Abstract
Drosophila tracheal fusion cells play multiple important roles in guiding and facilitating tracheal branch fusion. Mechanistic understanding of how fusion cells function during development requires deciphering their transcriptional circuitry. In this paper, three genes with distinct patterns of fusion cell expression were dissected by transgenic analysis to identify the cis regulatory modules that mediate their transcription. Bioinformatic analysis involving phylogenetic comparisons coupled with mutational experiments were employed. The dysfusion bHLH-PAS gene was shown to have two fusion cell cis-regulatory modules; one driving initial expression and another autoregulatory module to enhance later transcription. Mutational dissection of the early module identified at least four distinct inputs, and included putative binding sites for ETS and POU homeodomain proteins. The ETS transcription factor Pointed mediates the transcriptional output of the branchless/breathless signaling pathway, suggesting that this pathway directly controls dysfusion expression. Fusion cell cis-regulatory modules of CG13196 and CG15252 require two Dysfusion:Tango binding sites, but additional sequences modulate the breadth of activation in different fusion cell classes. These results begin to decode the regulatory circuitry that guides transcriptional activation of genes required for fusion cell morphogenesis.
Keywords: bHLH-PAS, cis-regulatory module, CG13196, CG15252, Drosophila, dysfusion, fusion cell, trachea, transcription
1. Introduction
The Drosophila tracheal system is derived from an array of segmentally-repeated clusters of precursor cells. After the tracheal precursor cells divide and invaginate, they extend branches (Manning and Krasnow, 1993). Most branches in each metamere grow towards branches from neighboring segments, and then fuse to form the mature tracheal tree (Samakovlis et al., 1996). Each branch fusion event is mediated by two specialized fusion cells, one on each branch, that recognize each other. During branch migration, the fusion cells extend filopodia that likely sense guidance cues and steer the branch to its target. The opposing fusion cells recognize and adhere to each other, leading to branch fusion. After fusion, the fusion cells undergo a sequence of morphological changes leading to a connected tracheal tubule system (Lee and Kolodziej, 2002). Fusion cell development is characterized by transcriptional changes. These changes include both the upregulation and downregulation of fusion cell-expressed genes. Two transcription factors present in fusion cells are the Dysfusion (Dys) bHLH-PAS protein (Jiang and Crews, 2003; Jiang and Crews, 2006) and the Escargot (Esg) zinc finger protein (Samakovlis et al., 1996; Tanaka-Matakatsu et al., 1996). Phenotypically, both esg and dys promote tracheal fusion and inhibit branching, although dys is downstream of esg and requires esg function for fusion cell expression in all branches, except the dorsal trunk (DT). In this paper, we describe a detailed analysis of multiple tracheal fusion cell cis-regulatory modules (CRMs) that are regulated in diverse ways. The molecular dissection of fusion cell CRMs provides insight into the regulation of fusion cell development, and also provides fusion cell-specific Gal4 lines useful for the purification and genetic analysis of fusion cells.
The Dys protein is one of four Drosophila bHLH-PAS proteins that function in various aspects of tracheal development. The Trachealess (Trh) protein acts as a master regulator of tracheal development and is expressed in all tracheal cells (Isaac and Andrew, 1996; Wilk et al., 1996). Trh requires the Ventral veins lacking (Vvl, or Drifter) coactivator protein to activate tracheal gene expression (Zelzer and Shilo, 2000), although Vvl may regulate expression of some tracheal genes in the absence of Trh (Boube et al., 2000). During tracheal fusion, the Trh protein is downregulated in fusion cells by a dys-dependent, post-transcriptional mechanism (Jiang and Crews, 2006), in which the Archipelago protease degrades Trh (Mortimer and Moberg, 2007). The Similar (Sima) protein controls tracheal branching in response to hypoxia (Lavista-Llanos et al., 2002; Nambu et al., 1996). All three proteins: Dys, Sima, and Trh, utilize the Tango (Tgo) bHLH-PAS protein as a heterodimerization partner (Jiang and Crews, 2003; Jiang and Crews, 2007; Sonnenfeld et al., 1997). Dys is an important regulator of fusion cell transcription that is required for fusion cell recognition, adhesion, and potentially other functions involved in fusion cell morphogenesis. Previously, we identified 4 genes, CG13196, CG15252, members only (mbo), and shotgun (shg), whose transcription is dependent on Dys function (Jiang and Crews, 2006), and showed that CG13196 is a direct target of Dys:Tgo (Jiang and Crews, 2007).
Employing S2 cell transient transfection approaches, we demonstrated that Dys:Tgo efficiently binds multiple asymmetric E-Box sequences, including ACGTG, GCGTG, and TCGTG (Jiang and Crews, 2007), a result confirmed by in vitro biochemical approaches (Ooe et al., 2007). This promiscuous DNA binding specificity is evolutionarily-conserved, as the human Dys ortholog, NXF/Npas4, binds the same DNA sequences (Jiang and Crews, 2007; Ooe et al., 2004; Ooe et al., 2007). We identified a 1 kb upstream fragment of CG13196 that contained multiple TCGTG sequences, as well as ACGTG and GCGTG motifs. Mutational studies in vivo revealed that only the TCGTG sequences are required in vivo (Jiang and Crews, 2007). The importance of the TCGTG motifs was reinforced when a transgenic reporter containing a promoter fused to multimerized TCGTG sequences was shown to be expressed in fusion cells. Nevertheless, the generality of TCGTG sequences and fusion cell expression remains unknown, as do the identities of additional co-regulatory proteins and cis-control sequences that mediate fusion cell gene expression.
In this paper, we analyzed 3 genes with diverse patterns of fusion cell expression: (1) the dys gene, which is expressed early in fusion cell development, (2) CG13196, which is a Dys target gene expressed in all fusion cells, and (3) CG15252, which is a Dys target gene expressed only in DT fusion cells. These studies contribute to an understanding of how dys fusion cell expression is regulated, as well as conserved features of Dys-dependent regulation. CRMs that drive fusion cell expression were identified for each gene using transgenic approaches. These fragments of DNA were then scanned for phylogenetically-conserved sequence motifs. Such motifs usually consist of DNA sequences between 4 and 20 bp, and are binding sites for transcription factors. Conserved and repeated sequences, including putative Dys:Tgo binding sites, were identified and their significance tested by in vitro mutagenesis and in vivo transgenic analysis. In this manner, we identified a number of essential cis-regulatory elements that are required for fusion cell expression. Other than Dys:Tgo binding sites, we did not identify required sequences that were repeated in other fusion cell CRMs, suggesting that multiple distinct regulatory mechanisms are utilized to activate each gene.
2. Results
2.1. Embryonic dys expression is regulated by diverse cis-regulatory sequences
The dys gene is expressed in multiple embryonic cell types beginning at stage 12, including tracheal fusion cells, a subset of brain cells, epidermal leading edge cells, the foregut atrium, hindgut, and anal pad (Jiang and Crews, 2003). We decided to explore the dys regulatory sequences for multiple reasons: (1) dys is expressed relatively early in fusion cells and expression is maintained throughout embryogenesis; is this due to a single CRM or multiple temporally-distinct CRMs? (2) Similarly, does each branch type with fusion cells: dorsal branch (DB), DT, lateral trunk (LT), and ganglionic branch (GB), have distinct CRMs for fusion cell expression? (3) As dys expression is genetically-dependent on esg in all fusion cells, except DT and possibly, LT (further suggesting branch-specific inputs to dys expression) (Jiang and Crews, 2003), is dys a direct target of Esg? (4) Does dys fusion cell expression share similar transcriptional inputs or CRMs that control dys expression in other tissues? (5) dys fusion cell-specific fragments would be useful as a Gal4 line for analyzing fusion cell development and function, and for purification of fusion cells to be used in biochemical or in vitro studies.
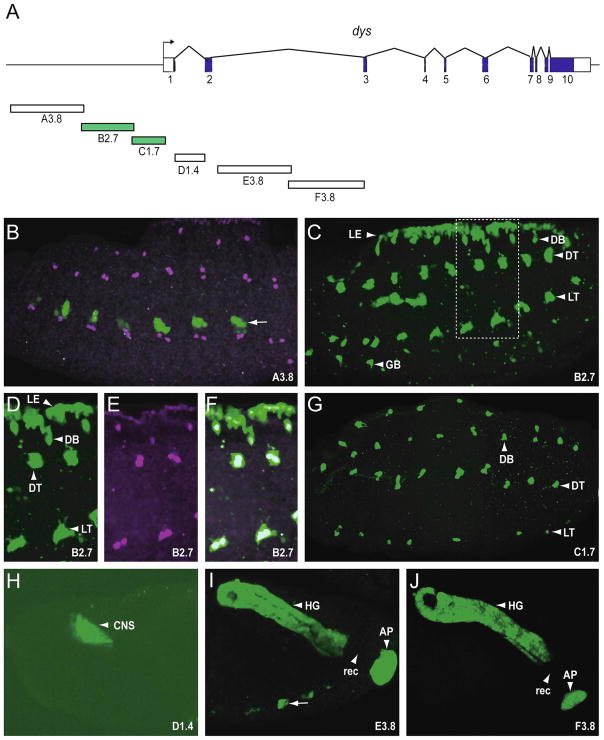
To identify CRMs that control dys fusion cell expression, we examined 1–4 kb size fragments of DNA from the 5′ flanking region and introns 1 and 2 using germline transformation (Fig. 1A). Fragments were PCR-amplified and cloned into either the pH-Stinger P-element vector (B2.7, C1.7) or into the pMintGate ΦC31 vector (A3.8, D1.4, E3.8, F3.8; Supplementary Fig. 1). Both vectors contain a nuclear GFP reporter gene. The pMintGate transgenes were all introduced into the attP2 site at 68A1-B2. Six transgenic reporter fly strains testing fragments A3.8, B2.7, C1.7, D1.4, E3.8, and F3.8 were generated and the embryonic expression of GFP analyzed. Immunostaining with anti-Dys was used to identify Dys+ cells. The A3.8 fragment drove GFP expression only in a subset of Dys− ectodermal cells (Fig 1B). This expression is likely due to sequences in the pMintGate vector or the integration site of the transgene, since they were commonly observed in pMintGate transformants. The B2.7 fragment drove GFP expression in all tracheal fusion cells, including those from DB, DT, LT, and GB (Fig. 1C), as well as leading edge cells. The presence of GFP in fusion cells was demonstrated by overlap with anti-Dys stained cells (Fig. D–F). The dynamics of B2.7 GFP expression (Supplementary Fig. 2A-I; P155, a B2.7 derivative is shown) is consistent with expression of endogenous dys from stages 12 through 17.
Fig. 1.
Transgenic analysis of the dys regulatory region. (A) Schematic illustrating 32 kb of the genomic region encompassing the dys gene. The 10 dys exons are indicated as blocks: coding sequences are filled and untranslated regions unfilled. The direction of transcription is indicated by the arrow. Fragments analyzed as reporter transgenes are labeled A–F with the size of the fragment in kb included. Fragments colored green indicate they drove expression in fusion cells. Stage 15 or 16 embryos from transgenic reporter-GFP strains were stained with anti-GFP (green) and anti-Dys (magenta) antibodies. All images depict sagittal views and anterior is to the left. (B) A3.8 drove GFP expression in subsets of ectodermal cells (arrow: here and throughout) that did not overlap with Dys+ cells. These cells were commonly observed with pMintGate transgenes that were introduced at the attP2 site at 68A1-B2, independent of the fragments being tested. (C) B2.7 drove GFP expression in leading edge (LE) cells and all tracheal fusion cells, including DB, DT, LT, and GB. (D–F) Enlarged image of the rectangle shown in C, indicating anti-GFP reactivity overlapping with anti-Dys reactivity in fusion cells and leading edge; (D) GFP only, (E) Dys only, (F) merge. (G) C1.7 drove GFP expression in all fusion cells. (H) D1.4 drove GFP expression in CNS brain cells. (I–J) Both E3.8 and F3.8 drove expression in hindgut (HG) and anal pad (AP). There was an absence of expression in the rectum (rec). This pattern of expression is identical to endogenous dys.
Expression in all tracheal fusion cells was also observed with the C1.7 fragment (Fig. 1G). However, C1.7 fusion cell expression was only observed from stages 14–17 (Supplementary Fig. 2J-O; T523, a C1.7 derivative is shown). These results indicated that the dys gene has at least two independent CRMs, both in the 5′-flanking region, that control fusion cell expression in temporally distinct ways. The C1.7 fragment overlaps with B2.7 by 20 bp, but subsequent experiments (Fig. 2) indicated that the tracheal fusion cell CRM within B2.7 does not overlap with C1.7. The D1.4, E3.8, and F3.8 transgenes were not expressed in tracheal fusion cells, but each was expressed in a Dys+ embryonic cell type, including brain (D1.4), hindgut and anal pad (E3.8, F3.8) (Fig. 1H–J). Of the 17.2 kb of the dys gene examined, all embryonic Dys+ embryonic cell types were observed, except the foregut atrium. In addition, the tracheal fusion cells, hindgut, and anal pad had at least two distinct CRMs controlling expression.
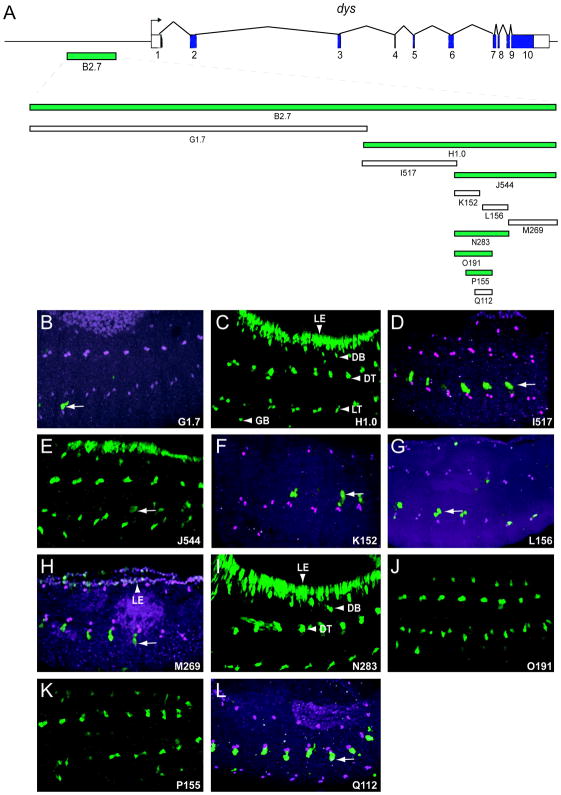
Fig. 2.
Reduction of the dys B2.7 fusion cell CRM to a 155 bp fragment. (A) Schematic of the dys gene and derivative fragments tested in vivo. Fragments labeled G–Q with associated number indicating kb or bp. Fragments colored green drove fusion cell expression. (B–L) Fragments H1.0, J544, and N283 had fusion cell and leading edge expression. Fragments O191 and P155 were expressed in only fusion cells. Fragment M269 had weak leading edge expression, and the other fragments (G1.7, I517, K152, L156, and Q112) had no expression in fusion cells or leading edge. Arrows indicate fragment-independent ectodermal expression.
2.2. Identification of fusion cell-specific dys CRMs
To isolate the dys minimal CRMs from other functional sequences, the B2.7 and C1.7 fragments were further subdivided, and the fragments tested for fusion cell expression (Fig. 2,3). B2.7 was divided into the G1.7 and H1.0 fragments (Fig. 2A). G1.7 was not expressed in fusion cells or leading edge (Fig. 2B), whereas H1.0 expression was observed in both cell types (Fig. 2C). Bisection of H1.0 yielded a non-expressing subfragment, I517 (Fig. 2D), and J544, which drove expression in fusion cells and leading edge (Fig. 2E). When J544 was subdivided into 3 fragments: K152, L156, and M269, none showed expression in fusion cells (Fig. 2F–H), although M269 had weak leading edge expression (Fig. 2H). These results suggested that fusion cell and leading edge expression were separable, and that we had disrupted the fusion cell CRM. Another fragment, N283, which encompassed K152 and L156, was analyzed and had strong fusion cell and leading edge expression (Fig. 2I). When 3 deletion fragments of N283 were tested, two of them, O191 and P155, drove expression in all fusion cells, but were unable to drive leading edge expression (Fig. 2J,K). In contrast, no expression was observed from Q112 (Fig. 2L). Thus, we were able to identify a small fragment of dys, P155, which was sufficient for fusion cell expression, and independent of leading edge expression. The O191 and P155 fusion cell specific fragments were also each inserted into a ΦC31-based Gal4 vector, and both transgenic lines were able to drive reporter gene expression specifically in fusion cells (data not shown), thus providing a fusion cell-specific driver strain for genetic and cell purification experiments.
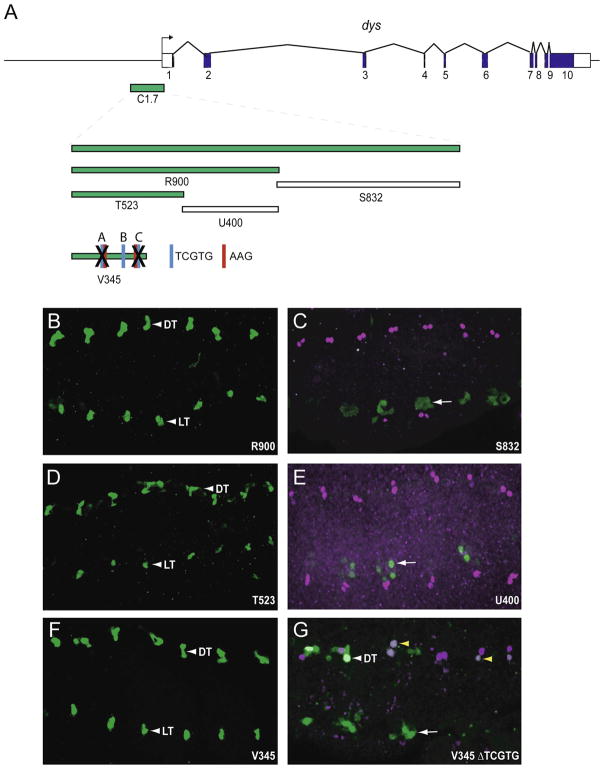
Fig. 3.
Identification of the dys C1.7 fusion cell CRM. (A) Schematic of the dys gene and fragments tested. C1.7 is expressed in fusion cells (see Fig. 1). V345 contains 3 TCGTG Dys:Tgo binding sites, labeled A–C. Two of the TCGTG sites are preceded by AAG. The X over TCGTG-A and TCGTG-C indicates that those sites were deleted in V345. (B–F) Fragments R900, T523, and V345 were expressed in all fusion cells, while fragments S832 and U400 were not. (G) The TCGTG-A and TCGTG-C sites were deleted in V345. GFP expression at wild-type levels was occasionally observed in DT (white arrowhead), but was generally highly reduced (yellow arrowheads) or absent. No expression was observed in DB, LT, or GB.
All dys fragments with fusion cell expression showed expression in all fusion cells, suggesting that the regulatory inputs responsible for activating expression in the B2.7 CRM act in all fusion cells. Analysis of the deletion derivatives of the fusion cell-expressing O191 fragment (K152, P155, Q112) indicated that there were at least two separable elements required for fusion cell expression. These reside in O191 at 36–79 and 152–191. Leading edge expression was observed from 2 adjacent fragments: M269 (weak) and N283 (strong), but the N283 deletions did not further localize activity. This suggests that multiple elements also contribute to leading edge expression.
To identify the minimal fusion cell specific enhancer within C1.7, it was dissected into two fragments: R900 and S832 (Fig. 3A). R900 drove GFP expression in all fusion cells (Fig. 3B), whereas S832 did not have fusion cell expression. R900 was subdivided into the T523, U400, and V345 fragments. T523 and V345 drove fusion cell expression (Fig. 3D,F), whereas U400 did not (Fig. 3E). Thus, we identified a second dys fusion cell CRM in a 345 bp stretch of DNA, and like the B2.7 CRM, the C1.7 CRM is expressed in all fusion cells.
2.3. Bioinformatic identification and in vivo functional analysis of dys CRM sequence motifs
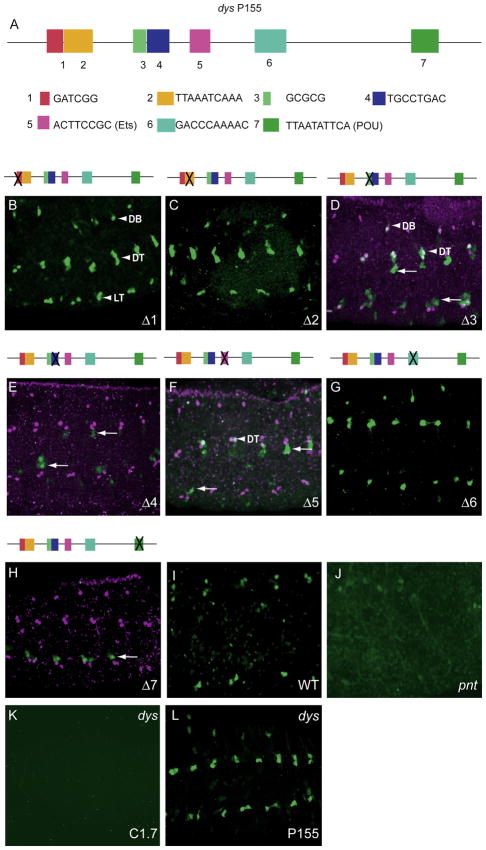
The P155 dys fragment was compared to orthologous sequences in other drosophilids, and seven conserved regions were selected, deleted individually in P155, and tested for fusion cell expression by germline transformation. These sequences were: 1 (GATCGG), 2 (TTAAATCAAA), 3 (GCGCG), 4 (TGCCTGAC), 5 (ACTTCCGC), 6 (GACCCAAAAC), and 7 (TTAATATTCA) (Fig. 4A, Supplementary Fig. 3). The Δ1, Δ2 and Δ6 deletion transgenes showed fusion cell expression (Fig. 4B,C,G) similar to the unmutated P155 fragment. In contrast, Δ3 and Δ5 resulted in reduced GFP expression, present in only a few DT and DB fusion cells (Fig. 4D,F). The sequence corresponding to Δ5 (ACTTCCG) resembles a consensus ETS binding site: (A/G)(C/T)(A/T)TCC(G/T) (Sharrocks et al., 1997). Previous work showed that the Pointed (Pnt) ETS protein plays a prominent role in tracheal development as a downstream effector of breathless signaling (Myat et al., 2005; Ohshiro et al., 2002). We note that, similar to Δ5, dys expression was reduced in pnt mutant embryos (Fig. 4I,J), consistent with Pnt directly regulating dys expression. The Δ4 and Δ7 deletions were required for dys expression, since deletion completely abolished GFP expression in fusion cells (Fig. 4E,H). The sequence corresponding to Δ7 (TTAATATTCA) resembles binding sites for POU-homeodomain proteins (consensus: TTAAAATTCA) (Okamoto et al., 1990) as determined by an unbiased search of P155 using TESS (Schug, 2008). The vvl gene encodes a POU-homeodomain protein gene (Anderson et al., 1995) that can regulate tracheal gene expression either in conjunction with the Trh bHLH-PAS protein (Zelzer and Shilo, 2000) or independently of Trh (Boube et al., 2000). If Vvl binds to the Δ7 sequence or some other sequence in P155, it does so without the partnership of Trh:Tgo, since there is no ACGTG Trh:Tgo binding site present in P155.
Fig. 4.
Mutational analysis of dys P155 fragment identified several sites required for fusion cell expression. (A) Schematic of P155 showing the conserved sequences (1–7) that were deleted and assayed for fusion cell expression. (B–H) The Δ1, Δ2, and Δ6 deletions had no effect on fusion cell expression, whereas the Δ4 and Δ7 deletions abolished expression, and the Δ3 and Δ5 deletions resulted in GFP expression in only a few DB and DT cells (arrowheads). Arrows indicate GFP-expressing non-fusion cells. (I,J) Expression of dys (anti-Dys; magenta) in fusion cells (I; wild-type control) was reduced in (J) pntΔ88 mutant embryos. The gain was increased in (J) to show that GFP was weakly present (and not absent) in pntΔ88 DT. (K,L) GFP expression of dys C1.7 was abolished in a dys1 mutant, whereas expression of dys P155 was unaffected.
The analysis of dys deletions (Fig. 2) indicated that there were at least two regions of P155 required for fusion cell expression, 1–42 and 117–155. The Δ3 deletion resulted in reduced fusion cell expression; it overlaps nucleotides 1–42 in P155, and its disruption in Q112 could contribute to, but not fully explain, the complete loss of expression observed with Q112 (Fig. 2L). The Δ7 deletion, which abolished expression, is present in P155 nucleotides 117–155, and its role is consistent with the absence of expression observed with K152 (Fig. 2F), which lacks 117–155.
The Esg zing-finger transcription factor is expressed in all fusion cells, and mutants in esg results in an absence of dys expression in DB and GB, but not DT (LT dies in esg mutants). One reported binding site for Esg is (G/A)CAGGTG (Fuse et al., 1994). We did not find closely–related sequences in dys P155. Consistent with the results on endogenous dys, the expression of dys P155 and T523 were absent in DB, LT, and GB in esg mutant embryos (Supplementary Fig. 4A–D). Thus, either Esg does not directly regulate dys, or else we are unable to identify Esg binding sites bioinformatically.
Sequence analysis of the dys T523 fragment, which drives late fusion cell expression, showed that it has five Dys:Tgo binding sites (TCGTG) (Supplementary Fig. 5), suggesting the possibility that T523 is autoregulated by dys. Consistent with this observation, C1.7 expression was abolished in dys mutant embryos (Fig. 4K) (T523 is derived from C1.7). The 5′-most 345 bp of T523 is well-conserved in the 12 Drosophila species compared, and the 3′-most 178 bp poorly-conserved (Supplementary Fig. 5). The V345 fragment includes the 5′-most conserved residues and was able to drive GFP expression in all fusion cells (Fig. 3F). Three of the TCGTG sites (A–C) are present in V345; TCGTG-C is conserved in all 12 species and TCGTG-A and TCGTG-B are conserved in 11/12 species. When TCGTG-A and TCGTG-C were deleted together in V345, only weak GFP expression was observed in fusion cells (Fig. 3G). These data indicated that the V345 CRM is autoregulated by dys. In contrast, the P155 dys CRM does not have any TCGTG sequences, and its expression was not altered in dys mutant embryos (Fig. 4L). Thus, the V345 CRM appears to amplify late dys expression by autoregulation.
2.4. CG13196 fusion cell transcription requires multiple Dys:Tgo binding sites and additional cis-control elements
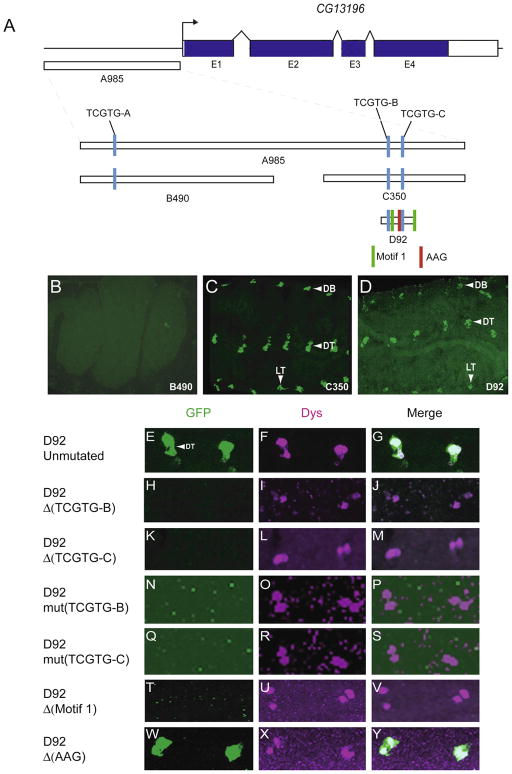
The CG13196 gene is expressed specifically in tracheal fusion cells in the embryo (Jiang and Crews, 2006), and is a direct target of Dys:Tgo (Jiang and Crews, 2007). CG13196 encodes a member of the zona pellucida (ZP) family of membrane proteins (Jazwinska and Affolter, 2004). Its function is unknown, but overexpression of CG13196 in trachea promotes ectopic branch fusion, indicating that CG13196 may play a role in membrane adhesion (Jiang and Crews, 2006). Previously, we identified a 985 bp fragment (A985; Fig. 5A) in the CG13196 5′-flanking region that drove fusion cell expression (Jiang and Crews, 2007). This fragment contains 3 TCGTG (Dys:Tgo) sequences. Mutation of all 3 sites resulted in loss of fusion cell expression, indicating that Dys:Tgo directly regulates CG13196. However, this experiment did not address whether all 3 TCGTG sequences were required for fusion cell expression, and if not, which ones are important. The A985 fragment was first subdivided into two fragments: B490 that contains TCGTG-A, and C350 that contain TCGTG-B and TCGTG-C (Fig. 5A). These fragments were tested for fusion cell expression in vivo. B490 was unable to drive GFP expression in fusion cells (Fig 5B), whereas the C350 transgene drove GFP expression in all fusion cells (Fig 5C). This indicated that TCGTG-A is not required for CG13196 fusion cell expression, nor is it able to drive fusion cell expression by itself. D92 is a 92 bp derivative of C350 that contains both TCGTG-B and TCGTG-C. It was able to promote fusion cell expression (Fig 5D), and its expression relatively late in development (Supplementary Fig. 2P–T) mimicked endogenous CG13196. Both TCGTG motifs were deleted individually in the context of D92 and tested for fusion cell activity. Deletion of either TCGTG-B or TCGTG-C resulted in an absence of fusion cell expression (Fig. 5E–M). To rule out the possibility that the deletions affected the arrangement along the DNA of nearby cis-control elements other than TCGTG motif, the D92 TCGTG motifs were individually mutated to CAATG. Mutation of either TCGTG-B or TCGTG-C also resulted in the absence of fusion cell expression (Fig. 5N–S). These results indicated that both Dys:Tgo binding sites are required for fusion cell expression.
Fig. 5.
Two Dys:Tgo sites and Motif 1 sites are required for fusion cell transcription of CG13196. (A) Schematic of the 3.3 kb genomic region encompassing the CG13196 gene, and fragments analyzed in vivo. Shown are the three TCGTG sequences, two Motif 1 sequences, and an AAG sequence adjacent to TCGTG-C. (B–D) The C350 and D92 fragments drove GFP expression in all fusion cells, whereas fusion cell expression was absent in the B490 transgenic strain. (E–Y) Shown are DT fusion cells for (E–G) unaltered and (H–Y) altered versions of D92. Both (H–M) deletion and (N–S) mutation of each TCGTG site abolished fusion cell expression. Deletion of Motif 1 sites also abolished expression (T–V), whereas deletion of the AAG sequence adjacent to TCGTG-C had no effect (W–Y).
Bioinformatic analysis of the CG13196 D92 fragment revealed strong conservation among the 12 Drosophila species. There were 4 major blocks of conservation: two corresponding to the two Dys:Tgo TCGTG sequences, and two to a repeated 11 bp motif (CCATGGAAAGT and CCATTAAAAGT) referred to as Motif 1 (Supplementary Fig. 6). When both instances of Motif 1 were deleted in D92, no fusion cell expression was observed (Fig. 5T–V). Thus, these sequences may be sites for a fusion cell coactivator that functions in conjunction with Dys:Tgo. However, Motif 1 sequences by themselves are unable to drive fusion cell expression, since when the TCGTG sequences were mutated in D92, all fusion cell expression was lost, despite the presence of the Motif 1 sequences. One of the Dys:Tgo binding sites (TCGTG-B) was immediately preceded by AAG. It was also noticed that AAG precedes two of the 5 TCGTG sequences in dys T523 (Supplementary Fig. 5) and both TCGTGs important for CG15252 expression (see below; Supplementary Fig. 7), suggesting this motif may be important for fusion cell expression. However, deletion of the CG13196 TCGTG-C AAG motif did not affect GFP expression in fusion cells (Fig 5W–Y).
2.5. The DT-specific fusion cell-expressed gene CG15252 is directly regulated by Dys:Tgo
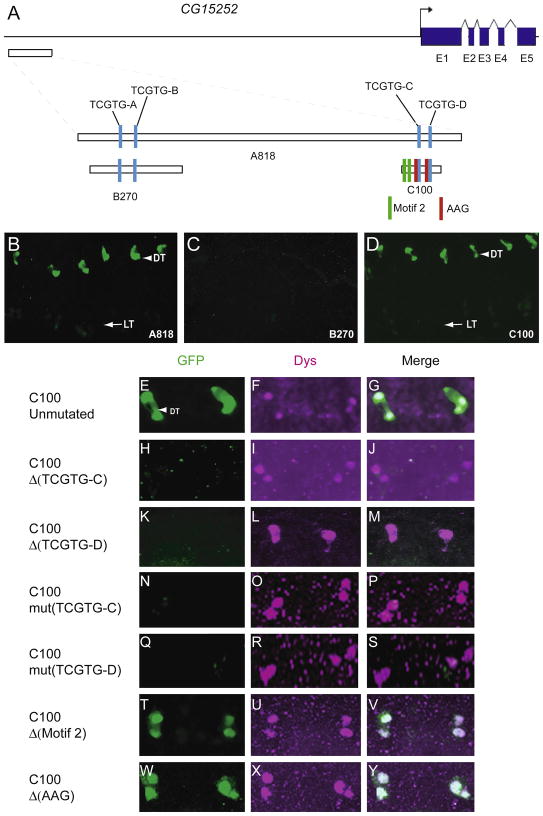
The CG15252 gene is expressed only in DT fusion cells, and its expression requires dys function (Jiang and Crews, 2006). The function of CG15252 is unknown, but it does share sequence homology with the bacterial FtsI protein that is a transpeptidase involved in cell wall peptidoglycan synthesis and cell division (Errington et al., 2003). The CG15252 gene has 9.9 kb of 5′-flanking sequence and 4 relatively small introns. We hypothesized that Dys:Tgo directly regulates CG15252 expression in DT, and selected an 818 bp fragment that contains 4 TCGTG sequences to analyze in vivo for fusion cell expression (Fig. 6A). This fragment is 6.8 kb upstream of the 5′ end of the longest cDNA clone. The A818 fragment drove GFP expression only in DT fusion cells (Fig 6B), similar to endogenous CG15252. To test which TCGTG sites are required for CG15252 fusion cell expression, fragment B270, containing the TCGTG-A and TCGTG-B sites, and fragment C100, containing the TCGTG-C and TCGTG-D sites, were analyzed. B270 was unable to drive GFP expression in DT fusion cells (Fig 6C), whereas the C100 transgenic line had expression in DT fusion cells (Fig 6D), and was expressed late in development similar to CG15252 (Supplementary Fig. 2U,V). Individually deleting or mutating both TCGTG-C and TCGTG-D resulted in a loss of fusion cell GFP expression (Fig. 6E–S). Thus, similar to CG13196, tracheal fusion cell expression requires two clustered Dys:Tgo binding sites, both of which are essential for expression.
Fig. 6.
Two Dys:Tgo sites are required for fusion cell transcription of CG15252. (A) Schematic of 11.5 kb of DNA surrounding the CG15252 gene. Shown are the 4 TCGTG motifs, 2 Motif 2 sequences, and the AAG motifs adjacent to TCGTG-C and TCGTG-D. (B–D) Fragments A818 and C100 drove GFP expression in DT cells, whereas B270 did not. (E–Y) DT expression of unaltered (E–G) and (H–Y) altered versions of C100 are shown. Both (H–M) deletion and (K–P) mutation of each TCGTG abolished DT fusion cell expression. Deletion of both Motif 2 sites (T–V) or both AAG sites adjacent to the TCGTG sites (W–Y) had no effect on DT expression.
Bioinformatic analysis of the C100 sequence revealed strong conservation among 4 species closely related to D. melanogaster (Supplementary Fig. 7). However, significant sequence similarity was not found in 7 other sequenced Drosophila species. Among the 5 related species, the C100 sequence had two instances of a conserved 6 bp motif, named Motif 2. Deletion of both Motif 2 sequences in C100 had no effect on fusion cell expression (Fig. 6T–V). Both of the conserved TCGTG sequences in C100 are preceded by a conserved AAG motif. Deletion of both AAG sequences together had no effect on C100 fusion cell GFP expression (Fig 6W–Y).
2.6. CG13196 has coactivator sequences that promote expression in DB, LT, and GB
Despite the similarity in the requirement of two adjacent Dys:Tgo binding sites for fusion cell expression, CG13196 is expressed in all fusion cells, whereas CG15252 is only expressed in DT. Both Dys and Tgo are present in all fusion cells. There are four general models that could explain the expression differences: (1) Dys:Tgo is able to activate transcription through the presence of one or more binding sites in all fusion cells, and CG15152 has sequences that repress expression in DB, LT, and GB; (2) Dys:Tgo is only able to activate expression in DT by itself, but CG13196 has sequences for a coactivator that allows expression in DB, LT, and GB; (3) Dys:Tgo can activate transcription in fusion cells only in cooperation with additional coactivators, including one functioning in all fusion cells (CG13196; dys autoregulation), and another in only DT (CG15252); and (4) Differential activation depending on the spacing/orientation of Dys:Tgo sequences.
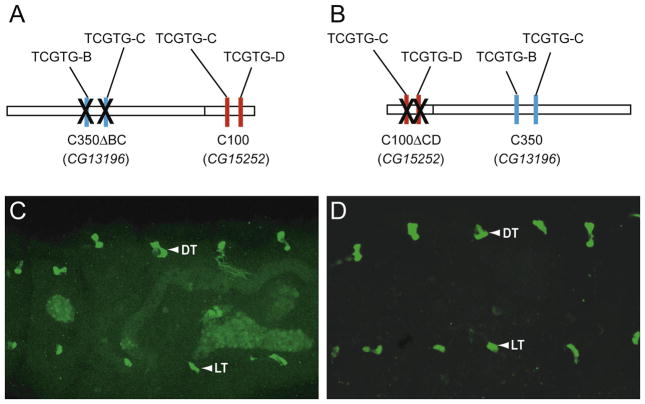
An initial test of these hypotheses was carried-out by fusing fragments containing fusion cell CRMs of CG13196 and CG15252 to each other, and testing their effects in vivo. To test the hypothesis that coactivators are required to turn on CG13196 in fusion cells other than DT, a chimeric enhancer containing CG13196 C350 with deleted TCGTG sites was fused with CG15252 C100 with both TCGTG sites intact (Fig. 7A). If the hypothesis that CG13196 has coactivator sequences for DB, LT, and GB (and possibly, DT) expression is correct, they may work in conjunction with the CG15252 TCGTG sequences to activate expression in all fusion cells. This is what was observed (Fig. 7C). The complementary experiment involved fusing CG15252 C100 with both TCGTG binding sites deleted to CG13196 C350 that had both TCGTG sequences intact (Fig. 7B). If CG15252 has corepressor sequences that restrict its expression to DT, then these repressive sequences could act in combination with the CG13196 TCGTG sequences to limit expression to DT. This was not observed; the chimeric transgene was expressed in all fusion cells (Fig. 7D). Thus, these experiments provide evidence for sequences in CG13196 that work in conjunction with Dys:Tgo binding sites to allow expression in all fusion cells. In contrast, we did not find evidence for sequences in CG15252 that restrict expression to DT. However, this may still be the case, since it is possible that they cannot function in the context of CG13196 C350.
Fig. 7.
CG13196 C350 fragment has sequences that promote transcription in all fusion cells. Schematics of (A) CG13196-A350ΔBC:CG15252-C100 and (B) CG15252-C100ΔCD:CG13196-A350 constructs. (X) indicates that the TCGTG motifs were deleted. (C,D) Both constructs were expressed in all fusion cells (DT and LT are shown).
3. Discussion
Tracheal fusion cells are a complex cell type that direct tracheal cell migration, recognition, adhesion, and cellular remodeling upon fusion. It is expected that these morphogenetic events will be accompanied by sophisticated regulatory pathways. The major goal of this work is to mechanistically explore the transcriptional circuitry that governs tracheal fusion cell function. We selected 3 genes to investigate, since each has a unique mode of fusion cell expression, while also sharing common features. Both dys and CG13196 are expressed in all fusion cells, but dys precedes CG13196 in developmental expression. CG13196 and CG15252 are target genes of dys, but CG15252 is only expressed in DT fusion cells. Employing transgenic reporter analysis of fragments of 5′-flanking and intronic sequences, we were able to identify fusion cell CRMs for each gene. After reducing the size of each fragment by deletional analysis, phylogenetic-assisted bioinformatics was employed to identify DNA sequences that are potential binding sites for transcription factors. Mutational analysis was carried-out and demonstrated the functionality of several motifs. An additional goal, which was successful, was to generate fusion cell-specific Gal4 lines that could be used for fusion cell purification or molecular analyses.
3.1. The dys gene contains multiple fusion cell CRMs
The dys gene is expressed in fusion cells from stages 12–17, and we identified two distinct CRMs that drive dys fusion cell expression. Additional embryonic pattern elements associated with dys expression were identified, including leading edge cells, hindgut, anal pad, and brain. The P155 fragment drove reporter gene expression from stages 12–17, and likely represents the CRM governing initial dys expression. The T523 CRM contains a dys autoregulatory CRM expressed from stages 14–17, since its expression was absent in dys mutant embryos and strongly reduced when two Dys:Tgo binding sites were deleted. Given the potential stability of GFP, it is unclear whether expression of P155 is maintained during later stages, and overlaps with the T523 CRM. However, previous work indicated that null dys mutant embryos possessed substantial dys RNA in late stage embryos (Jiang and Crews, 2003). This suggests that dys-independent transcription, likely derived from the P155 CRM, is still able to contribute detectable levels of dys RNA in a dys mutant. Thus, the dys autoregulatory CRM likely amplifies dys transcription (Crews and Pearson, 2009), but is not solely responsible for late embryonic expression.
Little is known regarding how dys expression is initiated in fusion cells. One potential regulator is the Esg zinc finger protein. Esg is present in all fusion cells, and precedes initial dys expression. Analysis of esg mutant embryos indicated that esg was required for dys expression in DB and GB, but not DT (Jiang and Crews, 2003). Since LT undergoes apoptosis in esg mutants (Samakovlis et al., 1996), it is unclear whether LT dys expression requires esg function. These results are consistent with Esg directly regulating dys in some fusion cells, but not others. By narrowing the limits of the dys stage 12–17 fusion cell CRM to 155 bp (P155), bioinformatic analyses were carried-out to potentially identify transcription factor binding sites, followed by mutagenesis and assaying function in vivo.
Seven conserved sequences from 5 to 10 bp in length were deleted in P155. The results of the deletion experiments indicated that deletions 1, 2, and 6 had no effect on expression; deletions 3 and 5 showed a reduction in fusion cell expression; and deletions 4 and 7 were required for all fusion cell expression. These results were consistent with the deletional analysis of the dys B2.7 fragment (Fig. 2) that indicated at least two separate regions were required for dys expression. One contained sequences 1–3 and the other sequence 7, and individual deletions Δ3 and Δ7 had expression defects. The consensus Esg binding site is: (G/A)CAGGTG, and no sequence strongly resembled this motif in P155. In addition, no deletion tested resulted in altered P155 GFP expression resembling the esg mutant phenotype (loss of expression in DB and GB, but no effect on DT). Instead, all mutants affected all 4 fusion cell types. Thus Esg either does not directly regulate dys transcription, or regulates transcription via binding to sites in P155 not clearly related to the known binding site sequence and not overlapping with Δs1–7. Future work employing in vitro binding studies with Esg protein or use of chromatin immunoprecipitation (ChIP) techniques from embryonic cells would help resolve this issue.
Two of the deleted sites required for high levels of expression are related to known transcription factor binding sites: the Δ5 site corresponds to an ETS protein binding site, and Δ7 to a POU homeodomain binding site. One ETS family protein, Pointed, is the transcriptional effector of the branchless/breathless signaling pathway required for tracheal branching (Myat et al., 2005; Ohshiro et al., 2002; Sutherland et al., 1996). Embryos mutant for pnt show a strong reduction of dys tracheal expression. While this does not prove that breathless signaling and Pointed directly regulate the sequence affected by the Δ5 deletion, the effects of both deleting the ETS site and pnt mutants support that possibility. It is possible that high levels of dys expression in fusion cells require continual signaling from branchless. One prominent tracheal-expressed POU homeodomain protein is Vvl. Mutants in vvl result in an early failure of tracheal branching and general absence of differentiation (Anderson et al., 1995; Llimargas and Casanova, 1997). One study proposed that Vvl is a coactivator of tracheal transcription along with the Trh:Tgo heterodimeric tracheal master regulator (Zelzer and Shilo, 2000). While the Δ7 sequence resembles a POU homeodomain sequence, this site does not strongly resemble known Vvl binding sites. Similarly, there are no Trh:Tgo binding sites (ACGTG) within P155, so positive evidence of dys direct regulation by Trh and Vvl is lacking. Thus, if D7 binds Vvl, then it may function independently of Trh, as was observed for regulation of thick veins, a tracheal-expressed genetic target of vvl (Boube et al., 2000; Llimargas and Casanova, 1997). In summary, these data suggest multiple distinct binding sites (at least 4) contribute to dys expression, and one of the regulators may be Pnt.
One model for dys expression is that separate CRMs drive expression in fusion cells corresponding to each branch. In the most extreme case, there would be 4 CRMs, each controlling DB, DT, LT, and GB fusion cell expression. Alternatively, there may be a single CRM that responds to the same transcription factors present in each fusion cell type. None of the deletions of dys B2.7 or the P155 deletions revealed evidence of branch-specific CRMs – all fusion cells were affected. In addition, none of the deletions resulted in temporal differences of expression: in each case expression was affected similarly from stages 12–17. Thus, while there may exist subtle differences regarding how different fusion cell types are regulated by dys B2.7, at a gross level the current data suggest that the same CRM responds to the same or similar transcription factors in each fusion cell type.
3.2. Analysis of CG13196 fusion cell expression reveals the existence of a Dys:Tgo coactivator
The CG13196 gene is expressed in all tracheal fusion cells, and is directly regulated by Dys:Tgo. The A985 bp fragment of CG13196 is sufficient for expression in fusion cells, and contains 3 TCGTG Dys:Tgo binding sites. Previously, it was demonstrated that mutation of all 3 sites together resulted in loss of fusion cell expression (Jiang and Crews, 2007). In this paper, we individually deleted each site, and showed that TCGTG-A is not required for fusion cell expression, but both TCGTG-B and TCGTG-C are. Furthermore, bioinformatic analysis revealed the presence of two related sequence motifs. When these sequences (referred to as Motif 1) were deleted together, there was a complete absence of expression in all fusion cells. This suggests that an additional transcription factor, possibly binding to the Motif 1 sequences, acts as a transcriptional coactivator with Dys:Tgo. Further evidence for an additional coactivator in the CG13196 CRM came from analysis of a hybrid CRM in which the CG15252 CRM that is expressed in only DT fusion cells was fused to CG13196 C350 with both TCGTG sequences deleted. This chimeric CRM was expressed in all fusion cells, indicating that sequences in CG13196 C350 (unknown, but possibly Motif 1) in addition to the Dys:Tgo binding sites are important for driving expression in all fusion cells. Most likely, the CG13196 ΔTCGTG fusion cell CRM requires the CG15252 TCGTG sequences for proper fusion cell expression.
3.3. CG15252 is a direct target of Dys:Tgo
The CG15252 gene is expressed in only the DT fusion cells, and genetically requires dys. The A818 fragment has 4 TCGTG sequences. Deletion of TCGTG-A and TCGTG-B had no effect on expression. However, deletion of TCGTG-C and TCGTG-D resulted in an absence of fusion cell expression, indicating that CG15252 is a direct target of Dys:Tgo. In both in vitro DNA binding and S2 cell transient transfection experiments, Dys:Tgo was able to bind to and activate transcription from ACGTG, GCGTG, and TCGTG sequences (Jiang and Crews, 2007). However, analysis of CG13196 indicated that only the TCGTG motif was required for expression in vivo (Jiang and Crews, 2007). The CG15252 C100 fragment has two TCGTG sequences shown to be important for fusion cell transcription, and no ACGTG or GCGTG sequences are present. Similarly, the T523 putative dys autoregulatory element has 5 TCGTG motifs but no ACGTG or GCGTG motifs. Thus, in vivo evidence to date indicates that Dys:Tgo activates transcription via TCGTG, and not other NCGTG sequences.
It was noted that both CG15252 TCGTG sequences were preceded by AAG, as was one of the two CG13196 TCGTG sequences, and two of five dys T523 TCGTGs. One hypothesis tested was that this variant AAGTCGTG motif may have special significance regarding fusion cell expression. However, deletion of the AAG sequences in both CG13196 and CG15252 had no effect on expression. Thus, the core TCGTG sequence is critical, but the precise sequence of flanking residues less so. This is similar to results observed with other bHLH-PAS proteins (Single-minded, Trh, Hypoxia Inducible Factor-1α/Sima, Aryl hydrocarbon receptor/Spineless) that form heterodimers with Tgo/Arnt, in which a core NCGTG sequence is invariant but flanking residues much less conserved (Semenza et al., 1996; Swanson et al., 1995; Wharton et al., 1994). We identified a repeated motif (Motif 2 in C100) in CG15252, but deletion of both sites together had no effect on expression.
One of the major issues is why CG15252 is expressed in only DT. One model is that Dys:Tgo binding sites are sufficient for only DT expression, and additional expression in all fusion cells requires binding sites for another activator: thus CG13196 would possess these additional coactivator binding sites and CG15252 would not. This seems unlikely since multimerization of only TCGTGs results in expression in all fusion cells (Jiang and Crews, 2007). While different arrangements of TCGTG sequences between CG13196 and CG15252 are a possibility, both genes have a relatively similar arrangement: two required TCGTG sequences within 40 bp of each other. Another possibility is that the CG15252 CRM contains a repressor of DB, LT, and GB, so that in combination with multiple Dys:Tgo binding sites, restriction to DT occurs. There are no data that contradict this model, but no data to support it. The Motif 2 sequences are unlikely to be corepressor binding sites, since their deletion had no effect on expression, but other, untested sequences in CG15252 may carry-out this role. When the CG15252 C100 fragment with deleted TCGTG sequences was fused to CG13196 C350, the hybrid fragment was able to drive expression in all fusion cells. This indicated that if a repressor sequence exists on C100 it is unable to repress transcription of the CG13196 fragment. Another model is that Dys:Tgo is able to activate transcription in fusion cells only in the presence of specific coactivators. In this case, CG13196 would include an activator present in all fusion cells, which is consistent with the Motif 1 sequences. CG15252 would require a coactivator present or functional in only DT cells. If so, we have not yet identified sites corresponding to this coactivator. We note that the knirps, knirps-like (also knirps-related), and spalt genes play important roles in defining tracheal branch-specific gene regulation (Chen et al., 1998; Franch-Marro and Casanova, 2002; Kuhnlein and Schuh, 1996; Ohshiro et al., 2002), and it is possible that these genes either indirectly or directly play a role in CG15252 DT-specific expression.
3.4. Search for a fusion cell regulatory code
One of the major goals of the research described in this paper is to begin to identify the CRMs and corresponding transcription factors that govern fusion cell gene regulation. Since fusion cells undergo a variety of developmental changes, an important aspect of mechanistically understanding how these changes occur concerns understanding transcriptional control. The dys gene plays an important role in controlling fusion cell development. One of the key results emerging from the analysis of the dys gene is that its expression requires multiple inputs. In particular, the finding of an essential ETS binding site provides unexpected evidence that direct input from the branchless/breathless/pointed signaling pathway may be required for dys expression. We have identified three direct targets of dys action, including dys itself as an autoregulatory target, CG13196, and CG15252. Each of these genes possesses multiple binding sites for Dys:Tgo. However, searching the genome for two TCGTG sites within 40 bp (the arrangement for CG13196 and CG15252) yields 6,673 hits (Markstein et al., 2002), which is unlikely to be helpful in identifying fusion-expressed genes. We have also provided evidence for an additional coactivator required for expression of CG13196 in all fusion cells, possibly corresponding to Motif 1. However, we currently do not know the transcription factor corresponding to this coactivator, or whether similar sequences are present in other target genes of dys expressed in all fusion cells, including the dys autoregulatory element. Another important issue, unresolved in this paper, concerns how branch-specific fusion cell transcription is regulated. The answers to this issue and others will require molecular, genetic, and biochemical analyses of additional fusion cell-expressed genes.
4. Materials and methods
4.1. Mutant Drosophila strains
The null mutant strains of Drosophila utilized were: dys1 (Jiang and Crews, 2006), esgG66 (Samakovlis et al., 1996), and pntΔ88 (Scholz et al., 1993).
4.2. Construction of the pMintGate ΦC31 transformation vector
The Drosophila Gateway-compatible ΦC31 transformation vector, pBPGw (Pfeiffer et al., 2008), was digested with BglII and SpeI to excise the Gal4 reporter gene. pH-Stinger-GFP (Barolo et al., 2000) was digested with BglII and SpeI, and a fragment containing an Hsp70 minimal promoter, GFP coding sequence with nuclear localization sequence, 3′-UTR, and poly(A) addition site was ligated into pBPGw. This vector, named pMintGate, is ΦC31-compatible with a nuclear-GFP reporter and Gateway sites for cloning fragments adjacent to the Hsp70 promoter (Supplementary Fig. 1).
4.3. Generation of transgenic strains
dys transgenic enhancer tester strains
DNA fragments containing sequences from the dys gene 5′-flanking region and introns 1 and 2 of the dys gene were PCR-amplified, and initially cloned into pCR8/GW/TOPO (Invitrogen) or pGEM-T Easy (Promega). The primer pairs employed for PCR are shown in Supplementary Table 1. The inserts in pCR8 were then cloned into pMintGate using Gateway LR Clonase II plus (Invitrogen). The inserts in pGEM-T Easy were removed by restriction digestion with KpnI and BamHI, and then ligated into the KpnI and BamHI sites of pH-Stinger. Each pMintGate construct was microinjected into Drosophila embryos that express germline-localized φC31 integrase and contain the φC31 genomic destination site attP2 (68A1-B2) (Groth et al., 2004). The pH-Stinger constructs were introduced into germline DNA by microinjection using standard techniques. Three independent lines of each transgene were analyzed for embryonic GFP expression.
dys deletion strains
The dys P155 fragment, which can drive tracheal fusion cell expression, was used to delete conserved sequences for testing functional significance in vivo. The following individual deletions were generated using QuickChange II XL site-mutagenesis kit (Stratagene): Δ1 (GATCGG); Δ2 (TTAAATCAAA); Δ3 (GCGCG); Δ4 (TGCCTGAC); Δ5 (ACTTCCGC); Δ6 (GACCCAAAAC); and Δ7 (TTAATATTCA). The dys V345 TCGTG-A and TCGTG-C sequences were deleted together. The altered fragments were cloned into pMintGate for introduction at attP2 (68A1-B2).
CG13196 transgenic strains
DNA fragments containing 5′-flanking sequences of CG13196 were PCR-amplified using the primers shown in Supplementary Table 1. CG13196 D92, which drives expression in all fusion cells, contains two TCGTG sites, TCGTG-B and TCGTG-C, two instances of Motif 1 (CCATGGAAAGT and CCATTAAAAGT), and one AAG site adjacent to TCGTG-C. Four mutant constructs were generated in which each TCGTG site was individually deleted or individually mutated to CAATG. The AAG site was individually deleted, and the two Motif 1 sites were deleted together. Mutated fragments were cloned into pMintGate and integrated at attP2 (68A1-B2).
CG15252 transgenic strains
The DNA fragments containing 5′-flanking sequences of CG15252 were PCR-amplified using the primers indicated in Supplementary Table 1. The CG15252 C100 fragment, which drives DT expression, was used to test the function of motifs. CG15252 C100 has two TCGTG sequences, TCGTG-C and TCGTG-D, and each was individually deleted and individually mutated to CAATG. The two Motif 2 sites present in C100 were deleted together, and the two AAG sites adjacent to both TCGTGs were deleted together.
Chimeric CG13196-CG15252 transgenic strains. CG13196-C350ΔBC:CG15252-C100
CG15252 C100 was cloned into pCR8/GW/TOPO using TA cloning. CG13196 C350 with deleted TCGTG sites was PCR-amplified using: AAGCTTTGGCAAGTGATTTGTGGGACA and AAGCTTGATTGGGCCGCAAGTGATATG (HindIII sites are underlined), and this fragment was cloned into the Hind III site of pCR8/GW/TOPO-CG15252-C100 to yield CG13196-C350ΔBC:CG15252-C100. CG15252-C100ΔCD:CG13196-C350. This construct was generated similar to CG13196-C350ΔBC:CG15252-C100, except that C350 was first cloned into pCR8/GW/TOPO and C100 with deleted TCGTGs added.
4.4 Immunostaining of embryos
Whole-mount embryos were immunostained using standard techniques (Patel et al., 1987). The following antibodies were used for immunostaining: rat anti-Dys (1:200), rabbit anti-GFP (1:1000; Abcam), Alexa Fluor 488-labeled anti-rabbit IgG (1:200; Invitrogen), and Cy3-labeled anti-rat IgG (1:200; PerkinElmer).
4.5 Bioinformatic analysis of regulatory regions
For each minimal fusion cell-expressed fragment (dys P155, dys T523, CG13196 D92, CG15252 C100), orthologous sequences from the 12 sequenced drosophilids were retrieved using the UCSC genome browser, and aligned by T-Coffee (Notredame et al., 2000). Local sub-alignments were hand-corrected using Lalign (Huang and Miller, 1991) to identify short regions of homology. Statistically over-represented motifs were identified within and between co-expressed fusion cell CRMs using PhyloGibbs (Siddharthan et al., 2005; Siddharthan, 2008). WinDotter (Sonnhammer and Durbin, 1995) was used to search for repeated motifs and palindromes within each enhancer sequence. Identified motifs were compared to consensus binding sites for known fusion cell-specific transcription factors and to matrices in the JASPAR and TRANSFAC databases using the TESS motif search program (Schug, 2008). GenePalette (Rebeiz and Posakony, 2004) was used to annotate genomic DNA sequences and to visualize putative transcription factor binding site locations within each fragment. FlyEnhancer (Markstein et al., 2002) was used to search for clusters of Dys:Tgo consensus sites in the D. melanogaster genome.
Supplementary Material
Supplementary Fig. 1 – pMintGate vector. This derivative of pBPGw contains a Gateway AttR destination site for cloning inserts, Hsp70 promoter, nuclear GFP reporter, mini-white, ΦC31 attP site for integration into the Drosophila genome, and ApR ampicllin resistance gene. The CmR and ccdB genes are components of the Gateway cloning system.
Supplementary Fig. 2 – Temporal dynamics of GFP expression driven by various fusion cell enhancers. Embryonic stages (S11–16) are indicated at top. The fusion cell branch imaged (DT or LT) is indicated at right adjacent to the gene and fragment shown. (A–I) The dys P155 fragment, which is derived from B2.7, drove expression in DT from stages 12–16 (A–E) and LT from stages 14–16 (F–I). This mimics endogenous dys, which is expressed in DT before LT. (J–O) The dys T523 fragment, derived from C1.7, drove expression in DT from stages 14–16 (J–L), and in LT from stages 15–16 (M–O). (P–T) The CG13196 fragment D92 fragment drove DT expression from stages 15–16 (P–R) and LT expression at stage 16 (S–T). (U–V) The CG15252 C100 fragment drove expression in DT at stage 16.
Supplementary Fig. 3 – The dys P155 fragment was aligned for 12 Drosophila species (see http://flybase.org/blast for species names and information). The sequence positions are shown at left. Each nucleotide type is colored: A (green), C (blue), G (black), and T (red). If a residue is conserved in >50% of the species it is depicted as a colored box. Seven conserved sequences were deleted by in vitro mutagenesis (Δ1–7), and tested for fusion cell expression in vivo. The deleted sequences are indicated by dashes bounded by vertical lines. The endpoints of the dys L156, K152, and Q112 fragments are indicated.
Supplementary Fig. 4 – Fusion cell expression of dys PT523 and P155 fragments is altered in esg mutant embryos. The dys (A) P155 and (C) T523 transgenes in wild-type (WT) backgrounds express GFP in all fusion cells (DT and LT are shown), whereas embryos mutant for esgG66 (B,D) show expression only in DT fusion cells, but not DB, LT, or GB. This is identical to dys expression in esg mutant embryos (Jiang and Crews, 2003).
Supplementary Fig. 5 – The dys T523 fragment was aligned for 12 Drosophila species. The positions of 5 Dys:Tgo sites are indicated, and two of the sites have an adjacent AAG sequence. The endpoint of the dys V345 fragment is indicated.
Supplementary Fig. 6 – The CG13196 D92 fragment was aligned for 12 Drosophila species and regions of conservation indicated. Two Dys:Tgo sites were indicated and one has an adjacent AAG sequence. Motif 1 is a sequence motif repeated twice in D92. The vertical lines indicate the boundaries of the region deleted by in vitro mutagenesis and tested in vivo. The dashes outside the vertical lines indicate the full extent of the conserved motifs.
Supplementary Fig. 7 – The CG15252 C100 fragment was aligned for 5 Drosophila species; no related sequences were found in the other 7 species examined. Dys:Tgo sites are indicated, and both have an adjacent 5′ AAG sequence. Motif 2 is a sequence motif repeated twice in C100, and tested by deletional analysis.
Acknowledgments
This project was funded by grants from the National Science Foundation (Developmental Mechanisms), NICHD, and the National Center for Research Resources (National Institutes of Health) to STC. The UNC Developmental Biology NIH training grant provided support to JCP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson MG, Perkins GL, Chittick P, Shrigley RJ, Johnson WA. drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 1995;9:123–137. doi: 10.1101/gad.9.1.123. [DOI] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Boube M, Llimargas M, Casanova J. Cross-regulatory interactions among tracheal genes support a co-operative model for the induction of tracheal fates in the Drosophila embryo. Mech Dev. 2000;91:271–278. doi: 10.1016/s0925-4773(99)00315-9. [DOI] [PubMed] [Google Scholar]
- Chen CK, Kuhnlein RP, Eulenberg KG, Vincent S, Affolter M, Schuh R. The transcription factors KNIRPS and KNIRPS RELATED control cell migration and branch morphogenesis during Drosophila tracheal development. Development. 1998;125:4959–68. doi: 10.1242/dev.125.24.4959. [DOI] [PubMed] [Google Scholar]
- Crews ST, Pearson JC. Transcriptional autoregulation in development. Curr Biol. 2009;19:R241–6. doi: 10.1016/j.cub.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch-Marro X, Casanova J. spalt-induced specification of distinct dorsal and ventral domains is required for Drosophila tracheal patterning. Dev Biol. 2002;250:374–82. [PubMed] [Google Scholar]
- Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Affolter M. A family of genes encoding zona pellucida (ZP) domain proteins is expressed in various epithelial tissues during Drosophila embryogenesis. Gene Expr Patterns. 2004;4:413–421. doi: 10.1016/j.modgep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Jiang L, Crews ST. The Drosophila dysfusion basic helix-loop-helix (bHLH)-PAS gene controls tracheal fusion and levels of the trachealess bHLH-PAS protein. Mol Cell Biol. 2003;23:5625–5637. doi: 10.1128/MCB.23.16.5625-5637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Crews ST. Dysfusion transcriptional control of Drosophila tracheal migration, adhesion, and fusion. Mol Cell Biol. 2006;26:6547–6556. doi: 10.1128/MCB.00284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Crews ST. Transcriptional specificity of Drosophila dysfusion and the control of tracheal fusion cell gene expression. J Biol Chem. 2007;282:28659–28668. doi: 10.1074/jbc.M703803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnlein RP, Schuh R. Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development. 1996;122:2215–23. doi: 10.1242/dev.122.7.2215. [DOI] [PubMed] [Google Scholar]
- Lavista-Llanos S, Centanin L, Irisarri M, Russo DM, Gleadle JM, Bocca SN, Muzzopappa M, Ratcliffe PJ, Wappner P. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol Cell Biol. 2002;22:6842–53. doi: 10.1128/MCB.22.19.6842-6853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kolodziej PA. The plakin Short Stop and the RhoA GTPase are required for E-cadherin-dependent apical surface remodeling during tracheal tube fusion. Development. 2002;129:1509–20. doi: 10.1242/dev.129.6.1509. [DOI] [PubMed] [Google Scholar]
- Llimargas M, Casanova J. ventral veinless, a POU domain transcription factor, regulates different transduction pathways required for tracheal branching in Drosophila. Development. 1997;124:3273–3281. doi: 10.1242/dev.124.17.3273. [DOI] [PubMed] [Google Scholar]
- Manning G, Krasnow MA. Development of the Drosophila Tracheal System. In: Bate M, Martinez Arias A, editors. The Development of Drosophila Melanogaster. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 1993. pp. 609–685. [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer NT, Moberg KH. The Drosophila F-box protein Archipelago controls levels of the Trachealess transcription factor in the embryonic tracheal system. Dev Biol. 2007;312:560–571. doi: 10.1016/j.ydbio.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat MM, Lightfoot H, Wang P, Andrew DJ. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev Biol. 2005;281:38–52. doi: 10.1016/j.ydbio.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu JR, Chen W, Hu S, Crews ST. The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human Hypoxia-inducible factor 1αDrosophila Single-minded. Gene. 1996;172:249–254. doi: 10.1016/0378-1119(96)00060-1. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ohshiro T, Emori Y, Saigo K. Ligand-dependent activation of breathless FGF receptor gene in Drosophila developing trachea. Mech Dev. 2002;114:3. doi: 10.1016/s0925-4773(02)00042-4. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol Cell Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooe N, Saito K, Oeda K, Nakatuka I, Kaneko H. Characterization of Drosophila and Caenorhabditis elegans NXF-like-factors, putative homologs of mammalian NXF. Gene. 2007;400:122–130. doi: 10.1016/j.gene.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Posakony JW. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev Biol. 2004;271:431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development. 1996;122:3531–6. doi: 10.1242/dev.122.11.3531. [DOI] [PubMed] [Google Scholar]
- Scholz H, Deatrick J, Klaes A, Klambt C. Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics. 1993;135:455–468. doi: 10.1093/genetics/135.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008;Chapter 2(Unit 2.6) doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- Siddharthan R. PhyloGibbs-MP: module prediction and discriminative motif-finding by Gibbs sampling. PLoS Comput Biol. 2008;4:e1000156.1–e1000156.15. doi: 10.1371/journal.pcbi.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharthan R, Siggia ED, van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput Biol. 2005;1:e67.0534–e67.0556. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4583–4594. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development. 1996;122:3697–705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- Wharton JKA, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- Wilk R, Weizman I, Glazer L, Shilo B. trachealess encodes a bHLH-PAS protein and is a master regulator gene in the Drosophila tracheal system. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Shilo B. Interaction between the bHLH-PAS protein Trachealess and the POU-domain protein Drifter, specifies tracheal cell fates. Mech Dev. 2000;19:163–173. doi: 10.1016/s0925-4773(99)00295-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 – pMintGate vector. This derivative of pBPGw contains a Gateway AttR destination site for cloning inserts, Hsp70 promoter, nuclear GFP reporter, mini-white, ΦC31 attP site for integration into the Drosophila genome, and ApR ampicllin resistance gene. The CmR and ccdB genes are components of the Gateway cloning system.
Supplementary Fig. 2 – Temporal dynamics of GFP expression driven by various fusion cell enhancers. Embryonic stages (S11–16) are indicated at top. The fusion cell branch imaged (DT or LT) is indicated at right adjacent to the gene and fragment shown. (A–I) The dys P155 fragment, which is derived from B2.7, drove expression in DT from stages 12–16 (A–E) and LT from stages 14–16 (F–I). This mimics endogenous dys, which is expressed in DT before LT. (J–O) The dys T523 fragment, derived from C1.7, drove expression in DT from stages 14–16 (J–L), and in LT from stages 15–16 (M–O). (P–T) The CG13196 fragment D92 fragment drove DT expression from stages 15–16 (P–R) and LT expression at stage 16 (S–T). (U–V) The CG15252 C100 fragment drove expression in DT at stage 16.
Supplementary Fig. 3 – The dys P155 fragment was aligned for 12 Drosophila species (see http://flybase.org/blast for species names and information). The sequence positions are shown at left. Each nucleotide type is colored: A (green), C (blue), G (black), and T (red). If a residue is conserved in >50% of the species it is depicted as a colored box. Seven conserved sequences were deleted by in vitro mutagenesis (Δ1–7), and tested for fusion cell expression in vivo. The deleted sequences are indicated by dashes bounded by vertical lines. The endpoints of the dys L156, K152, and Q112 fragments are indicated.
Supplementary Fig. 4 – Fusion cell expression of dys PT523 and P155 fragments is altered in esg mutant embryos. The dys (A) P155 and (C) T523 transgenes in wild-type (WT) backgrounds express GFP in all fusion cells (DT and LT are shown), whereas embryos mutant for esgG66 (B,D) show expression only in DT fusion cells, but not DB, LT, or GB. This is identical to dys expression in esg mutant embryos (Jiang and Crews, 2003).
Supplementary Fig. 5 – The dys T523 fragment was aligned for 12 Drosophila species. The positions of 5 Dys:Tgo sites are indicated, and two of the sites have an adjacent AAG sequence. The endpoint of the dys V345 fragment is indicated.
Supplementary Fig. 6 – The CG13196 D92 fragment was aligned for 12 Drosophila species and regions of conservation indicated. Two Dys:Tgo sites were indicated and one has an adjacent AAG sequence. Motif 1 is a sequence motif repeated twice in D92. The vertical lines indicate the boundaries of the region deleted by in vitro mutagenesis and tested in vivo. The dashes outside the vertical lines indicate the full extent of the conserved motifs.
Supplementary Fig. 7 – The CG15252 C100 fragment was aligned for 5 Drosophila species; no related sequences were found in the other 7 species examined. Dys:Tgo sites are indicated, and both have an adjacent 5′ AAG sequence. Motif 2 is a sequence motif repeated twice in C100, and tested by deletional analysis.