Abstract
Functional neuroimaging has evolved into an indispensable tool for noninvasively investigating brain function. A recent development of such methodology is the creation of connectivity models for brain regions and related networks, efforts that have been inhibited by notable limitations. We present a new method for ascertaining functional connectivity of specific brain structures using metaanalytic connectivity modeling (MACM), along with validation of our method using a nonhuman primate database. Drawing from decades of neuroimaging research and spanning multiple behavioral domains, the method overcomes many weaknesses of conventional connectivity analyses and provides a simple, automated alternative to developing accurate and robust models of anatomically‐defined human functional connectivity. Applying MACM to the amygdala, a small structure of the brain with a complex network of connections, we found high coherence with anatomical studies in nonhuman primates as well as human‐based theoretical models of emotive‐cognitive integration, providing evidence for this novel method's utility. Hum Brain Mapp, 2010. © 2009 Wiley‐Liss, Inc.
Keywords: meta‐analysis, brainmap, fMRI, PET, CoCoMac
INTRODUCTION
Functional brain imaging methods, including positron emission tomography (PET) and functional magnetic resonance imaging (fMRI), have been propelling neuroscience research for more than 25 years. A more recent, but highly productive, application of these methods is the use of interregional temporal covariances to develop connectivity models of brain regions and related networks. The popularity of connectivity modeling is not surprising, given the necessity of such information in our understanding of any given neural system. However, no methodological approach to date has taken full advantage of the resources and tools that have been developed and are readily available as the result of decades of neuroimaging research.
Three types of connectivity are readily accepted in the field: structural, functional, and effective. Structural connectivity refers to the organization of axonal fibers within the brain, and has largely been derived from anatomical nonhuman primate data and human‐based diffusion tensor imaging (DTI). Functional connectivity, on the other hand, is aimed at identifying coactivated regions of the brain, and relies heavily on interregion coherence, or correlation. These models account for both direct and indirect neural influence, a significant advantage over models of structural connectivity, which cannot account for the dynamic nature of the brain. Effective connectivity models focus on identifying the flow of information from one brain region to another within a given network, deeming models of functional connectivity, which specify brain regions to be included in such models, critically important for their development. Therefore, accurate and robust models of functional connectivity become a necessary precursor if we are to begin developing empirically driven and realistic representations of how information is processed in the human brain.
Multimodal neuroimaging methods have been the most powerful attempt at delineating functional connectivity of the human brain. Diffusion‐weighted MRI data, a noninvasive procedure that provides information about anatomical connectivity, has been used in conjunction with fMRI data in attempts to discern the relationship between a given brain structure and other brain regions that have an anatomical relationship and exhibit concomitant activity during a specified task [Cohen et al., 2008]. This method is hindered by the limits of diffusion‐weighted imaging (e.g., biased towards highly myelinated structures and confounded by crossing fibers) and relies on the performance of a specific task, making generalizability difficult. Resting state fMRI has been used in efforts to overcome task‐specificity, though these data have been difficult to replicate, likely because of the nature of the acquisition (e.g., low signal‐to‐noise ratio [SNR]). Alternatively, focal, electrical stimulation of the brain (e.g., transcranial magnetic stimulation [TMS] or deep brain stimulation [DBS]) in combination with neuroimaging techniques represents a powerful method in which researchers can examine interregional connectivity in a task‐independent manner. Studies employing such methodology have yielded convincing results, showing high concordance with nonhuman primate data in simple networks such as motor system connectivity [Fox et al., 1997] and frontal eye field connectivity [Paus et al., 1997]. However, the same characteristics that make these techniques useful (i.e., each imaging techniques uniqueness) also limit its applicability. For example, deep brain stimulation in humans is extremely invasive, and is generally limited to individuals undergoing the operation for symptom remission of a neurological or psychiatric condition. TMS allows for noninvasive, discrete, focal changes, though its application is limited to cortical regions. Thus, examining subcortical structures (e.g., amygdalae) and their related networks (e.g., the limbic system) is not currently possible.
Coactivation patterns are becoming an increasingly popular venue for functional connectivity, a strategy pioneered by Friston [ 1994] and refined to include the numerous benefits of metaanalysis by Koski and Paus [ 2000]. The latter created a database spanning 3 years of PET imaging research, and searched the frontal cortex for areas showing parallel increases in blood flow with the anterior cingulate. The approach was novel, although the time commitment and lack of automaticity coupled with the exponentially increasing number of neuroimaging studies in both PET and fMRI limited the possibility of this method becoming a routine approach towards dissociating functional connectivity.
The conceptual framework has since been refined because of resources that have become available over the years (e.g., large neuroimaging databases) which provide immensely rich and immediately‐available datasets (e.g., sets of coordinates) acquired from various behavioral and cognitive domains spanning decades of neuroimaging research. As an example of one such resource's versatility, Toro et al. [ 2008] used the BrainMap [Laird et al., 2005b] database to examine whole‐brain functional connectivity of the human brain using a metaanalytic approach and confirmed the presence of important functional networks, including the fronto‐parietal attention network [Fox et al., 2005], the resting state network [Greicius et al., 2003], and the motor network [Paus et al., 1998]. While whole‐system metaanalyses have been shown to be an elegant approach to establishing highly likely connectivity patterns, efforts to examine the connectivity of a specific brain region have been less elegant. Existing models have been severely limited by task‐specificity [Stein et al., 2007], methodological approaches employed, acquisition limitations, nonanatomically driven regions of interest (ROIs) (e.g., regularly shaped ROIs that are cuboidal or spherical), and single hemisphere analyses. To date, no method exists to examine task‐independent coactivation patterns of any given anatomically‐defined ROI.
Here, we present a novel methodology, metaanalytic connectivity modeling (MACM), which can be used to examine the functional connectivity of a specific brain region by identifying patterns of coactivation across thousands of subjects, an improvement which has implications for generalizability, robustness, and power [Fox et al., 1998]. Though computationally different, the methodological ideas presented by Toro et al. [ 2008] are conceptually similar. We used a probability atlas to define the amygdala in each hemisphere of the human brain, seeded the ROIs within the BrainMap database, and queried all archived neuroimaging papers to determine which had reported activation within the ROIs. Coordinates of activation from these papers were then downloaded, and metaanalytic statistics were computed to determine regions of the brain that were coactivated. The use of metaanalytic techniques, in combination with the advancement of carefully coded databases such as BrainMap [Fox and Lancaster, 2002; Laird et al., 2005b], has been an exciting addition to the neuroscience community that holds great promise for discovering accurate models of functional connectivity. We present a simple, easily adaptable method for structure‐specific functional connectivity modeling and provide proof of concept using one of the most complex and dynamic structures of the human brain: the amygdala. We also provide validation of our method using the largest searchable nonhuman primate literature database, CoCoMac [Stephan et al., 2001] in combination with recently developed connectivity and visualization tools [Kotter, 2004].
Functional Connectivity of the Amygdala
The limbic system is evolutionarily advantageous as it is responsible for emotional processing and the evaluation of threatening or dangerous elements in the environment [Amaral and Price, 1984; LeDoux, 2007]. The critical brain structure driving these processes is the amygdala, which has been under scientific scrutiny for decades. A PubMed search on “amygdala” yields 20,760 papers. This is not surprising given the amygdala's diversity of involvement in a wide range of cognitive and behavioral tasks including fear conditioning [Adolphs et al., 2005], memory formation [Packard and Cahill, 2001; Phelps, 2004] and learning [Phelps et al., 2004], social processing [Anderson and Phelps, 2000; Hariri et al., 2002], and affective processing including recognition and regulation of emotions [Goldin et al., 2008; Ochsner et al., 2004]. In addition to the role it plays in healthy behavioral processes, aberrant amygdalar function is implicated in a host of psychiatric disorders [Abercrombie et al., 1998; Davidson et al., 1999], as well as in populations that are genetically at risk for such illnesses [Glahn et al., 2007; Hariri et al., 2005]. Improving our understanding of amygdala function and how this region interacts with other brain regions has the potential to significantly advance our understanding of healthy and aberrant cognitive and affective processing.
Amygdalae do not work in isolation, but rather are thought to serve as a node within multiple neural networks [Papez, 1995; Pessoa, 2008]. While anatomical connectivity of the amygdala has been largely elucidated with the assistance of nonhuman primate studies [Amaral and Price, 1984; Barbas and De Olmos, 1990; Ghashghaei and Barbas, 2002], delineating the functional connectivity of the amygdala in humans has been a challenge plagued by limitations that, until recently, have been impossible to overcome. With the advent of noninvasive neuroimaging techniques, such as fMRI, a surge of research was directed towards this aim [Cohen et al., 2008; Lawrie et al., 2003; Roy et al., 2009; Stein et al., 2007]. Despite the technological advances, generalizability remains constrained due to the methodologies (e.g., resting‐state fMRI which has low SNR, task‐specific fMRI which is not generalizable, or DTI which is biased to heavily myelinated regions and fails where fibers cross) being employed. Thus, empirical evidence is lacking as to how the amygdalae are functionally connected to the rest of the brain, allowing for their versatility and necessity in healthy neural functioning.
METHODS
MACM was employed to assess amygdala functional connectivity. Below, we describe methods for ROI selection as well as employing MACM using the amygdala as an example.
ROI Selection
Bilateral amygdala ROIs were defined using the Harvard‐Oxford Structural Probability Atlas (thresholded at 70% probability; Fig. 1) distributed with FSL neuroimaging analysis software (http://www.fmrib.ox.ac.uk/fsl/fslview/atlas-descriptions.html#ho) [Smith et al., 2004]. Using anatomically driven (i.e., irregular) ROIs represents an improvement over current connectivity (and other neuroimaging) studies which typically use regular (i.e., spherical or cuboidal) ROIs [Stein et al., 2007], ROIs derived from functional activations within a given study [Gianaros et al., 2008; Mohanty et al., 2007], or use less developed (i.e., based on a single brain) automatic labeling systems [Tzourio‐Mazoyer et al., 2002; Williams et al., 2006]. Using probabilistic, anatomically defined regions eliminates differences in placement of the geometrically or functionally driven ROIs, and allows for consistency across studies. This issue becomes critically important with brain structures that are large and heterogeneous in nature (e.g., cingulate cortex). Utilizing a probability atlas in which partitions of such structures are better defined (e.g., anterior versus posterior cingulate) will undoubtedly improve future research. An additional benefit that is more pertinent to the current study is the assurance that the ROI captures and confines the brain structure of interest, and can easily be described to the neuroimaging community. In the case of the amygdala, the mean probability for the left (M ± SD: 84.85% ± 6.76%) and right amygdala (87.11% ± 7.46%) was over 80%, and the centroid for each was over 90% (left: 94% at Talairach coordinates [x,y,z] −21.5, −6.0, −14.6; right: 98% at Talairach coordinates 23.0, −4.9, −14.9). The total volume for the left amygdala was 469 voxels, and for the right 644 voxels. Centroid, average threshold, and volume statistics were gathered using Mango's ROI and histogram capabilities (http://ric.uthscsa.edu/mango).
Figure 1.
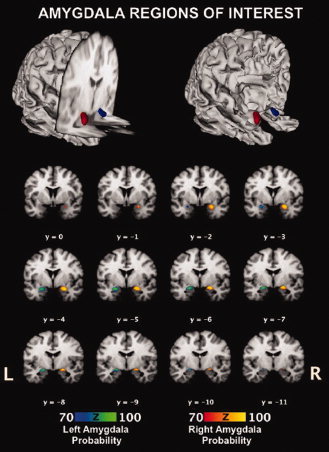
Anatomical 3D‐renderings of the amygdala ROIs used for the meta‐analysis. The right amygdala is represented in red and the left amygdala is represented in blue. Figure created using Mango (http://ric.uthscsa.edu/mango).
BrainMap Metaanalysis Methods
The left and right amygdala ROIs were input into the BrainMap database separately, to search for all studies that reported activation within each ROI boundary. Whole‐brain coordinates of activations from the isolated contrasts were then downloaded (left amygdala = 124 papers, 170 contrasts, 2,077 locations; right amygdala = 116 papers, 156 contrasts, 1,765 locations). The total number of subjects in all studies reporting activation in the left amygdala was 1,873, and for the right amygdala 2,140. Papers were drawn from multiple behavioral domains including cognition, emotion, action, and perception (Fig. 2B), with emotion representing the majority of studies followed by cognition for both the left and right amygdala. Within the emotion domain, a variety of affective states were represented (Fig. 2C).
Figure 2.
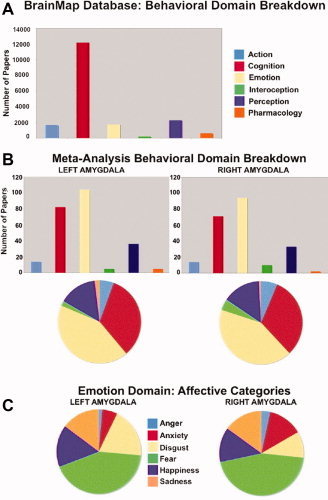
Papers reporting activation within the left and right amygdala ROIs were drawn from the BrainMap Database for subsequent metaanalysis. The graphs below demonstrate the number of papers coded in each behavioral domain in the entire BrainMap database (a), as well as the behavioral domain breakdown of the papers included in the amygdala‐specific meta‐analysis (b). From the emotion domain, which contained the highest percentage of papers, a variety of affective states were represented (c).
Activation likelihood estimation (ALE) metaanalyses [Laird et al., 2005a; Turkeltaub et al., 2002] were performed on the sets of coordinates identified as coactivated during left and right amygdala activation, to identify regions of convergence. ALE capitalizes on the nature of voxel‐wise studies that are commonly reported in a standard stereotaxic space (x, y, z) by pooling 3D coordinates from like studies, and providing the probability of an event occurring at each brain voxel. Resultant ALE maps from the present study were thresholded conservatively (P < 0.001, corrected for multiple comparisons and false discovery rate). The method of seeding an anatomically driven ROI and performing ALE metaanalysis will be referred to as metaanalytic connectivity modeling (MACM).
Nonhuman Primate Connectivity Analysis
We used the CoCoMac‐Paxinos3D Viewer (http://134.95.56.239/WWW/paxinos3D/index.html) to query the CoCoMac [Stephan et al., 2001] database for any afferent or efferent connections to the nonhuman primate amygdala. The CoCoMac‐Paxinos3D Viewer uses manually drawn cortical, striatal, thalamic, and amygdaloid structures from the “Rhesus Monkey Brain in Stereotactic Coordinates” by Paxinos et al. [ 2000]. The program allows an individual to select a brain structure from any of the 151 slices of the monkey brain. We chose every amygdaloid structure in the atlas at the present time, which included the following: amygdalohippocampal area (magnocellular and posteriomedial parts), anterior amygdaloid area, amygdaloid intramedullary gray area, amygdalopiriform transition area, the central amygdaloid nucleus (lateral and medial divisions), the medial amygdaloid nucleus, the paralaminar amygdaloid nucleus, the ventral amygdaloid nucleus, the basolateral amygdaloid nucleus, and the basomedial amygdaloid nucleus.
RESULTS
We found strong support for functional connectivity of both left and right amygdala to regions of the left posterior cingulate (BA23), anterior cingulate (BA32), inferior (BA47) and medial frontal (BA9) gyri, and thalamus (Table I). Amygdalae showed convergence in bilateral culmen, insula, parahippocampal gyri (i.e., each amygdala ROI is functionally connected to the contralateral amygdala), and right middle temporal gyrus (BA37). While significant overlap was observed for the two ROIs (Figs. 3 and 4), some hemisphere‐specific effects emerged from the cluster analysis. Specifically, the left amygdala was functionally connected to a unique region of the anterior cingulate (BA32), right occipital gyri (BA19), left fusiform gyrus (BA 19/37), and left middle temporal gyrus (BA 21/39), while results for the right amygdala demonstrated unique functional connectivity patterns to the right inferior frontal gyrus (BA9), precuneus (BA31), thalamus, and right caudate (Table I; Fig. 3).
Table I.
Clusters shared for both left and right amygdala connectivity analyses
| Left amygdala connectivity clusters | Right amygdala connectivity clusters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lobe | Region | BA | ALE | x | y | z | ALE | x | y | z |
| Anterior | Left culmen | 0.052 | −36 | −42 | −18 | 0.035 | −32 | −34 | −20 | |
| Anterior | Right culmen | 0.033 | 36 | −46 | −18 | 0.031 | 36 | −40 | −20 | |
| Frontal | Left inferior frontal gyrus | 47 | 0.038 | −44 | 26 | 0 | 0.029 | −44 | 26 | 0 |
| 0.027 | −34 | 26 | −12 | |||||||
| Frontal | Left medial frontal gyrus | 9 | 0.034 | −6 | 16 | 44 | 0.025 | −6 | 48 | 22 |
| Limbic | Left anterior cingulate | 32 | 0.047 | −2 | 32 | −6 | 0.028 | 0 | 46 | 4 |
| 24 | 0.041 | −2 | 32 | 0 | ||||||
| Limbic | Left parahippocampal gyrus | Amygdala | 0.314 | −22 | −6 | −14 | 0.144 | −22 | −6 | −14 |
| Limbic | Right parahippocampal gyrus | Amygdala | 0.137 | 22 | −4 | −14 | 0.255 | 22 | −6 | −14 |
| Limbic | Left posterior cingulate | 23 | 0.036 | −2 | −52 | 22 | 0.038 | −4 | −54 | 20 |
| Sub‐lobar | Left thalamus | 0.037 | −6 | −12 | 8 | 0.035 | −8 | −14 | 4 | |
| Sub‐lobar | Left insula | 13 | 0.042 | −34 | 16 | 10 | 0.038 | −32 | 14 | 4 |
| Sub‐lobar | Right insula | 13 | 0.036 | 38 | 20 | 2 | 0.039 | 36 | 16 | 0 |
| Temporal | Right middle temporal gyrus | 37 | 0.032 | 44 | −68 | 16 | 0.027 | 46 | −62 | −2 |
| 0.036 | 52 | −62 | 6 | |||||||
| Hemisphere specific clusters | ||||||||||
| Frontal | Left superior frontal gyrus | 9 | 0.037 | −4 | 52 | 22 | ||||
| Limbic | Right anterior cingulate | 32 | 0.031 | 2 | 46 | 2 | ||||
| Occipital | Left fusiform gyrus | 19 | 0.043 | −40 | −66 | −14 | ||||
| Occipital | Right inferior occipital gyrus | 19 | 0.034 | 40 | −74 | −8 | ||||
| Occipital | Right middle occipital gyrus | 19 | 0.027 | 34 | −78 | 12 | ||||
| Temporal | Left fusiform gyrus | 37 | 0.040 | −42 | −58 | −14 | ||||
| Temporal | Left middle temporal gyrus | 21 | 0.028 | −56 | −56 | 4 | ||||
| Temporal | Left middle temporal gyrus | 39 | 0.037 | −44 | −66 | 18 | ||||
| Frontal | Right inferior frontal gyrus | 9 | 0.030 | 44 | 8 | 30 | ||||
| Parietal | Left precuneus | 31 | 0.038 | −2 | −46 | 30 | ||||
| Sub‐lobar | Left thalamus | 0.029 | 0 | −4 | 10 | |||||
| Sub‐lobar | Right caudate | 0.028 | 8 | 4 | 10 | |||||
Local maxima of clusters for the left and right amygdala meta‐analysis (P < 0.001). Coordinates are reported in Talairach space.
Figure 3.
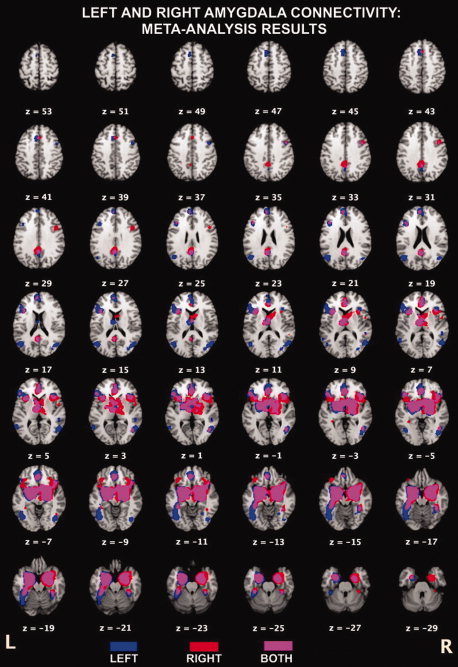
Coactivation patterns obtained from meta‐analysis with the left (blue) and right (red) amygdala. Regions of the brain that coactivated with both left and right amygdala are indicated in purple. Maps thresholded at P < 0.001. Figure created using Mango (http://ric.uthscsa.edu/mango).
Figure 4.
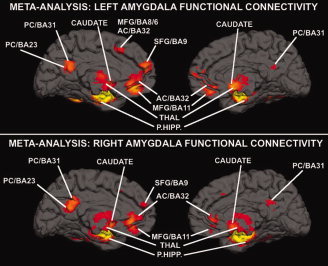
The 3D renderings of coactivation patterns for the left (top panel) and right (bottom panel) amygdala. AC = anterior cingulate; BA = Brodmann area; MFG = medial frontal gyrus; PC = posterior cingulate cortex; PHIPP = parahippocampal gyrus; SFG = superior frontal gyrus; THAL = thalamus.
Nonhuman Primate Results
Analysis of the nonhuman primate literature revealed efferent connections from the amygdala to the entorhinal cortex, the mediodorsal thalamic nucleus, temporal area TE (occipital part), and cortex regions 6, 11, 13, 24, 25, 32, 35, and 46 (Table II; visually presented in Fig. 5). Strong connections were anatomically supported in cortex Regions 24 and 25. Afferent connections were less prevalent with the only evidence emerging from projections originating in temporal area TF projecting to the central medial amygdaloid nucleus (Fig. 5; Panel F).
Table II.
Results of the CoCoMac‐Paxinos3D viewer connectivity queries
| Region | Anterior amygdaloid area | Amygdalopiriform transition area | Basolateral amygdaloid nucleus | Basomedial amygdaloid nucleus | Central medial amygdaloid nucleus | Net result |
|---|---|---|---|---|---|---|
| Afferent connections | ||||||
| Pul | − | − | 0 | 0 | − | No |
| Re | − | − | 0 | 0 | − | No |
| TF | − | − | − | − | − | Yes; unknown |
| 23 | 0 | − | 0 | 0 | − | No |
| 24 | 0 | − | 0 | 0 | − | No |
| 32 | 0 | − | − | 0 | − | No |
| Efferent connections | ||||||
| AD | − | − | 0 | 0 | − | No |
| AL | − | − | 0 | 0 | − | No |
| AM | − | − | 0 | 0 | − | No |
| AV | − | − | 0 | 0 | − | No |
| B | − | − | 0 | 0 | − | No |
| CM | − | − | 0 | − | − | No |
| EC | − | − | 1 | − | − | Weak |
| MD | − | − | − | X | 0 | Yes; unknown |
| Pall | − | − | 0 | 0 | − | No |
| PC | − | − | 0 | − | − | No |
| Pul | − | − | 0 | 0 | − | No |
| R | − | − | 0 | 0 | − | No |
| SG | − | − | 0 | 0 | − | No |
| TEO | − | − | 1 | − | − | Weak |
| TF | − | − | − | − | − | No |
| TH | − | − | 0 | − | − | No |
| VP | − | X | 0 | 0 | − | No |
| 1 | − | − | 0 | − | 0 | No |
| 2 | − | X | 0 | − | 0 | No |
| 4 | − | − | 0 | − | − | No |
| 6 | − | − | X | 0 | − | Yes; unknown |
| 8A | − | − | 0 | 0 | − | No |
| 10 | − | − | 0 | 0 | 0 | No |
| 11 | − | 0 | 1 | X | 0 | Weak/unknown |
| 11m | − | 0 | − | − | 0 | No |
| 13 | − | − | 1 | X | 0 | Weak/unknown |
| 13a | − | − | 0 | − | − | No |
| 23 | − | − | 0 | − | − | No |
| 24 | − | − | 3 | − | − | Strong |
| 24b | − | − | − | − | − | No |
| 25 | − | X | 3 | X | − | Strong |
| 32 | − | X | X | X | − | Yes; unknown |
| 35 | − | X | 0 | X | − | Yes; unknown |
| 45 | − | 0 | 0 | 0 | − | No |
| 46 | − | 0 | X | 0 | − | Yes; unknown |
Twelve amydaloid regions were included in the analysis, but only those that had results are presented. A full list of all abbreviations can be found in Table 4. 0 = Evidence for no direct anatomical connectivity, 1 = Weak anatomical connectivity, 3 = Strong anatomical connectivity, − = No data presented for this region in the query, X = Connection of unknown density. All connectivity definitions are described fully in CocoMac.
Figure 5.
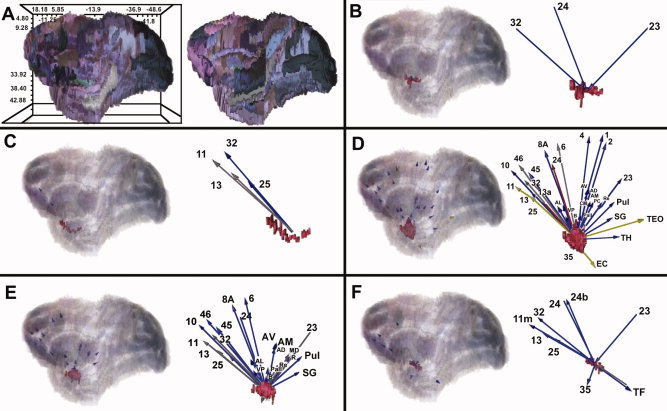
Images were constructed using the CoCoMac‐Paxinos3D Viewer. Panel A shows all 151 slices of the atlas in stereotaxic space. Within each of the panels B–F, the brain region is displayed in this space (left side of each panel) in addition to a magnified image of the connectivity results. Please see Table IV for a list of all abbreviations. B = Anterior amygdaloid area, C = Amygdalopiriform transition area, D = Basolateral amygdaloid nucleus, E = Basomedial amygdaloid nucleus, F = Central medial amygdaloid nucleus, medial division. Blue arrows indicate evidence for no anatomical connectivity, gray represents anatomical connectivity of unknown density, yellow is weak anatomical connectivity, and red indicates strong anatomical connections.
Compared to the human‐based MACM results, there was high concordance among many key regions of the limbic system (Table III; please see Table IV for abbreviations used in nonhuman primate literature). Evidence of connectivity with the prefrontal and orbitofrontal cortices, anterior cingulate, insula, and thalamus are highly consistent in the nonhuman primate anatomical connectivity literature as well as the human limbic system literature. Some notable differences emerged, particularly with regard to the posterior cingulate, the parahippocampus, the culmen, and the caudate nucleus. These regions were found to be functionally connected in our MACM model, despite evidence against, or no data supporting, their existence in the nonhuman primate literature. Some, but not all of these structures, were also found to be functionally connected to the amygdala in the resting state [Roy et al., 2009]. This highlights the utility of MACM and the robustness of the method in capturing the dynamic interactions between a particular region and other brain areas that may not be anatomically supported, but are heavily involved through indirect neural influences.
Table III.
Comparison between human‐based MACM results and the nonhuman primate literature as queried with the CoCoMac database
| Connection with the amygdala | Human (MACM) | Nonhuman primate |
|---|---|---|
| Prefrontal cortex (BA9/46) | √ | √ |
| Medial/superior frontal gyrus (BA6) | √ | √ |
| Orbitofrontal cortex (BA11) | √ | √ |
| Anterior cingulate | − | − |
| Subgenual (BA25) | √ | √ |
| Supragenual (BA32) | √ | √ |
| Dorsal cingulate (BA24) | √ | √ |
| Posterior cingulate (BA23) | √ | Absent |
| Middle temporal gyrus (BA21/37/39) | √ | √ |
| Entorhinal cortex | √ | √ |
| Parahippocampal gyrus | √ | No data |
| Insula (BA13) | √ | √ |
| Thalamus | √ | √ |
| Caudate | √ | No data |
| Culmen | √ | No data |
Table IV.
List of abbreviations used in the nonhuman primate connectivity query
| Abbreviation | Structure | Abbreviation | Structure |
|---|---|---|---|
| 1 | Area 1 of cortex (somatosensory) | AD | Anterodorsal thalamic nucleus |
| 2 | Area 2 of cortex (somatosensory) | Al | Alar nucleus |
| 4 | Area 4 of cortex (primary motor) | AM | Anteromedial thalamic nucleus |
| 6 | Area 6 of cortex | AV | Anteroventral thalamic nucleus |
| 10 | Area 10 of cortex | B | Basal nucleus (Meynert) |
| 11 | Area 11 of cortex | CM | Central medial thalamic nucleus |
| 13 | Area 13 of cortex | EC | Entorhinal cortex |
| 23 | Area 23 of cortex | MD | Mediodorsal thalamic nucleus |
| 24 | Area 24 of cortex | Pall | Parainsular cortex, lateral division |
| 25 | Area 25 of cortex | PC | Paracentral thalamic nucleus |
| 32 | Area 32 of cortex | Pul | Pulvinar nuclei |
| 35 | Area 35 of cortex | R | Red nucleus |
| 45 | Area 45 of cortex | Re | Reuniens thalamic nucleus |
| 46 | Area 46 of cortex | SG | Suprageniculate thalamic nucleus |
| 11m | Area 11 of cortex, medial part | TEO | Temporal area TE, occipital part |
| 13a | Area 13a of cortex | TF | Temporal area TF |
| 24b | Area 24b of cortex | TH | Temporal area TH |
| 8A | Area 8A of cortex | VP | Ventral pallidum |
DISCUSSION
Amygdala functional connectivity was mapped in the human using a novel method (MACM) that capitalizes on the statistical advantages of metaanalysis, and the brain mapping database development initiatives that have occurred. We provide evidence of validity for our method using the most prominent nonhuman primate database, CoCoMac [Stephan et al., 2001]. Our data provide support for the use of MACM for modeling functional connectivity, as it identifies brain regions that are likely to be indirectly connected to our region of interest, the amygdala. Thus, reliance on an entirely structural model could lead to the absence of crucial brain regions (i.e., posterior cingulate) within a neural network model, making it imperative for the discovery of accurate and robust models of functional connectivity.
The consistency of MACM results with the nonhuman primate literature is a testament to the robustness of the method. Nonhuman primate studies have played a key role in establishing a foundation with which to build models of human neural connectivity, making it essential to validate our results with those obtained from these hallmark studies. Anatomical connectivity studies examining the afferent projections from the primate amygdala indicated direct connections to the orbitofrontal cortex, anterior cingulate gyrus, subcallosal gyrus, temporal pole, superior temporal gyrus, inferotemporal gyrus, and the insula [Aggleton et al., 1980]. Subcortical findings included pathways to the hypothalamus and thalamus, all of which have been amply replicated [Amaral and Price, 1984; Barbas and De Olmos, 1990; Ghashghaei and Barbas, 2002; Young et al., 1994], with some regions, such as the anterior cingulate, accumulating indisputable evidence for connectivity both anatomically and functionally [Amaral and Price, 1984; Stephan et al., 2000]. We performed an extensive search using all 12 amygdala regions listed throughout the 151 slices of the CoCoMac‐Paxinos3D Viewer, which contains an atlas based on the meticulous efforts to put the rhesus monkey brain in stereotaxic space by Paxinos et al. [ 2000]. We found high concordance, with strong anatomical connections being noted to the anterior cingulate from the basolateral amygdaloid nucleus, and weaker connections to regions of the primate cortex that are thought to have similar functionality as their analogous regions in the human. The significant overlap between the nonhuman primate literature and the findings of functional connectivity in the human amygdalae provide support for the validation of MACM methodology.
However, the foundation that nonhuman primate research has provided for the development of human connectivity models is inadequate when examining complex cognitive systems. Human models have included structures that are not as well‐supported in the nonhuman primate literature but appear consistently in studies examining limbic system function, indicating either a direct or indirect relationship with one or both amygdalae. Stein et al. [ 2007] used the posterior cingulate as a potential node in their model of effective connectivity of the human amygdala based largely on support from activation studies in humans [Mayberg et al., 1999; Meyer‐Lindenberg et al., 2005] and using structural equation modeling, concluded that the inclusion of this structure improved the proposed model fit substantially. Our data also support the notion that this is a critically relevant region that should be included in models of amygdala connectivity, as it likely represents an indirect yet functional pathway, though we did not rely on task‐dependent data. Further connectivity modeling utilizing MACM may not only lend support for the inclusion of other brain regions not typically implied by anatomical models, but also how these additional structures interact in an indirect fashion with the target ROI.
A recent delineation of the functional connectivity of the amygdala using resting state fMRI also showed consistent results across many brain regions including the right caudate, bilateral insula, left inferior frontal gyrus, and left precuneus for the right amygdala and the left middle temporal gyrus, bilateral insula, left superior frontal gyrus, right cingulated gyrus, and the right hippocampus for the left amygdala [Roy et al., 2009]. However, MACM identified several regions that the resting state analysis did not identify for both left and right amygdalae. The right amygdala was determined to be functionally connected to the posterior cingulate (BA23) and bilateral parahippocampus as well as bilateral culmen using MACM and the left amygdala was functionally connected to the left anterior and posterior cingulate in addition to bilateral parahippocampus and bilateral culmen. While resting state methods are common and represent a reasonable approach to functional connectivity during the nontask state, they are difficult to interpret with certainty based on low signal‐to‐noise ratios, poor replicability, and competing network activity (i.e., default mode network). More importantly, using MACM to identify functional connectivity provides additional information regarding connectivity that was not otherwise reported, and in a much less costly and time‐consuming fashion.
The versatility of MACM allows for the testing of developing theoretical models. Recent acknowledgement of the neural interconnectedness of emotionally‐ and cognitively‐based processes has led to the development of theoretical models of which the amygdala plays a central role, and which, to date, have not been empirically tested [Pessoa, 2008; Phelps, 2006]. Using MACM, we found support for the theoretical model in which the “emotional brain” incorporates key structures involved in cognitive processes that have long been thought to be influenced by the limbic system. Based on the behavioral domains from which the papers were drawn in this metaanalysis, it is clear that the amygdala plays a vital role in both emotion and cognition [Damasio, 1994], providing strong support for Pessoa's [ 2008] argument of an integrative emotional‐cognitive brain. MACM is an approach that is multidisciplinary as it crosses the boundaries of all behavioral domains, while still maintaining the flexibility to be limited to a specific domain. Thus, the method is a strong indicator of functional connectivity for any given ROI.
MACM also provides a starting point for the development of additional models using structural equation modeling (SEM) and path analysis that will allow investigators to draw distinct conclusions about the interactions between brain structures, and in doing so, develop more accurate models of effective connectivity [Protzner and McIntosh, 2006], which has eluded the neuroimaging community to date. Our research did not apply an a priori model, nor did we apply restraints on any potential paths, but rather we allowed the data to drive the model, increasing the robustness of the analysis. This represents a significant improvement in methodology that will allow for SEM in which connectivity models can be assessed between populations of interest (e.g., depressed patients versus healthy controls) where deficits are prominent in portions of any given neural circuitry (e.g., affective). Such an analysis may be instrumentally useful in combination with other neuroimaging techniques such as diffusion tensor imaging, in which neural fiber tracts can be quantified accurately with new techniques that are being implemented [Behrens et al., 2003; McNab et al., 2009; Ramnani et al., 2004], and fMRI or PET, in which blood‐oxygen‐level‐dependent (BOLD) signal or oxygen metabolism can be deduced during a resting (basal) state or during a task requiring cognitive and/or affective processing.
Here, we used one of the smallest and most complex structures as proof of concept for MACM and to identify the potential caveats of utilizing this method. While research has suggested potential relationships between the human amygdala and various brain structures, this is the first paper empirically supporting functional connectivity in the human utilizing decades worth of neuroimaging data. We have demonstrated great coherence among our results and the nonhuman primate literature, as well as the human functional neuroimaging literature, and have provided avenues for further investigation including subsequent structural equation modeling that will allow the neuroimaging community to develop refined models of effective and functional connectivity among any human brain structure. We emphasize the utility of MACM methods for identifying brain regions which are functionally connected, but may be part of an indirect network or lack the robustness necessary to show up in typical connectivity methods based on resting state data, or limited to the average‐sized functional neuroimaging study. Our method relies entirely on an objective, statistically‐driven, task‐independent approach that utilizes human data collected across multiple behavioral domains. MACM is less complex, and easier to implement than previous whole‐brain metaanalysis based connectivity methods [Toro et al., 2008]. It does not require that computations continually be recalculated, and instantly uses the most up‐to‐date data that is available in the BrainMap database. Ultimately, MACM could provide nodes to a network that would otherwise go overlooked when using traditional methods of connectivity because ROIs are defined subjectively or based on author‐specific operational definitions, or because of methodological issues such as task‐specificity or power. MACM overcomes these issues by providing a framework for identifying models of functional connectivity that are nontask specific for any region of the human brain that capitalizes on the diversity and quantity of the neuroimaging community's work as a whole.
Although novel, unique, and solid as an emerging technique for developing models of functional connectivity, we identified a couple of issues that should be addressed with future studies. Rerunning the metaanalysis using an amygdala probability threshold of 40% (rather an 70%) yielded results that were very similar both in the resultant maps, and also in the number of manuscripts represented. This may be due to the Gaussian smoothing applied as part of the ALE analysis, or could reflect the tightness of amygdala activation clusters. We speculate that the latter is the case, but future metaanalytic connectivity modeling in other structures ranging in size and complexity will help elucidate this issue, and may lead to refining the existing methodology presented in this article. Additionally, metaanalytic techniques lose task‐specific information. In the future, studies should be conducted in which a global (i.e., all domains) analysis is conducted as well as MACM models within each behavioral domain (e.g., only emotion) to try and recapture some of this information. As the database continues to grow, behavioral domains will naturally become more complete, and further refinement to models may be necessary.
REFERENCES
- Abercrombie HC,Schaefer SM,Larson CL,Oakes TR,Lindgren KA,Holden JE,Perlman SB,Turski PA,Krahn DD,Benca RM,Davidson RJ ( 1998): Metabolic rate in the right amygdala predicts negative affect in depressed patients. Neuroreport 9: 3301–3307. [DOI] [PubMed] [Google Scholar]
- Adolphs R,Gosselin F,Buchanan TW,Tranel D,Schyns P,Damasio AR ( 2005): A mechanism for impaired fear recognition after amygdala damage. Nature 433: 68–72. [DOI] [PubMed] [Google Scholar]
- Aggleton JP,Burton MJ,Passingham RE ( 1980): Cortical and subcortical afferents to the amygdala of the rhesus monkey (macaca mulatta). Brain Res 190: 347–368. [DOI] [PubMed] [Google Scholar]
- Amaral DG,Price JL ( 1984): Amygdalo‐cortical projections in the monkey (Macaca fascicularis). J Comp Neurol 230: 465–496. [DOI] [PubMed] [Google Scholar]
- Anderson AK,Phelps EA ( 2000): Expression without recognition: Contributions of the human amygdala to emotional communication. Psychol Sci 11: 106–111. [DOI] [PubMed] [Google Scholar]
- Barbas H,De Olmos J ( 1990): Projections from the amygdala to basoventral and mediodorsal prefrontal regions in the rhesus monkey. J Comp Neurol 300: 549–571. [DOI] [PubMed] [Google Scholar]
- Behrens TE,Woolrich MW,Jenkinson M,Johansen‐Berg H,Nunes RG,Clare S,Matthews PM,Brady JM,Smith SM ( 2003): Characterization and propagation of uncertainty in diffusion‐weighted MR imaging. Magn Reson Med 50: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Cohen MX,Elger CE,Weber B ( 2008): Amygdala tractography predicts functional connectivity and learning during feedback‐guided decision‐making. NeuroImage 39: 1396. [DOI] [PubMed] [Google Scholar]
- Damasio AR ( 1994): Descartes' Error: Emotion, Reason, and the Human Brain. New York: G.P. Putnam's Sons. [Google Scholar]
- Davidson RJ,Abercrombie H,Nitschke JB,Putnam K ( 1999): Regional brain function, emotion and disorders of emotion. Curr Opin Psychiatry 9: 228–234. [DOI] [PubMed] [Google Scholar]
- Fox MD,Snyder AZ,Vincent JL,Corbetta M,Van Essen DC,Raichle ME ( 2005): The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA 102: 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT,Lancaster JL ( 2002): Opinion: Mapping context and content: The BrainMap model. Nat Rev Neurosci 3: 319–321. [DOI] [PubMed] [Google Scholar]
- Fox PT,Ingham R,George M,Mayberg H,Ingham J,Roby J,Martin C,Jerabek P ( 1997): Imaging human intra‐cerebral connectivity by PET during TMS. Neuroreport 8: 2787–2791. [DOI] [PubMed] [Google Scholar]
- Fox PT,Parsons LM,Lancaster JL ( 1998): Beyond the single study: Function/location metanalysis in cognitive neuroimaging. Curr Opin Neurobiol 8: 178–187. [DOI] [PubMed] [Google Scholar]
- Friston KJ ( 1994): Functional and effective connectivity in neuroimaging: A synthesis. Hum Brain Mapp 2: 56–78. [Google Scholar]
- Ghashghaei HT,Barbas H ( 2002): Pathways for emotion: Interactions of prefrontal and anterior temporal pathways in the amygdala of the rhesus monkey. Neuroscience 115: 1261. [DOI] [PubMed] [Google Scholar]
- Gianaros PJ,Sheu LK,Matthews KA,Jennings JR,Manuck SB,Hariri AR ( 2008): Individual differences in stressor‐evoked blood pressure reactivity vary with activation, volume, and functional connectivity of the amygdala. J Neurosci 28: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC,Lovallo WR,Fox PT ( 2007): Reduced amygdala activation in young adults at high risk of alcoholism: Studies from the Oklahoma family health patterns project. Biol Psychiatry 61: 1306–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin PR,McRae K,Ramel W,Gross JJ ( 2008): The neural bases of emotion regulation: Reappraisal and suppression of negative emotion. Biol Psychiatry 63: 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD,Krasnow B,Reiss AL,Menon V ( 2003): Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc Natl Acad Sci USA 100: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR,Tessitore A,Mattay VS,Fera F,Weinberger DR, Clinical Brain Disorders Branch IRPNIoMHNIoHBMUSAhinng ( 2002): The amygdala response to emotional stimuli: A comparison of faces and scenes. Neuroimage 17: 317–323. [DOI] [PubMed] [Google Scholar]
- Hariri AR,Drabant EM,Munoz KE,Kolachana BS,Mattay VS,Egan MF,Weinberger DR ( 2005): A susceptibility gene for affective disorders and the response of the human amygdala. Arch Gen Psychiatry 62: 146–152. [DOI] [PubMed] [Google Scholar]
- Koski L,Paus T ( 2000): Functional connectivity of the anterior cingulate cortex within the human frontal lobe: A brain‐mapping meta‐analysis. Exp Brain Res 133: 55–65. [DOI] [PubMed] [Google Scholar]
- Kotter R ( 2004): Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics 2: 127–144. [DOI] [PubMed] [Google Scholar]
- Laird AR,Fox PM,Price CJ,Glahn DC,Uecker AM,Lancaster JL,Turkeltaub PE,Kochunov P,Fox PT ( 2005a): ALE meta‐analysis: Controlling the false discovery rate and performing statistical contrasts. Hum Brain Mapp 25: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR,Lancaster JL,Fox PT ( 2005b): BrainMap: The social evolution of a human brain mapping database. Neuroinformatics 3: 65–78. [DOI] [PubMed] [Google Scholar]
- Lawrie SM,Whalley HC,Job DE,Johnstone EC ( 2003): Structural and functional abnormalities of the amygdala in schizophrenia. Ann N Y Acad Sci 985: 445–460. [DOI] [PubMed] [Google Scholar]
- LeDoux J ( 2007): The amygdala. Curr Biol 17: R868. [DOI] [PubMed] [Google Scholar]
- Mayberg HS,Liotti M,Brannan SK,McGinnis S,Mahurin RK,Jerabek PA,Silva JA,Tekell JL,Martin CC,Lancaster JL, et al. ( 1999): Reciprocal limbic‐cortical function and negative mood: Converging PET findings in depression and normal sadness. Am J Psychiatry 156: 675–682. [DOI] [PubMed] [Google Scholar]
- McNab JA,Jbabdi Sd,Deoni SCL,Douaud G,Behrens TEJ,Miller KL ( 2009): High resolution diffusion‐weighted imaging in fixed human brain using diffusion‐weighted steady state free precession. NeuroImage 46: 775–785. [DOI] [PubMed] [Google Scholar]
- Meyer‐Lindenberg A,Hariri AR,Munoz KE,Mervis CB,Mattay VS,Morris CA,Berman KF ( 2005): Neural correlates of genetically abnormal social cognition in Williams syndrome. Nat Neurosci 8: 991. [DOI] [PubMed] [Google Scholar]
- Mohanty A,Engels AS,Herrington JD,Heller W,Ringo Ho M‐H,Banich MT,Webb AG,Warren SL,Miller GA ( 2007): Differential engagement of anterior cingulate cortex subdivisions for cognitive and emotional function. Psychophysiology 44: 343–351. [DOI] [PubMed] [Google Scholar]
- Ochsner KN,Ray RD,Cooper JC,Robertson ER,Chopra S,Gabrieli JDE,Gross JJ ( 2004): For better or for worse: Neural systems supporting the cognitive down‐ and up‐regulation of negative emotion. NeuroImage 23: 483. [DOI] [PubMed] [Google Scholar]
- Packard MG,Cahill L ( 2001): Affective modulation of multiple memory systems. Curr Opin Neurobiol 11: 752. [DOI] [PubMed] [Google Scholar]
- Papez JW ( 1995): A proposed mechanism of emotion. J Neuropsychiatry Clin Neurosci 7: 103–112. [DOI] [PubMed] [Google Scholar]
- Paus T,Jech R,Thompson CJ,Comeau R,Peters T,Evans AC ( 1997): Transcranial magnetic stimulation during positron emission tomography: A new method for studying connectivity of the human cerebral cortex. J Neurosci 17: 3178–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T,Jech R,Thompson CJ,Comeau R,Peters T,Evans AC ( 1998): Dose‐dependent reduction of cerebral blood flow during rapid‐rate transcranial magnetic stimulation of the human sensorimotor cortex. J Neurophysiol 79: 1102–1107. [DOI] [PubMed] [Google Scholar]
- Paxinos G,Huang X,Toga AW ( 2000): The rhesus monkey brain in stereotaxic coordinates. In: Paxinos G, editor. London, UK: Academic Press. [Google Scholar]
- Pessoa L ( 2008): On the relationship between emotion and cognition. Nat Rev Neurosci 9: 148. [DOI] [PubMed] [Google Scholar]
- Phelps EA ( 2004): Human emotion and memory: Interactions of the amygdala and hippocampal complex. Curr Opin Neurobiol 14: 198. [DOI] [PubMed] [Google Scholar]
- Phelps EA ( 2006): Emotion and cognition: Insights from studies of the human amygdala. Annu Rev Psychol 57: 27–53. [DOI] [PubMed] [Google Scholar]
- Phelps EA,Delgado MR,Nearing KI,LeDoux JE ( 2004): Extinction learning in humans: Role of the amygdala and vmPFC. Neuron 43: 897. [DOI] [PubMed] [Google Scholar]
- Protzner AB,McIntosh AR ( 2006): Testing effective connectivity changes with structural equation modeling: What does a bad model tell us? Hum Brain Mapp 27: 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnani N,Behrens TEJ,Penny W,Matthews PM ( 2004): New approaches for exploring anatomical and functional connectivity in the human brain. Biol Psychiatry 56: 613–619. [DOI] [PubMed] [Google Scholar]
- Roy AK,Shehzad Z,Margulies DS,Kelly AMC,Uddin LQ,Gotimer K,Biswal BB,Castellanos FX,Milham MP ( 2009): Functional connectivity of the human amygdala using resting state fMRI. NeuroImage 45: 614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM,Jenkinson M,Woolrich MW,Beckmann CF,Behrens TE,Johansen‐Berg H,Bannister PR,De Luca M,Drobnjak I,Flitney DE, et al. ( 2004): Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 ( Suppl 1): S208–S219. [DOI] [PubMed] [Google Scholar]
- Stein JL,Wiedholz LM,Bassett DS,Weinberger DR,Zink CF,Mattay VS,Meyer‐Lindenberg A ( 2007): A validated network of effective amygdala connectivity. NeuroImage 36: 736. [DOI] [PubMed] [Google Scholar]
- Stephan KE,Hilgetag CC,Burns GAPC,O'Neill MA,Young MP,Kotter R ( 2000): Computational analysis of functional connectivity between areas of primate cerebral cortex. Philos Trans R Soc B Biol Sci 355: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE,Kamper L,Bozkurt A,Burns GA,Young MP,Kotter R ( 2001): Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Philos Trans R Soc Lond B Biol Sci 356: 1159–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R,Fox PT,Paus T ( 2008): Functional coactivation map of the human brain. Cereb Cortex 18: 2553–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub PE,Eden GF,Jones KM,Zeffiro TA ( 2002): Metaanalysis of the functional neuroanatomy of single‐word reading: Method and validation. NeuroImage 16( 3, Part 1): 765. [DOI] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N,Landeau B,Papathanassiou D,Crivello F,Etard O,Delcroix N,Mazoyer B,Joliot M ( 2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. NeuroImage 15: 273. [DOI] [PubMed] [Google Scholar]
- Williams LM,Liddell BJ,Kemp AH,Bryant RA,Meares RA,Peduto AS,Gordon E ( 2006): Amygdala‐prefrontal dissociation of subliminal and supraliminal fear. Hum Brain Mapp 27: 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MP,Scannell JW,Burns GA,Blakemore C ( 1994): Analysis of connectivity: Neural systems in the cerebral cortex. Rev Neurosci 5: 227–250. [DOI] [PubMed] [Google Scholar]


