Abstract
MicroRNAs (miRNAs) constitute an evolutionarily conserved class of small non-coding RNAs that are endogenously expressed with crucial functions in fundamental cellular processes such as cell cycle, apoptosis and differentiation. Disturbance of miRNA expression and function leads to deregulation of basic cellular processes leading to tumorigenesis. A growing body of experimental evidence suggests that human tumors have deregulated expression of microRNAs, which have been proposed as novel oncogenes or tumor suppressors. Recent studies have shown that microRNA expression patterns serve as phenotypic signatures of different cancers and could be used as diagnostic, prognostic and therapeutic tools. A few studies have analyzed global microRNA expression profiles or the functional role of microRNAs in prostate cancer. Here we have reviewed the role of microRNAs in prostate carcinogenesis by summarizing the findings from such studies. In addition, recent evidence indicates that dietary factors play an important role in the process of carcinogenesis through modulation of miRNA expression, though such studies are lacking in regards to prostate cancer. It has been proposed that dietary modulation of miRNA expression may contribute to the cancer-protective effects of dietary components. In this review, we have summarized findings from studies on the effect of dietary agents on miRNA expression and function.
KEY WORDS: diet, microRNAs, prostate cancer
INTRODUCTION
MicroRNAs are small (∼22 nucleotides) endogenously expressed non-coding RNAs that regulate gene expression. It is being increasingly recognized that microRNAs constitute an important class of gene modulators that play crucial roles in almost every cellular process that has been investigated, including the cell cycle, apoptosis, development, differentiation and metabolism. It has been estimated that miRNAs regulate ∼30% of the human genome (1). Given the crucial role of these small RNAs in fundamental cellular processes, it is not surprising that the deregulated expression of microRNAs leads to various disease states, including cancer.
Environmental and dietary factors are believed to contribute to differences in cancer incidence among populations with different dietary habits. A growing body of literature suggests that dietary factors play a role in carcinogenesis. Several studies suggest that dietary components act as chemopreventive agents in prostate carcinogenesis (2). Prostate cancer statistics point to differences in cancer incidences amongst populations with different dietary habits. Dietary components potentially influence fundamental cellular processes involved in carcinogenesis, including apoptosis, cell-cycle control, angiogenesis, inflammation and DNA repair. Since miRNAs also play a central role in controlling these cellular processes, it is intuitive that diet and microRNAs co-operatively regulate the process of tumorigenesis. A few reports discussed below suggest that dietary modulation of miRNA expression may contribute to the cancer-protective effects of dietary components. However, this is an area that is largely underexplored, in particular in prostate cancer, and warrants further investigation.
MICRORNAs: MASTER REGULATORS OF GENE EXPRESSION
MicroRNAs regulate the expression of protein-coding genes largely at the post-transcriptional level. It has been estimated that miRNAs regulate ∼30% of the human genome (1). MicroRNA genes are evolutionarily conserved and are located within the introns or exons of protein-coding genes (70%) or in intergenic regions (30%) (3). Most of the intronic or exonic miRNAs are oriented in sense with their host gene, suggesting that their transcription occurs in parallel with the transcription of the host gene. Additionally, some miRNAs are clustered in polycistronic transcripts, enabling a coordinated expression. The miRNAs in intergenic regions or in isolated regions of the genome are transcribed independently (3). MicroRNAs are preferentially transcribed by RNA polymerase II into large precursor RNAs, often several kilobases in length, called primary miRNAs (pre-miRNAs) (4–6). These pri-miRNAs of long nucleotide sequence are usually capped at the 5’end and polyadenylated at the 3’ regions (5). Pri-miRNAs form specific hairpin-shaped stem-loop secondary structures and are cleaved by nuclear RNase III Drosha to release a miRNA precursor (pre-miRNA) that is about 60–75 nucleotides in length (7,8). Drosha also requires a protein cofactor, DGCR8 or Pasha (9). In humans, Drosha and Pasha form a large complex known as the microprocessor complex (10). The pre-miRNAs are then exported to the cytoplasm by exportin-5 (11,12) where they are further processed by the enzyme Dicer, a second RNase III enodnuclease, resulting in a mature double-stranded miRNA (19–24 nucleotides) (13,14). The functional strand of this mature miRNA is incorporated into an effector complex, called the RNA-induced silencing complex (RISC), which functions to silence gene expression (14,15). The opposite strand is eliminated by cleavage. MicroRNAs regulate their targets by direct cleavage of the mRNA or by inhibition of protein synthesis. Perfect or nearly perfect complementarity between the miRNA and its target 3’UTR induces RISC to cleave the target mRNA, whereas imperfect base matching induces mainly translational silencing of the target gene. MicroRNAs can also direct deadenylation of the target mRNA that leads to either mRNA decay or reduces its level (14).
Recent studies from our laboratory have provided a novel concept that miRNAs and non-coding double-stranded RNAs (dsRNAs) can activate various tumor suppressor genes (16–18). By scanning gene promoters in silico for sequences complementary to known miRNAs, we identified a putative miR-373 target site in the promoter of E-cadherin. Transfection of miR-373 or its precursor hairpin RNA (pre-miR-373) into prostate cancer PC-3 cells readily induced E-cadherin expression (17) by targeting its specific site in the gene promoter. Another report suggests that several microRNAs, including Let7, induce translational upregulation of target mRNAs on cell-cycle arrest, yet they repress translation in proliferating cells (19). Another study supports the concept that microRNA can directly bind to promoter regions in cis and mediate transcriptional gene silencing (20). Additionally, specific miRNAs that carry a distinct hexanucleotide terminal motif, such as miR-29b, were found enriched in the nucleus, suggesting extra miRNA functions in distinct subcellular compartments (21). The ability for miRNAs to act through pleiotropic mechanisms points to the fundamental importance of miRNAs in regulating gene expression as crucial regulatory elements in fundamental biological processes. Recently, proteomic studies were used to study the impact of a single miRNA on global changes in protein expression, and it was found that a single miRNA can impact on hundreds of targets (22,23).
ROLE OF MICRORNA IN CANCER: A PLETHORA OF MECHANISMS
Disturbances in the expression of miRNAs, processing of miRNA precursors or mutations in the sequence of the miRNA, its precursor, or its target mRNA, may have detrimental effects on cellular function and have been associated with cancer (24). Strikingly, half of the known miRNA genes are located inside or close to fragile genomic sites and in minimal regions of loss of heterozygosity, minimal regions of amplifications, and common breakpoints associated with cancer (25). It has been proposed that microRNAs may regulate tumorigenesis through a plethora of possible oncogenic mechanisms (26). Genomic deletion or epigenetic silencing of a miRNA that normally represses expression of one or more oncogenes might lead to increased oncogenic expression. Alternatively, amplification, overexpression, or loss of epigenetic silencing of a gene encoding an miRNA that targets one or more tumor suppressor genes could inhibit the activity of an anti-oncogenic pathway (26). In addition, mutations affecting the sequence of the mature miRNA or target mRNA could alter binding of the miRNA to its cognate targets leading to alterations in the balance of critical growth regulatory proteins. MicroRNAs can act as oncogenes or tumor suppressor genes (27). Examination of tumor-specific miRNA expression profiles has revealed widespread dysregulation of these molecules in diverse cancers. Over-expressed miRNAs in cancers, such as mir-17-92 cluster, may function as oncogenes and promote cancer development by negatively regulating tumor suppressor genes and/or genes that control cell differentiation or apoptosis. The onco-microRNA expression profiling of human malignancies has also identified a number of diagnostic and prognostic cancer signatures (28). Also, some microRNAs are downregulated in cancer and act as bona fide tumor suppressor genes, such as let-7 and miR-34a. The widespread differential expression of miRNA genes between malignant and normal cells is a complex phenomenon and may involve multiple mechanisms, including miRNA transcriptional control by tumour suppressor genes, oncogenes, epigenetic mechanisms and genomic abnormalities (26,27). For example, the tumour suppressor miR-34a is transactivated by the tumor suppressor p53, is kept in check by MYC, is silenced by aberrant CpG methylation, and is located at 1p36, a chromosomal region that is frequently lost in neuroblastomas (29–34).
MicroRNA: Cancer Initiation and Progression
Cancer initiation and progression can involve miRNAs, and their expression profiles can be used for the classification, diagnosis, and prognosis of human malignancies. Loss or amplification of miRNA genes has been reported in a variety of cancers, and altered patterns of miRNA expression may affect cell cycle and survival programs. Germ-line and somatic mutations in miRNAs or polymorphisms in the mRNAs targeted by miRNAs may also contribute to cancer predisposition and progression. It has been proposed that alterations in miRNA genes play a critical role in the pathophysiology of many, perhaps all, human cancers (35).
MicroRNAs: Tumor Invasion and Metastasis
A role for miRNAs has been established in the later steps of tumorigenesis, progression, and metastasis. More than 20 miRNAs have been shown to impact critical steps in the metastatic cascade, such as epithelial-mesenchymal transition (EMT), apoptosis, and angiogenesis, by acting on multiple signaling pathways and targeting various proteins that are major players in this process. Eleven different miRNAs have been directly shown to promote or inhibit metastasis in experimental models of various cancers (36). Furthermore, several clinical studies have identified correlations between miRNA expression and recurrence, development of metastases and/or survival (36,37). As an example, miR-10b seems to play an important role in metastasis. miR-10b is highly expressed in metastatic breast cancer cells and positively regulates cell migration and invasion. (38). Further, expression of miR-10b is induced by the transcription factor Twist that binds directly to the putative promoter of miR-10b and transcriptionally activates this miR which further leads to repression of homeobox D10 (HOXD10). This results in increased expression of a well-characterized pro-metastatic gene, RHOC. Significantly, the level of miR-10b expression in primary breast carcinomas correlates with clinical progression (38). These findings exemplify a complex regulatory pathway, in which a pleiotropic transcription factor induces expression of a specific microRNA, which suppresses its direct target and in turn activates another pro-metastatic gene, leading to tumor cell invasion and metastasis.
In summary, studies suggest that miRNAs are involved in the initiation, progression and metastasis of various cancers.
MICRORNA AND PROSTATE CANCER
Prostate cancer (PCa) is the most common male malignancy and the second leading cause of cancer death among men in the United States with an estimated 192, 280 new cases and 27,360 deaths in 2009. The precise molecular mechanisms underlying the development and progression of prostate cancer remain poorly understood. It is now recognized that the process of prostate cancer development is a consequence of genetic and epigenetic alterations that transform normal glandular epithelium to preneoplastic lesions that lead to invasive carcinoma (39,40). Clinically, prostate cancer is diagnosed as local or advanced, and treatments include surveillance, radiotherapy, radical prostatectomy or androgen-deprivation. Surgery and radiation are generally effective against clinically localized PCa; however, metastatic prostate cancer invariably remains incurable. Androgen ablation therapy for advanced prostate cancer reduces symptoms in a majority of patients, but most tumors relapse within 2 years to an incurable hormone-independent state causing mortality (41). The underlying regulatory mechanisms that cause this transition remain largely unknown, and, at present, there is no effective therapy for metastatic prostate cancer. Also, there is an urgent need to develop better diagnostic and prognostic indicators for predicting the severity of the disease. This emphasizes the need for a better understanding of the molecular pathogenesis of prostate cancer which could lead to better diagnostic and therapeutic interventions for the disease. Recent studies have shown that microRNAs are significantly altered in prostate cancer, suggesting that miRNAs act as key regulators of prostate carcinogenesis. Several studies have been conducted to identify the PCa-specific miRNA signature. However, no consensus has been reached on which miRNAs are relevant for development and progression of this malignancy (42,43). Below, we summarize the results obtained from miRNA expression profiling studies and single miRNA-focused functional studies carried out in prostate cancer.
MICRORNA PROFILING STUDIES IN PROSTATE CANCER
Lu et al. (2005) performed a systematic expression analysis of 217 mammalian miRNAs from 334 samples, including multiple human cancers using a bead-based flow cytometric expression profiling method and observed a general downregulation of miRNAs in tumors compared with normal tissues (44). Volinia et al. (2006) conducted a large-scale microarray analysis on six solid tumors (including lung, breast, stomach, prostate, colon, and pancreatic tumors) and identified a common miRNA signature for solid tumors largely composed of overexpressed miRNAs, such as miR-17-5p, miR-20a, miR-21, miR-92, miR-106a, and miR-155 (45). In the prostate cancer subset, 56 tumor tissues and 7 tissues from non-cancerous subjects were analysed. This analysis showed that 39 miRs were upegulated and 6 downregulated in prostate cancer, suggesting that gain of miRNA expression rather than loss is a more frequent event in prostate tumorigenesis (45).
Porkka et al. (2007) first identified the miRNA signature specific for prostate cancer in a study on 6 prostate cancer cell lines, 9 prostate cancer xenografts, 4 benign prostatic hyperplasia (BPH), and 9 prostate carcinoma samples (5 untreated and 4 hormone-refractory) using an oligonucleotide array hybridization method (46) Differential expression of 51 individual miRNAs (14 upregulated and 37 downregulated) was found between non-malignant tissues and carcinomas. Hierarchical clustering of the tumor samples by their miRNA expression accurately separated the carcinomas from the BPH samples and also further classified the carcinoma tumors according to their androgen dependence (hormone naïve versus hormone refractory), indicating the potential of miRNAs as a novel diagnostic and prognostic tool for prostate cancer. Also, there was a significant trend between the expression of miRNAs and miRNA locus copy number determined by array comparative genomic hybridization, indicating that genetic aberrations may target miRNAs (46). In another study, Ozen et al. (2008) analyzed expression of 480 human miRNAs in 10 benign peripheral zone tissues and 16 prostate cancer tissues using microarray-based profiling and found widespread downregulation of miRNAs in clinically localized prostate cancer relative to the benign peripheral zone tissue (47). Expression data for select miRNAs, including miR-125b, miR-145 and let-7c, was confirmed by real-time RT-PCR assays. The downregulated miRNAs included several with proven target mRNAs whose proteins have been previously shown to be increased in prostate cancer by immunohistochemistry, including RAS, E2F3, BCL-2 (47).
Ambs et al. (2008) performed a genome-wide expression profiling of microRNAs and mRNAs in 60 primary prostate tumors and 16 non-tumor prostate tissues (48). The mRNA analysis revealed that key components of microRNA processing and several genes harboring miRNA coding sequences in their introns, e.g., MCM7 and C9orf5, were significantly upregulated in prostate tumors. Consistent with these findings, tumors expressed the miR-106b-25 cluster, which maps to intron 13 of MCM7, and miR-32, which maps to intron 14 of C9orf5, at significantly higher levels than non-tumor prostate. Additional differences in microRNA abundance were found between organ-confined tumors and those with extraprostatic disease extension (48). Also, this study provided evidence that some microRNAs are androgen-regulated. Those included miR-338 and miR-126 and the miR-181b-1, miR-181c, miR-221 clusters, among others. A motif search showed that these microRNAs have putative androgen receptor binding sites in their flanking regions (48). Also, miRNA expression profiling of tumors with perineural (PNI) invasion showed differential expression of 19 microRNAs in PNI tumors than in non-PNI tumors, where miR-224 was reported to be the most differently expressed microRNA (49).
In another effort, Tong et al. (2009) conducted paired miRNA profiling analysis of normal and microdissected malignant tissues of prostatectomy specimens from 40 patients diagnosed with stage T2 prostate cancer to characterize the miRNA expression profile in early disease (50). This study showed that five miRNAs (miR-23b, -100, -145, -221 and -222) were significantly downregulated in malignant tissues. Further, differential miRNA expression (miR-194 and -135b overexpression) was found in patients with early chemical relapse (<2 years post-prostatectomy) and those without clinical relapse for a period of more than 10 years (48). This suggests that altered miRNA expression accompanies the prostate oncogenic process, with the early relapse subset earmarked by additional aberrant miRNA features prior to clinical relapse.
A recent study implemented a systems biology approach to identify microRNAs and gene regulatory networks that contribute to the aggressive phenotype of prostate cancer and evaluated transcriptional profiles of 125 lymphoblastoid cell lines derived from 62 aggressive (Gleason grade ≥ 8) and 63 nonaggressive (Gleason grade ≤ 5) prostate cancer patients. This study identified a set of cell cycle-related genes and miRNAs whose expression levels are associated with prostate cancer aggressiveness. From 273 miRNAs tested, seven different miRNAs, including miR-222, miR-221, miR-331-3p, miR-16, miR-145, miR-9*, and miR-551a, showed a significant association with prostate cancer grade (51). Ribas et al. (2009) identified a set of 16 androgen-responsive miRNAs using high-throughput microarray analysis on two AR-positive prostate cancer (CaP) cell lines LNCaP and LAPC-4 (52).
SINGLE-MIRNA FOCUSED STUDIES IN PROSTATE CANCER
There have been a few single-miRNA focused studies in prostate cancer, where the functional role of specific miRNAs has been thoroughly investigated. Based on these studies, a small number of miRNAs have been characterized as potential tumor suppressors or oncogenes in prostate cancer as reviewed in the following sections. The key findings from such studies are summarized in Fig. 1.
Fig. 1.
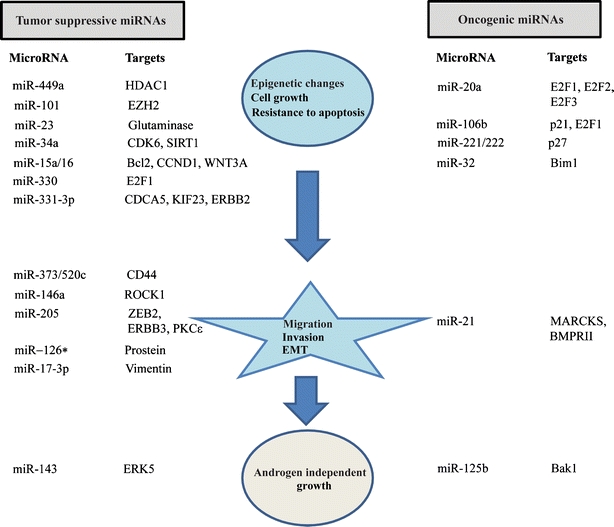
Summarized results from single-miRNA focused functional studies in prostate cancer. A schematic representation of microRNAs and their targets that have been directly shown to affect epigenetic reprogramming, cell growth, apoptosis, invasion, migration, EMT (epithelial-to-mesenchymal transition) and androgen-independent growth in prostate cancer. MicroRNAs are defined as tumor-suppressive or oncogenic based on their expression patterns and functional targets in prostate cancer.
Tumor Suppressor MicroRNA in Prostate Cancer
Bonci et al. (2008) reported the frequent downregulation of miR-15a/miR-16 levels in prostate carcinomas (53). Q-PCR analysis of 20 prostate tumor-derived primary cultures compared to their normal counterparts, and in situ hybridization analysis of 15 prostate tumor biopsies showed that miR-15a/miR-16 levels are downregulated in the vast majority of the cases (up to 85% of the samples). Delivery of antagomirs specific for miR-15a and miR-16 to normal mouse prostate results in marked hyperplasia, and knockdown of miR-15a and miR-16 promotes survival, proliferation and invasiveness of untransformed prostate cells, which become tumorigenic in immunodeficient NOD-SCID mice. These observations were, at least in part, explained by the evidence that miR-15a/miR-16 are able to post-transcriptionally repress the expression of Bcl2, CCND1 (i.e. Cyclin D1) and WNT3A, thus interfering with multiple oncogenic activities (53). Also, a recent report implicates a role of miR-15a/miR-16 in the control of angiogenesis by regulating VEGF levels (54).
miR-23a and miR-23b are significantly decreased in human prostate cancer as compared with normal prostate tissue (46). miR-23a and miR-23b are transcriptionally repressed by c-Myc in PC3 prostate cancer cells, resulting in upregulated expression of their target protein, mitochondrial glutaminase (55). This leads to upregulation of glutamine catabolism, conferring a growth advantage to cancer cells.
miR-34a has been implicated as a target of the tumor suppressor p53 (29,30) and represents a tumor suppressor gene which is inactivated by CpG methylation and subsequent transcriptional silencing in a broad range of tumors, including prostate cancer (33). In prostate cancer cell lines, miR-34a expression is markedly reduced in p53-null PC3 cells and p53-mutated DU145 cells compared with LNCaP cells expressing wild-type p53 (56). In PC3 cells, ectopic miR-34a expression resulted in cell-cycle arrest, growth inhibition, senescence and attenuated chemoresistance to anticancer drug camptothecin by inducing apoptosis, suggesting a potential role of miR-34a for the treatment of p53-defective prostate cancer. Cyclin dependent kinase 6 (CDK6) and deacetylase sirtuin SIRT1 have been identified as targets of miR-34a (33,56). SIRT1 regulates p53-dependent apoptosis through deacetylating and stabilizing p53. It was found that SIRT1 mediates miR-34a activation of apoptosis by regulating p53 activity, suggesting a positive feedback loop in which p53 induces expression of miR-34a, which suppresses SIRT1, increasing p53 activity (57,58).
miR-101 expression decreases during prostate cancer progression due to loss of one or both of the two genomic loci encoding miR-101 (59). Appoximately 37.5% of clinically localized prostate cancers (6/16) and 66.7% of metastatic disease (22/33) showed a genomic loss of miR-101. miR-101 inhibits the expression and function of a polycomb group member, enhancer of zeste homolog 2 (EZH2), in prostate cancer, suggesting that genomic loss of miR-101 in cancer leads to overexpression of EZH2 and concomitant dysregulation of epigenetic pathways, resulting in cancer progression (59). Studies show a tumor-suppressive role for miR-449a as well (60). miR-449a is downregulated in prostate cancer tissues relative to patient-matched normal control tissues (60). Introduction of miR-449a into PC-3 prostate cancer cells resulted in cell-cycle arrest, apoptosis and a senescent-like phenotype. miR-449a directly targets HDAC1, which is frequently overexpressed in prostate cancer (60,61).
Expression of miR-100, miR-218 and miR-let7c has been reported to be markedly reduced in metastatic cancer compared with localized high-grade disease (62). A recent study shows that miR-143 levels are inversely correlated with advanced stages of prostate cancer (63). Rescue of miR-143 expression in prostate cancer cells leads to the inhibition of extracellular signal-regulated kinase-5 (ERK5) activity and concomitant growth arrest in vitro and in vivo (63).
Recent studies have shown that the miR-200 family regulates epithelial-mesenchymal transition (EMT) by directly targeting the E-cadherin transcriptional repressors ZEB1 (zinc-finger E-box binding homeobox 1) and ZEB2 (SMAD-interacting protein 1 (SIP1) (64). miR-200 is significantly downregulated in EMT induced by platelet-derived growth factor-D in PC-3 cells (65). miR-200b restoration led to reversal of this phenotype by suppressing ZEB1, ZEB2 and snail2 expression. A recent report documents a tumor-suppressive role of miR-205 in human prostate by counteracting epithelial-to-mesenchymal transition and reducing cell migration/invasion (66). Significantly lower miR-205 expression levels were found in cancer than in normal prostate cell lines as well as in tumor compared with matched normal prostate tissues, with a particularly pronounced reduction in carcinomas from patients with local-regionally disseminated disease. Restoring the expression of miR-205 in prostate cancer cells resulted in cell rearrangements consistent with a mesenchymal-to-epithelial transition, such as upregulation of E-cadherin and reduction of cell motility and invasion, and in the downregulation of several oncogenes known to be involved in disease progression (i.e., interleukin 6, caveolin-1, EZH2). These events are presumably driven by the concurrent repression of specific predicted miR-205 targets, namely N-chimaerin, ERBB3, E2F1, E2F5, ZEB2, and protein kinase Cε. Strikingly, protein kinase Cε seemed to play a direct role in regulating epithelial-to-mesenchymal transition (66).
Downregulation of miR-146a has also been reported in prostate cancer (67). Transfection of miR-146a in PC-3 prostate cancer cells suppressed the Rho-associated protein kinase ROCK1 and markedly reduced cell proliferation, invasion, and metastasis to human bone marrow endothelial cell monolayers (67). Similarly, miR-373 and miR-520c are downregulated in prostate cancer. These miRs target the adhesion receptor CD44 and enhance invasion of prostate cancer cells in vitro (68). Prostate cancer cells are deficient in an intronic miRNA- miR-126* as the expression of its host gene, EGF-like domain 7 (Egfl7), is attenuated in prostate cancer (69). miR-126* causes translational repression of prostein, a prostate-specific antigen that leads to reduced migratory and invasive potential of LNCaP cells (69).
miR-330 is another tumor suppressive miR that has been reported to be significantly under expressed in human prostate cancer. miR-330 targets E2F1 and induces apoptosis in prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation (70). Similarly, in vitro and in vivo studies showed that miR-17-3p is a prostate tumor suppressor (71). The expression of miR-17-3p is decreased in high-grade prostate tumors analyzed by laser capture microdissection-purified clinical human prostatectomy specimens. Downregulated expression of miR-17-3p correlates with increased vimentin expression and tumorigenicity (71). miR-331-3p expression is also decreased in prostate cancer tissue relative to normal adjacent prostate tissue (72). Reintroduction of miR-331-3p reduced ERBB-2 tyrosine kinase receptor that is frequently overexpressed in prostate cancer and is associated with disease progression and poor survival. Also, miR-331-3p transfection blocked the androgen receptor signaling pathway in prostate cancer cells, reducing activity of an androgen-stimulated prostate-specific antigen promoter and blocking prostate-specific antigen expression (72). Ectopic expression of miR-331-3p in prostate cancer cell lines LNCaP and VCaP induces apoptosis and growth arrest concomitant with a repression of Cell division cycle associated 2 (CDCA5) and Kinesin family member 23 (KIF23) genes (51).
Oncogenic MicroRNA in Prostate Cancer
miR-21 was identified as an androgen receptor-regulated miRNA whose levels are elevated in prostate cancer compared with adjacent normal tissue (52). Inhibition of miR-21 diminished androgen-induced PCa cell proliferation, whereas elevated expression of miR-21 promotes enhanced tumor growth and castration resistance in vivo (52). In a separate study, miR-21 levels were reported to be elevated in androgen-independent prostate cancer cell lines DU145 and PC-3 (73). Inactivation of miR-21 by antisense oligonucleotides in these cell lines resulted in enhanced sensitivity to apoptosis and inhibition of cell motility and invasion. miR-21 targets Myristoylated alanine-rich protein kinase c substrate (MARCKS), which plays key roles in cell motility and cell adhesion, suggesting that miR-21 could promote apoptosis resistance, motility, and invasion in prostate cancer cells (73). Also, it has been reported that miR-21 directly targets bone morphogenetic protein receptor II (BMPRII), suggesting that miR-21 may have a potential role in regulating the malignancy and metastatic abilities of prostate cancer cells (74).
Shi et al. (2007) reported that miR-125b is an androgen-regulated miR that is upregulated in androgen-independent prostate cancer cells (75). Furthermore, transfection of synthetic miR-125b in LNCaP cells stimulated androgen-independent growth and downregulated the expression of Bak1, a pro-apoptotic gene of the Bcl-2 family (75). Also, miR-32 is upregulated in prostate cancer and regulates the expression of Bim, a pro-apoptotic member of the Bcl-2 family. Downregulation of Bim can contribute to the resistance of tumor cells to death stimuli (48).
miR-221 and miR-222 are two highly homologous microRNAs whose upregulation has been recently described in various cancers, including prostate cancer (50,76–79). Galardi et al. (2007) reported that miR-221 and miR-222, encoded in tandem on chromosome X, are over-expressed in the PC3 cellular model of aggressive prostate carcinoma, as compared with LNCaP and 22Rv1 cell line models of slowly growing carcinomas (76). Over-expression of miR-221/222 in LNCaP cells enhanced the clonogenicity and growth potential in vitro and in vivo by inducing a G1 to S-phase shift in the cell cycle. Inhibition of miR-221 and miR-222 expression by antisense LNA oligonucleotides or antagomir treatment strongly reduced the clonogenicity and tumorigenicity of PC3 cells (76,77). miR-221 and miR-222 directly targets the cyclin-dependent kinase (CDK) inhibitor p27(Kip1) contributing to the oncogenesis and progression of prostate carcinoma through p27(Kip1) downregulation (76,77). Another study suggests the involvement of miR-221 and miR-222 in the development or maintenance of the castration-resistant prostate cancer phenotype (78). miR-221 and miR-222 expression levels were reported to be significantly increased in an androgen-independent cell-line LNCaP-Abl as compared with the androgen-dependent prostate cancer cell line LNCaP. Overexpression of miR-221 or miR-222 in androgen-dependent cell lines significantly reduced the level of the dihydrotestosterone (DHT), induced upregulation of prostate-specific antigen (PSA) expression and increased androgen-independent growth. Antagonism of miR-221 and miR-222 in the LNCaP-Abl cell line restored the response to the DHT induction of PSA transcription and also increased the growth response of the LNCaP-Abl cells to the androgen treatment (76,77). Contrary to these studies, a recent report suggests that expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis (80).
miR 17-92 cluster have been ascribed oncogenic roles in prostate cancer. Overexpression of a member of the mir-17-92 cluster, miR-20a, decreased apoptosis in a prostate cancer cell line, while inhibition of miR-20a by an antisense oligonucleotide resulted in increased cell death after doxorubicin treatment (81). miR-20a directly represses E2F1, E2F2 and E2F3 (45,81). E2F1 along with p21/WAF1 is also a target for miR-106b cluster that plays anti-apoptotic and growth-inhibitory roles in prostate cancer (48).
In summary, these studies suggest that miRNAs act as tumor suppressors as well as oncogenes and are involved in the etiology of prostate cancer.
DIET AND MICRORNA
The molecular pathology of prostate cancer is complex; not only are multiple genetic and epigenetic events involved in its pathogenesis, but additional environmental factors such as dietary components are also involved (2,39). Although epidemiologically CaP can be divided into hereditary and sporadic forms, most PCa seem to be sporadic with <10% inherited (2). Dietary factors are believed to contribute to differences in cancer incidence among populations with different dietary habits and life-styles. In the United States, the incidence of prostate cancer is highest in African-Americans, followed by Caucasians, Hispanics, Asian/Pacific Islanders, and American Indians (39). Dietary factors may influence the risk of prostate cancer, and some factors may possess chemopreventive and therapeutic potential in prostate cancer. Experimental evidence suggests that dietary factors play a crucial role in prostate carcinogenesis by affecting fundamental cellular processes involved in carcinogenesis, including apoptosis, cell-cycle control, angiogenesis, inflammation, and DNA repair (2). Since miRNAs influence these biological processes as well, it is not surprising that there is an interplay between dietary factors and microRNAs that ultimately affects the process of malignant transformation. A few reports discussed below suggest that dietary modulation of miRNA expression may contribute to the cancer-protective effects of dietary components. However, this area is still emerging and needs further investigations. To our knowledge, there are no reports on diet and miRNA in prostate cancer. Below we give a brief account of the reports in other cancers that gives a flavor of the miRNA-diet connection that advocates investigation in this direction in PCa.
Genistein
Epidemiological data suggest that the rates of clinically significant prostate cancer are 15-fold higher in men from the United States than in men from Asian countries (82), attributable to the fact that Asian diet comprises a significant portion of soybean-enriched foods. Genistein, an isoflavone isolated from soybean, has been found to be a potent antitumor agent. However, the precise mechanism by which genistein suppresses tumorigenesis remain unclear. Our group has been exploring the chemopreventive effects of genistein in prostate cancer and other malignancies. Several studies suggest that the inhibition of human cancer cell growth by genistein is mediated via the modulation of genes that are related to the control of cell cycle, apoptosis, angiogenesis, invasion, and metastasis (83,84). Genistein induces the p21WAF1/CIP, p16INK4a and other tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modifications (85–88). It has been shown that genistein inhibits the activation of NF-kappaB and Akt signaling pathways, both of which are known to maintain a homeostatic balance between cell survival and apoptosis. Moreover, genistein antagonizes estrogen- and androgen-mediated signaling pathways, possesses antioxidant properties, and is a potent inhibitor of angiogenesis and metastasis. Due to its pleiotropic anti-cancer activities, genistein is a promising agent for prostate cancer chemoprevention (83,84) .
Sun et al. (2009) explored the possible activity of genistein to inhibit human uveal melanoma cell growth and investigated the effect of genistein on microRNA-27a (miR-27a) and its cognate targets (89). The results suggested that genistein significantly inhibits uveal melanoma cell growth in a time- and dose-related manner in vitro and in vivo. Functional assays revealed that the levels of miR-27a and its target gene zinc finger and BTB domain containing 10 (ZBTB10) were significantly different based on the dose of genistein, suggesting a correlation between antitumor activity of genistein and miR-27a-mediated regulatory mechanism (89). Another study reports the modulation of miR-16 effects by genistein in a malignant B-1 cell line derived from a murine chronic lymphocytic leukemia (CLL) model (90). Alterations in the human 13q14 genomic region containing mir-15a and mir-16-1 are present in most human chronic lymphocytic leukemia (CLL) leading to a decrease in mature miR-16 and miR-15a levels. Augmentation of miR-16 leads to an accumulation of cells in the G1 phase of the cell cycle and increased apoptosis. Combining miR-16 with genistein resulted in a significant synergistic induction of apoptosis, suggesting that genistein can potentially modulate the biological effects of microRNAs (90).
Curcumin
Curcumin (diferuloylmethane), a naturally occurring flavanoid and proapoptotic compound derived from the rhizome of Curcuma longa, has been shown to have anti-cancer effect in various cancers, including prostate (91–98). In vitro and in vivo preclinical studies have shown that curcumin has antioxidant, anti-inflammatory, antiproliferative, and proapoptotic activities. Curcumin putatively downregulates the expression of gene products such as nuclear factor-κB, suppresses growth, induces apoptosis, and modulates various signal transduction pathways and the expression of many oncogenes (98,99). It has been found that curcumin downregulates the transactivation and expression of AR and AR-related molecules (AP-1 and NF-κB) in prostate cancer cell lines (98).
A recent study shows that curcumin alters the miRNA expression profiles in human pancreatic cancer cells (100,101). The potential effects of curcumin (10 μmol/L) on miRNA expression profiles were studied in a pancreatic cancer cell line, BxPC3 which showed that 11 miRNAs were significantly upregulated, whereas 18 were significantly downregulated by curcumin compared with the control. Among these altered miRNAs, miRNA-22 was up-regulated, whereas miRNA-199a* was downregulated by curcumin. miR-22 upregulation was accompanied by repression of its target genes SP1 transcription factor (SP1) and estrogen receptor 1 (ESR1), suggesting that modulation of miRNA expression may be an important mechanism underlying the biological effects of curcumin (101). Another study shows upregulation of miR- 15a and miR-16 expression in curcumin-treated MCF-7 breast cancer cells. Both miR-15a and miR-16 could inhibit the expression of Bcl-2, thereby inducing apoptosis in these cancer cells (102).
Retinoic Acid
Several reports on another dietary component, retinoic acid, suggest its ability to modulate miRNA expression as detailed below.
Acute promyelocytic leukemia (APL) is caused by a novel fusion protein (PML-RAR-α) resulting from the reciprocal translocation between the retinoic acid receptor-α (RAR-α) on chromosome 17 with the promyelocytic leukemia gene (PML) on chromosome 15 (103). In the absence of an adequate dose of retinoic acid (RA), this fusion protein interferes with myeloid differentiation by forming a corepressor complex that binds to target promoters, resulting in transcriptional repression of retinoic acid-responsive genes These effects get reversed by pharmacological doses of all-trans-retinoic acid (ATRA) (103). Recent studies suggest that these effects are mediated through miRNAs as summarized below.
Careccia et al. (2009) analysed the expression of 12 granulocytic differentiation signature miRNAs in a cohort of APL patients successfully treated with ATRA and chemotherapy and reported a downregulation of miR-181b and upregulation of miR-15b, -16, -107, -223, -342 and let-7c (104). Further, it was found that PML/RARα binds the regulatory sequences of the intragenic miR-342 and let-7c, and in response to ATRA, the PML/RARα is released, leading to the transcriptional activation of these microRNAs together with their host genes (104). Similarly, Garzon et al. (2007) analysed microRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia and found that ATRA causes upregulation of miR-15a, miR-15b, miR-16-1, let-7a-3, let-7c, let-7d, miR-223, miR-342 and miR-107, whereas miR-181b was downregulated (105). To understand the ATRA-mediated regulation of miRNAs, a search for ATRA-modulated transcription factor binding sites in the upstream genomic region of the let-7a-3/let-7b cluster identified several putative nuclear factor-kappa B (NF-κB) consensus elements. Functional assays could validate one proximal NF-κB binding site as essential for the ATRA-mediated transactivation of the let-7a-3/let-7b cluster. Also, it was shown that the downregulation of RAS and Bcl2 after ATRA treatment correlates with the activation of known miRNA regulators of those proteins, let-7a and miR-15a/miR-16-1, respectively (105). Another group identified miR-342 as one of the microRNAs upregulated by ATRA during APL differentiation, which is a direct transcriptional target of the critical hematopoietic transcription factors PU.1 and interferon regulatory factor (IRF)-1 and IRF-9 (106). IRF-1 maintains miR-342 at low levels, whereas the binding of PU.1 and IRF-9 in the promoter region following retinoic ATRA-mediated differentiation upregulates miR-342 expression. Enforced expression of miR-342 in APL cells stimulated ATRA-induced differentiation (106).
miR-10a overexpression is a key mediator of metastatic behavior in pancreatic cancer, which regulates metastasis via suppression of HOX genes, HOXB1 and HOXB3. miR-10a is a retinoic acid target, and retinoic acid receptor antagonists effectively repress miR-10a expression and completely block metastasis (107). miR-34a functions as a potential tumor suppressor in neuroblastoma cells, and retinoic acid-induced differentiation of the neuroblastoma cell line led to increased miR-34a expression and decreased expression of its target E2F3 (108).
miR-23 has a critical role in the RA-induced neuronal differentiation of human embryonal carcinoma NT2 cells (109). Retinoic acid downregulates several microRNAs (i.e., miR-9/9*, miR-124a, and miR-125B) to induce abnormal development of the spinal cord in a spina bifida rat model (107).
Folate and Methyl Deficiency
It has been proposed that DNA methylation may mediate some of the effects of environmental exposures and lifestyle factors on disease risk, as methyl-deficient diets influences global DNA methylation (110). A case-control study of 1,294 prostate cancer patients and 1,451 controls also supports a favorable role of dietary folate on prostate cancer risk (111).
Experimental rats fed a folate- methionine- and choline-deficient diet develop hepatocellular carcinoma (HCC) at 54 weeks of age in the absence of carcinogen treatment (112). miRNA expression analysis of livers from the animals fed the folate-/methyl-deficient diet showed let-7a, miR-21, miR-23, miR-130, miR-190, and miR-17-92 family of genes were upregulated, whilst miR-122, an abundant liver-specific miR, was downregulated in the tumors. The decrease in hepatic miR-122 was a tumor-specific event because it did not occur in rats switched to the folate- and methyl-adequate diet after 36 weeks on the deficient diet, which did not lead to hepatocarcinogenesis (112). In another study, it was reported that HCC induced by methyl deficiency was characterized by profound downregulation of tumor suppressor miR-34a, miR-16a, miR-181a, and miR-127. This was accompanied by alterations in protein levels of the E2F3 and BCL6, which are confirmed targets for miR-34a and miR-127, respectively (100,113). A recent study shows that feeding choline-deficient and amino acid-defined diet to experimental mice promotes hepatocarcinogenesis accompanied by altered microRNA expression profiles that includes an up-regulation of oncogenic miR-155, miR-221/222, and miR-21 and downregulation of miR-122 at early stages of hepatocarcinogenesis (114). Another study shows down-regulation of the microRNAs involved in the regulation of apoptosis, cell proliferation, cell-to-cell connection, and epithelial-mesenchymal transition, including miR-34a, miR-127, miR-16a and miR-200b, in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet (115).
Folate deficiency induced a pronounced global increase in miRNA expression in human lymphoblastoid cells (116). miR-222 was identified as significantly overexpressed under folate-deficient conditions in vitro, and this finding was confirmed in vivo in human peripheral blood from individuals with low folate intake, suggesting that miRNA expression might be potential biomarkers of nutritional status in humans (116).
Other Dietary Factors
Maternal high fat diet during pregnancy and lactation induced coordinated and long-lasting changes in the expression of insulin-like growth factor2 (Igf2) and several key miRNAs in the offspring (117). Another study explored the effect of n-3 polyunsaturated fatty acids on carcinogen-directed non-coding microRNA signatures in rat colon (118). Experimental rats were fed diets containing corn oil or fish oil with pectin or cellulose and injected with a colon-specific carcinogen, and effects on microRNA expression in colonic mucosa were analysed. At an early stage of cancer progression, five different miR (let-7d, miR-15b, miR-107, miR-191 and miR-324-5p) were significantly affected by diet-carcinogen interactions. Overall, fish-oil-fed animals exhibited the smallest number of differentially expressed microRNAs pointing to a novel role of fish oil in protecting the colon from carcinogen-induced microRNA dysregulation (118). Melkamu et al. (2009) studied the modulation of miR expression by the chemopreventive agent indole-3-carbinol in induced mouse lung tumors and found that the levels of miR-21, mir-31, miR-130a, miR-146b, and miR-377 were reduced in indole-3-carbinol-fed mice relative to the level in mice treated with the carcinogen alone (119).
CONCLUSIONS
Experimental evidence suggests a role for miRNAs in the etiology of prostate cancer. However, our current understanding in this area is still limited, and more focused functional studies are required to decipher the precise miRNA signatures for prostate cancer. Overall, a better understanding of miRNA-regulated pathways in prostate cancer will improve our knowledge of the pathogenesis of the disease and can potentially aid in developing miRNA-based diagnostic and therapeutic strategies for the management of prostate cancer. The potential application of miRNAs in prostate cancer is elegantly demonstrated in a study by Mitchell et al. (2008), where serum levels of miR-141 (a miRNA expressed in prostate cancer) can distinguish patients with prostate cancer from healthy controls (120). Also, it is being recognized that dietary components such as genistein, curcumin, folate and retinoids can affect carcinogenesis through modulation of miRNA expression patterns. Such studies are urgently needed in regards to prostate cancer given the fact that dietary components have been shown to play a crucial chemopreventive role in prostate cancer progression and metastasis.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Syed DN, Suh Y, Afaq F, Mukhtar H. Dietary agents for chemoprevention of prostate cancer. Cancer Lett. 2008;265:167–176. doi: 10.1016/j.canlet.2008.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 8.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 9.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 12.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 14.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 15.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitson JM, Noonan EJ, Pookot D, Place RF, Dahiya R. Double stranded-RNA-mediated activation of P21 gene induced apoptosis and cell cycle arrest in renal cell carcinoma. Int J Cancer. 2009;125:446–452. doi: 10.1002/ijc.24370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 22.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 24.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 28.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 30.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009 [DOI] [PubMed]
- 33.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 34.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, Khan J. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 36.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs–the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 38.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 39.Li LC. Epigenetics of prostate cancer. Front Biosci. 2007;12:3377–3397. doi: 10.2741/2320. [DOI] [PubMed] [Google Scholar]
- 40.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim Biophys Acta. 2004;1704:87–102. doi: 10.1016/j.bbcan.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Damber JE, Aus G. Prostate cancer. Lancet. 2008;371:1710–1721. doi: 10.1016/S0140-6736(08)60729-1. [DOI] [PubMed] [Google Scholar]
- 42.Coppola V, de Maria R, Bonci D. MicroRNAs and prostate cancer. Endocr Relat Cancer. 2009. [DOI] [PubMed]
- 43.Gandellini P, Folini M, Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: where are we now? Trends Mol Med. 2009;15:381–390. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 45.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 47.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 48.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, Croce CM. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, Yfantis HG, Lee DH, Stephens RM, Liu CG, Calin GA, Croce CM, Ambs S. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68:1152–1164. doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Tang H, Thayanithy V, Subramanian S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ, Thibodeau SN. Gene Networks and microRNAs Implicated in Aggressive Prostate Cancer. Cancer Res. 2009;69(24):9490–9497. doi: 10.1158/0008-5472.CAN-09-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, Mendell JT, Lupold SE. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 54.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249–254. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, Deguchi T, Ito M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 57.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 59.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 61.Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leite KR, Sousa-Canavez JM, Reis ST, Tomiyama AH, Camara-Lopes LH, Sanudo A, et al. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol Oncol. 2009. [DOI] [PubMed]
- 63.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, Avances C, Villalba M, Culine S, Fajas L. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS ONE. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, Valdagni R, Daidone MG, Zaffaroni N. miR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 67.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c Are Downregulated in Prostate Cancer, Suppress CD44 Translation and Enhance Invasion of Prostate Cancer Cells in vitro. Int J Clin Exp Pathol. 2009;2:361–369. [PMC free article] [PubMed] [Google Scholar]
- 69.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, Lu PJ. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene. 2009;28:3360–3370. doi: 10.1038/onc.2009.192. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Ladd A, Dragoescu E, Budd WT, Ware JL, Zehner ZE. MicroRNA-17-3p is a prostate tumor suppressor in vitro and in vivo, and is decreased in high grade prostate tumors analyzed by laser capture microdissection. Clin Exp Metastasis. 2009;26(8):965–979. doi: 10.1007/s10585-009-9287-2. [DOI] [PubMed] [Google Scholar]
- 72.Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696–24704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 74.Qin W, Zhao B, Shi Y, Yao C, Jin L, Jin Y. BMPRII is a direct target of miR-21. Acta Biochim Biophys Sin (Shanghai) 2009;41:618–623. doi: 10.1093/abbs/gmp049. [DOI] [PubMed] [Google Scholar]
- 75.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ. and R.W. deVere White. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 77.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, Spagnoli LG, Farace MG, Ciafre SA. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE. 2008;3:e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun T, Yang M, Kantoff P, Lee GS. Role of microRNA-221/-222 in cancer development and progression. Cell Cycle. 2009;8:2315–2316. doi: 10.4161/cc.8.15.9221. [DOI] [PubMed] [Google Scholar]
- 80.Spahn M, Kneitz S, Scholz CJ, Nico S, Rudiger T, Strobel P, Riedmiller H, Kneitz B. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2009;7(4):30–31. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 81.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 82.Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1998. CA Cancer J Clin. 1998;48:6–29. doi: 10.3322/canjclin.48.1.6. [DOI] [PubMed] [Google Scholar]
- 83.Banerjee S, Li Y, Wang Z, Sarkar FH. Multi-targeted therapy of cancer by genistein. Cancer Lett. 2008;269:226–242. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khan N, Afaq F, Mukhtar H. Cancer chemoprevention through dietary antioxidants: progress and promise. Antioxid Redox Signal. 2008;10:475–510. doi: 10.1089/ars.2007.1740. [DOI] [PubMed] [Google Scholar]
- 85.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, Majid S, Igawa M, Dahiya R. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 86.Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, Saini S, Tanaka Y, Dahiya AV, Khatri G, Dahiya R. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 2009. [DOI] [PMC free article] [PubMed]
- 88.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, Hirata H, Li LC, Zhao H, Okino ST, Place RF, Pookot D, Dahiya R. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 89.Sun Q, Cong R, Yan H, Gu H, Zeng Y, Liu N, Chen J, Wang B. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol Rep. 2009;22:563–567. doi: 10.3892/or_00000472. [DOI] [PubMed] [Google Scholar]
- 90.Salerno E, Scaglione BJ, Coffman FD, Brown BD, Baccarini A, Fernandes H, Marti G, Raveche ES. Correcting miR-15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol Cancer Ther. 2009;8:2684–2692. doi: 10.1158/1535-7163.MCT-09-0127. [DOI] [PubMed] [Google Scholar]
- 91.Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 92.Everett PC, Meyers JA, Makkinje A, Rabbi M, Lerner A. Preclinical assessment of curcumin as a potential therapy for B-CLL. Am J Hematol. 2007;82:23–30. doi: 10.1002/ajh.20757. [DOI] [PubMed] [Google Scholar]
- 93.Li L, Aggarwal BB, Shishodia S, Abbruzzese J, Kurzrock R. Nuclear factor-kappaB and IkappaB kinase are constitutively active in human pancreatic cells, and their down-regulation by curcumin (diferuloylmethane) is associated with the suppression of proliferation and the induction of apoptosis. Cancer. 2004;101:2351–2362. doi: 10.1002/cncr.20605. [DOI] [PubMed] [Google Scholar]
- 94.Li L, Ahmed B, Mehta K, Kurzrock R. Liposomal curcumin with and without oxaliplatin: effects on cell growth, apoptosis, and angiogenesis in colorectal cancer. Mol Cancer Ther. 2007;6:1276–1282. doi: 10.1158/1535-7163.MCT-06-0556. [DOI] [PubMed] [Google Scholar]
- 95.Li L, Braiteh FS, Kurzrock R. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer. 2005;104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 96.Li M, Zhang Z, Hill DL, Wang H, Zhang R. Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res. 2007;67:1988–1996. doi: 10.1158/0008-5472.CAN-06-3066. [DOI] [PubMed] [Google Scholar]
- 97.Liang T, Chen MJ, Zhou KY, Tang XD, Wang XG. Induction of apoptosis in human nasopharyngeal carcinoma cell line CNE-2Z by curcumin. Ai Zheng. 2004;23:1651–1654. [PubMed] [Google Scholar]
- 98.Nakamura K, Yasunaga Y, Segawa T, Ko D, Moul JW, Srivastava S, Rhim JS. Curcumin down-regulates AR gene expression and activation in prostate cancer cell lines. Int J Oncol. 2002;21:825–830. [PubMed] [Google Scholar]
- 99.Sarkar FH, Li Y, Wang Z, Kong D. Cellular signaling perturbation by natural products. Cell Signal. 2009;21:1541–1547. doi: 10.1016/j.cellsig.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008;66:477–482. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 101.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 102.Yang J, Cao Y, Sun J, Zhang Y. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med Oncol. 2009. [DOI] [PubMed]
- 103.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 104.Careccia S, Mainardi S, Pelosi A, Gurtner A, Diverio D, Riccioni R, Testa U, Pelosi E, Piaggio G, Sacchi A, Lavorgna S, Lo-Coco F, Blandino G, Levrero M, Rizzo MG. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene. 2009;28:4034–4040. doi: 10.1038/onc.2009.255. [DOI] [PubMed] [Google Scholar]
- 105.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, Calin GA, Liu CG, Andreeff M, Croce CM. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 106.De Marchis ML, Ballarino M, Salvatori B, Puzzolo MC, Bozzoni I, Fatica A. A new molecular network comprising PU.1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009;23:856–862. doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- 107.Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2145. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 108.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 109.Kawasaki H, Taira K. Functional analysis of microRNAs during the retinoic acid-induced neuronal differentiation of human NT2 cells. Nucleic Acids Res. 2003;3(1):243–244. doi: 10.1093/nass/3.1.243. [DOI] [PubMed] [Google Scholar]
- 110.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 111.Pelucchi C, Galeone C, Talamini R, Negri E, Parpinel M, Franceschi S, Montella M, La Vecchia C. Dietary folate and risk of prostate cancer in Italy. Cancer Epidemiol Biomarkers Prev. 2005;14:944–948. doi: 10.1158/1055-9965.EPI-04-0787. [DOI] [PubMed] [Google Scholar]
- 112.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 113.Pogribny IP, Tryndyak VP, Ross SA, Beland FA. Differential expression of microRNAs during hepatocarcinogenesis induced by methyl deficiency in rats. Nutr Rev. 2008;66(Suppl 1):S33–35. doi: 10.1111/j.1753-4887.2008.00064.x. [DOI] [PubMed] [Google Scholar]
- 114.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48:479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 116.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 117.Zhang J, Zhang F, Didelot X, Bruce KD, Cagampang FR, Vatish M, Hanson M, Lehnert H, Ceriello A, Byrne CD. Maternal high fat diet during pregnancy and lactation alters hepatic expression of insulin like growth factor-2 and key microRNAs in the adult offspring. BMC Genomics. 2009;10:478. doi: 10.1186/1471-2164-10-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.L.A. Davidson, N. Wang, M.S. Shah, J.R. Lupton, I. Ivanov, and R.S. Chapkin. n-3 polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis (2009). [DOI] [PMC free article] [PubMed]
- 119.Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl-carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2009. [DOI] [PubMed]
- 120.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]


