the sample sets required to investigate this may be daunting for studies of mental retardation and other complex disease phenotypes.
For many common human diseases most of the genetic basis of inherited susceptibility remains to be elucidated. Some may be encoded by common susceptibility alleles, which can be detected by association studies based on linkage disequilibrium maps. However, a significant proportion may be attributable to multiple rare variants, each of which accounts for a very small proportion of the familial risk of disease. In the relatively near future it will become possible to sequence the complete human genome, or biologically interesting components such as all protein coding exons, in large numbers of disease cases and controls to identify all variants, common and rare, disease-causing and innocuous. This systematic screen of the coding sequences of the X chromosome in XLMR illustrates some of the strengths of large-scale resequencing in disease gene identification, but also highlights its pitfalls and challenges.
METHODS
XLMR-affected families and controls
XLMR-affected families from the UK, United States, Australia, Europe and South Africa were included if there were two or more cases of mental retardation in males, predominant sparing of carrier females and no evidence of male-to-male transmission of mental retardation. Subjects were examined by a clinical geneticist with expertise in the field and the severity of the disease was categorized using DSM–IV or ICD-10 classifications (profound mental retardation was classified as severe). Where formal IQ testing using either the Stanford Binet or Wechsler IQ scoring was not performed, functional assessments of the degree of mental retardation were used. The individual entered into the sequencing screen was an affected male in 203 of 208 cases. Five screened individuals were obligate carrier females, all of whom were unaffected by mental retardation. Control DNAs were either from lymphoblastoid cell lines derived from adults of European ancestry from the UK without mental retardation or from filtered white cells from blood donors. XLMR-affected families and controls provided informed consent for research and the studies were reviewed and approved by ethics committees and institutional review boards of each collaborating institution.
Sequencing of X-chromosome coding exons
We included the full complement of 829 known protein-coding genes on the X chromosome in the Vega database of manually curated genes in the design. A number of genes on the X chromosome are present in multiple, highly similar copies. Where it was impossible to place PCR amplimers such that only a single copy was amplified, the gene was omitted from the screen. DNA from a single affected individual in each family was PCR-amplified to produce products of approximately 500 bp which were sequenced in both directions on ABI 3730 DNA analysers as previously described34. Sequence variants were called automatically using in-house analysis software, autoCSA35, and also by alignment of base-called data to the X-chromosome reference sequence. We verified all identified mutations by manual inspection of the trace files. All potential truncating variants were further investigated by resequencing of the appropriate sample from a newly amplified PCR product. Because of the large number of missense and synonymous base substitutions it was not practical to confirm these similarly. Therefore, to assess the overall reproducibility of base substitutions, we resequenced 60 and showed that 56 (93%) were confirmed. All missense and synonymous substitutions were therefore included in the analyses.
Statistical analyses
To evaluate the significance of potential disease-causing variants, we assessed cosegregation with disease using a method previously described36. We calculated lod scores using a model assuming 100% penetrance of mental retardation–causing variants, variant prevalence of 0.1% and an mental retardation sporadic rate of 1%, using the Genehunter program. An estimate of the proportion of variants that were deleterious was derived by calculating a heterogeneity lod score over all families, assuming that a proportion (alpha) of variants were associated with the disease risk, and maximizing over alpha.
To measure the evolutionary constraint on X-chromosomal genes for comparison between genes associated with XLMR, genes associated with diseases other than mental retardation, and genes not associated with disease, Ensembl macaque orthologs were identified and, where necessary, realigned using ClustalW2. We used DnaSP37 to determine the total and aligned length of each alignment, the number of synonymous positions and differences, and the number of nonsynonymous positions and differences. Genes for which a macaque ortholog was not found, or which showed an aligned length less than 90% of the total length, were omitted. From the remaining genes (72 genes associated with XLMR, 48 genes associated with other diseases and 393 genes not associated with disease, respectively), the ratio dN/dS (number of nonsynonymous changes per nonsynonymous site/number of synonymous changes per synonymous site) was calculated both for each individual gene and for all genes within each of the three categories concatenated (Supplementary Table 8 online). The significance of differences between the categories was evaluated using a Mann-Whitney test. To investigate whether any genes showed more missense variants within humans than expected from their evolutionary history, we used a McDonald-Kreitman test24.
To determine average conservation of X chromosome genes, we obtained Ensembl orthologs and MAFFT alignments for as many genes as possible. The conservation at every amino acid was determined using Scorecons, and the average protein conservation for each gene computed for each species. The value cited refers to a human-mouse comparison, but similar results were obtained for other species. Sequence lengths and exon information was obtained from Ensembl. Paralog comparison was based on Ensembl compara one-to-one paralogs. Brain expression of X genes was compared using data from the HugeIndex database38 and from previously published work39.
To determine the deleteriousness of missense variants, we identified orthologs for each gene containing a missense mutation using the Ensembl compara database (v. 47), requiring a one-to-one relationship and minimum 80% amino acid identity to ensure good alignments. The protein and its orthologs were aligned using MAFFT40 version 6.240, ‘linsi’ method, with 1,000 iterations. The alignment was scored at the missense position for conservation using Scorecons41, with scoring method ‘valdar01’, matrix ‘modified PET91’, and matrix transformation ‘karkinlike’. The conservation score for an amino acid affected by a missense change was calculated as the product of the Scorecons score, representing how identical the orthologs were at the given position, and the number of orthologs for the gene, giving an overall conservation score that is higher for amino acids that are highly conserved deeply in evolution. The efficacy of the deleteriousness score is illustrated by a comparison of nonrecurrent and recurrent missense variants (Supplementary Fig. 2 online) Additionally, a score for amino acid change was determined using a Grantham matrix42.
URLs
Mental retardation database, http://www.LOVD.nl/MR; LOVD gene ID, http://www.LOVD.nl/‘geneID’.
ACKNOWLEDGMENTS
We would like to express our gratitude to the many families with members with mental retardation or learning disability who agreed to participate in these studies performed by the IGOLD consortium. We would also like to thank many clinicians and researchers, including J.T. den Dunnen, L. Holloway, M. May, B. Kerr, J. Erikson, R. Smith, E. Fraser, P. Hodgins, I. Wilkinson, A.J. Gardener, D. Amor, D. Anderson, M. Edwards, V.J. Ajith, A. Barnicoat, P. Beales, K. Becker, A. Brady, C. Brewer, J. Christodoulou, J. Clayton-Smith, T. Cole, A. Collins, J. Cook, J. Craig, S. Davies, H. Dawkins, M. Delatycki, D. Donnai, M. Edwards, F. Essop, D. Fitzpatrick, F. Flinter, B. Franco, A. Fryer, T. Homfray, S. Garcia, C. Gardiner, R. Gardner, C. Garrett, R. Gibbons, A. Green, P. Guilbert, D. Halliday, B. Hamel, P. Harper, E. Hobson, S. Holder, E. Holinski-Feder, H. Hughes, J. Hurst, F. Kavalier, B. Kerr, U. Kini, T. Kleefstra, F. Kooy, K. Lachlan, W. Lam, M. Lees, S. Lindsay, C. Longman, S. Lynch, S. Mansour, C. Mercel, J. McGaughran, V. Murday, J. Nelson, G. Neri, R. Newbury-Ecob, C. Owens, M. Patton, M. Porteous, S. Price, A. Proctor, O. Quarrell, N. Rahman, W. Reardon, E. Rosser, F. Stewart, H. Stewart, S. Tompkins, D. Trump, B. de Vries, T. Webb, D. Wellesley, D. Williams, L. Wilson, R. Winter, N. Wood, and J. Yates, who referred families for research, and N. Rahman for comments on the manuscript. The work was supported by the European Community's Seventh Framework Programme-the GEN2PHEN Project, the New South Wales Department of Health, the Australian NHMRC, the SMILE foundation, the WCH Foundation, D. Harwood, EU grant QLG3-CT- 2002-01810 (EURO-MRX), US National Institutes of Health (HD26202) to C.E.S., the South Carolina Department of Disabilities and Special Needs (SCDDSN), Action Medical Research and the Wellcome Trust.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.World Health Organization . The ICD-10 classification of mental and behavioural disorders. World Health Organization; Geneva: 1992. [Google Scholar]
- 2.American Psychiatic Association . Diagnostic and statistical manual of mental disorders DSM-IV. American Psychiatric Association; Washington, D.C.: 1994. [Google Scholar]
- 3.Polder JJ, Meerding WJ, Bonneux L, van der Maas PJ. Healthcare costs of intellectual disability in the Netherlands: a cost-of-illness perspective. J. Intellect. Disabil. Res. 2002;46:168–178. doi: 10.1046/j.1365-2788.2002.00384.x. [DOI] [PubMed] [Google Scholar]
- 4.McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children's hospital. Am. J. Hum. Genet. 2004;74:121–127. doi: 10.1086/381053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laxova R, Ridler MA, Bowen-Bravery M. An etiological survey of the severely retarded Hertfordshire children who were born between January 1, 1965 and December 31, 1967. Am. J. Med. Genet. 1977;1:75–86. doi: 10.1002/ajmg.1320010109. [DOI] [PubMed] [Google Scholar]
- 6.Baird PA, Sadovnick AD. Mental retardation in over half-a-million consecutive livebirths: an epidemiological study. Am. J. Ment. Defic. 1985;89:323–330. [PubMed] [Google Scholar]
- 7.Chelly J, Khelfaoui M, Francis F, Cherif B, Bienvenu T. Genetics and pathophysiology of mental retardation. Eur. J. Hum. Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- 8.Inlow JK, Restifo LL. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–881. doi: 10.1093/genetics/166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vissers LE, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am. J. Hum. Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreppers-Tijdink GA, Curfs LM, Wiegers A, Kleczkowska A, Fryns JP. A systematic cytogenetic study of a population of 1170 mentally retarded and/or behaviourly disturbed patients including fragile X-screening. The Hondsberg experience. J. Genet. Hum. 1988;36:425–446. [PubMed] [Google Scholar]
- 11.Ravnan JB, et al. Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J. Med. Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. Eur. J. Hum. Genet. 2008;16:422–434. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- 13.Ropers HH, Hamel BC. X-linked mental retardation. Nat. Rev. Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- 14.de Brouwer AP, et al. Mutation frequencies of X-linked mental retardation genes in families from the EuroMRX consortium. Hum. Mutat. 2007;28:207–208. doi: 10.1002/humu.9482. [DOI] [PubMed] [Google Scholar]
- 15.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarpey PS, et al. Mutations in the gene encoding the sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X–linked mental retardation. Am. J. Hum. Genet. 2006;79:1119–1124. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarpey PS, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 2007;80:345–352. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond FL, et al. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X–linked mental retardation associated with a marfanoid habitus. Am. J. Hum. Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field M, et al. Mutations in the BRWD3 gene cause X–linked mental retardation associated with macrocephaly. Am. J. Hum. Genet. 2007;81:367–374. doi: 10.1086/520677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilfillan GD, et al. SLC9A6 Mutations Cause X–Linked Mental Retardation, Microcephaly, Epilepsy, and Ataxia, a Phenotype Mimicking Angelman Syndrome. Am. J. Hum. Genet. 2008;82:1003–1010. doi: 10.1016/j.ajhg.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najm J, et al. Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat. Genet. 2008;40:1065–1067. doi: 10.1038/ng.194. [DOI] [PubMed] [Google Scholar]
- 23.Hsueh YP. The role of the MAGUK protein CASK in neural development and synaptic function. Curr. Med. Chem. 2006;13:1915–1927. doi: 10.2174/092986706777585040. [DOI] [PubMed] [Google Scholar]
- 24.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 25.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 26.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, et al. Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol. 2004;5:R47. doi: 10.1186/gb-2004-5-7-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osada N. Inference of expression-dependent negative selection based on polymorphism and divergence in the human genome. Mol. Biol. Evol. 2007;24:1622–1626. doi: 10.1093/molbev/msm080. [DOI] [PubMed] [Google Scholar]
- 30.Wang HY, et al. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics. 2005;6:55. doi: 10.1186/1471-2105-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 33.Sakharkar MK, et al. A report on single exon genes (SEG) in eukaryotes. Front. Biosci. 2004;9:3262–3267. doi: 10.2741/1478. [DOI] [PubMed] [Google Scholar]
- 34.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dicks E, et al. AutoCSA, an algorithm for high throughput DNA sequence variant detection in cancer genomes. Bioinformatics. 2007;23:1689–1691. doi: 10.1093/bioinformatics/btm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson D, Easton DF, Goldgar DE. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am. J. Hum. Genet. 2003;73:652–655. doi: 10.1086/378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R, Dna SP. DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 38.Hsiao LL, et al. A compendium of gene expression in normal human tissues. Physiol. Genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- 39.Liang S, Li Y, Be X, Howes S, Liu W. Detecting and profiling tissue-selective genes. Physiol. Genomics. 2006;26:158–162. doi: 10.1152/physiolgenomics.00313.2005. [DOI] [PubMed] [Google Scholar]
- 40.Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 2005;16:22–33. [PubMed] [Google Scholar]
- 41.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 42.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]