Abstract
Glycosyltransferase enzymes play important roles in numerous cellular pathways. Despite their participation in many therapeutically-relevant pathways, there is a paucity of information on how to effectively inhibit this class of enzymes. Here we report that UDP-(5F)-GlcNAc acts as a slow-binding, competitive inhibitor of the retaining glycosyltransferase MshA from Corynebacterium glutamicum (Ki ~1.6 µM). The kinetic data are consistent with a single-step inhibition mechanism whose equilibration is slow relative to catalysis. We believe this is the first slow-onset inhibitor to be reported for the glycosyltransferase family of enzymes. The potent inhibition of the enzyme by the fluoro-substituted substrate is consistent with the involvement of an oxo-carbenium transition-state structure, which has been previously proposed for this family of enzymes. Additionally, although several members of the GT-B enzyme family, including MshA, have been shown to undergo a conformational change upon UDP-GlcNAc binding, the kinetic data are inconsistent with a two-step inhibition mechanism. This suggests that there may be other conformations of the enzyme that are useful for the design of inhibitors against the large family of GT-B glycosyltransferase enzymes.
Glycosyltransferase (GT) enzymes, which catalyze the transfer of a sugar moiety to an acceptor molecule, are involved in numerous cellular pathways including bacterial cell wall biosynthesis1, post-translational modification of proteins2, and signal transduction3. These enzymes can be classified as either retaining or inverting based on the stereochemical outcome of the anomeric carbon center of the donor sugar.4 Despite their participation in many therapeutically-relevant pathways, there is a paucity of information on how to effectively inhibit this class of enzymes. The lack of information can be traced to the observations that only a small percentage of GT enzymes have been characterized, and the affinity of the substrates for characterized enzymes tends to be low. We recently reported the three-dimensional structure and basic kinetic characterization of the retaining glycosyltransferase MshA from Corynebacterium glutamicum (CgMshA).5 MshA catalyzes the transfer of N-acetyl-glucosamine (GlcNAc) from UDP-GlcNAc to 1-L-myo-inositol-1-phosphate (L-I1P) in the first committed step in the biosynthesis of the small molecule reductant mycothiol (Scheme 1).
Scheme 1.
It is generally accepted that the chemical mechanism of glycosyltransferase enzymes involves the development of substantial oxo-carbenium character in the transition state, similar to glycosidase enzymes.6 In support of this hypothesis, sugar nucleotides with electron withdrawing substituents are inhibitors of glycosidases. Recently, the synthesis of UDP-(5F)-GlcNAc (Scheme 2) provided the ability to probe the mechanism of glycosyltransferase enzymes as well.7,8
Scheme 2.
Based on steady-state kinetic data, UDP-(5F)-GlcNAc acts as a competitive inhibitor of CgMshA versus UDP-GlcNAc with a Kis value of 1.4 ± 0.2 µM, approximately 150-fold lower than the Michaelis constant determined for UDP-GlcNAc (210 µM) (Figure S1).5 The 5-fluoro compound did not act as a viable donor substrate for the MshA-catalyzed reaction at concentrations up to 200 µM under conditions similar to those in the inhibition assays. These results are similar to previous studies demonstrating that the 5-fluoro compound will not act as a donor substrate, but acts as a functional acceptor substrate in glycosyltransferase reactions.8 The results shown here provide further support for the formation of substantial charge development on the donor sugar in glycosyltransferase reactions and demonstrate the power of 5-F sugars as mechanistic probes.
Unlike previous studies however, the time-courses of CgMshA activity obtained in the presence of UDP-(5F)-GlcNAc displayed non-linear kinetics described by a rapid initial rate followed by a slower steady-state rate (Figure 1A). The time-courses were fit using eq 1, where [P]t is the concentration of product formed, [E] is the total enzyme, vi is the initial velocity, vss is the steady-state velocity, t is time, kobs is the exponential rate constant for equilibration, and C is a finite intercept.9 The inhibition was demonstrated to be reversible by incubating the enzyme with the inhibitor prior to initiation of the reaction with UDP-GlcNAc; this resulted in a progress curve that consisted of a lag phase followed by reactivation of the enzyme comparable to levels seen in the absence of the inhibitor (Figure 1B).
| (1) |
Figure 1.
(A) Kinetics of time-dependent inhibition of CgMshA by UDP-(5F)-GlcNAc. Progress curves for MshA in the absence (blue) and presence (black) of UDP-(5F)-GlcNAc. The red lines are a fit to eq 1. Saturating substrate concentrations of 2 mM UDP-GlcNAc and 2 mM L-I1P were used in this experiment. (B) Effect of pre-incubation with UDP-(5F)-GlcNAc on CgMshA activity initiated with UDP-GlcNAc. A solution of 8 nM enzyme, 2 mM L-I1P, and 5 µM UDP-(5F)-GlcNAc was incubated at 25 °C for 10 minutes prior to the addition of 1 mM UDP-GlcNAc (black line). The volume of UDP-GlcNAc added was 1% of the reaction volume. The blue line represents the same experiment in the absence of UDP-(5F)-GlcNAc. See Supporting Information for full experimental details.
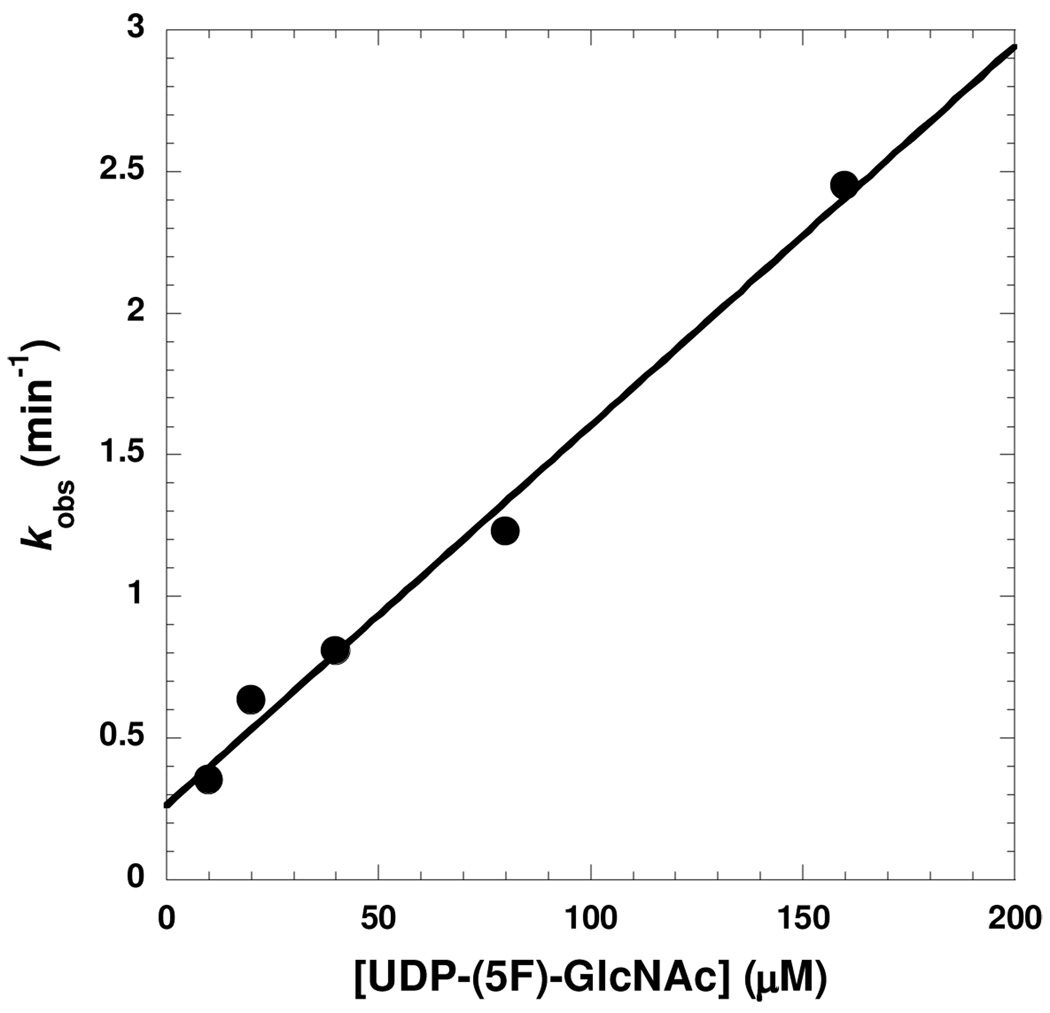
Two possible kinetic mechanisms that can explain non-linear kinetics are shown in Scheme 3.9 Mechanism A depicts a two-step mechanism where an initial inhibitory complex is formed followed by a slow equilibration to a more tightly-bound complex (EI*). Mechanism B depicts a single-step mechanism where the formation of the initial complex is the slow step relative to catalysis. A plot of kobs versus inhibitor concentration displays a linear dependence (Figure 2), consistent with the single-step mechanism depicted in mechanism B.
Scheme 3.
Figure 2.
Plot of kobs versus [UDP-(5F)-GlcNAc]. The values for kobs were determined by fits of the progress curves to equation 1. The solid black line is a fit of the data to equation 2.
The dependence of kobs on [UDP-(5F)-GlcNAc] can be fit to eq 2, where k1 and k2 are the rate constants shown in mechanism B, [I] is the concentration of the inhibitor, [S] is the concentration of UDP-GlcNAc, and Km is the Michaelis constant for UDP-GlcNAc. From the results of the fit, values of 0.14 ± 0.01 µM−1 min−1 and 0.26 ± 0.06 min−1 can be calculated for k1 and k2, respectively. Using these values derived from kobs, the calculated Ki value of 1.8 ± 0.4 µM is in excellent agreement with the Kis value determined from the steady-state inhibition experiments (1.4 ± 0.2 µM).
| (2) |
Structural studies demonstrate that CgMshA and other GT-B fold glycosyltransferases can adopt both open and closed forms. It has been proposed that a large conformational change takes place after the sugar nucleotide binds to the enzyme, creating the closed form of the enzyme and a binding site for the second substrate.5,10–13 One might expect the inhibitor to bind in a similar fashion, consistent with mechanism A. However, the kinetic data indicate that UDP-(5F)-GlcNAc inhibits the enzyme in a single step that occurs slowly relative to catalysis.
Based on steady-state kinetic data, CgMshA follows an ordered bi-bi sequential mechanism with UDP-GlcNAc binding first, followed by L-I1P.5 As UDP-(5F)-GlcNAc is a competitive inhibitor versus UDP-GlcNAc, it is expected that both compounds bind to the apo-form of the enzyme. Attempts to cocrystallize CgMshA with the 5-fluoro compound have been unsuccessful. One possible explanation for the slow-onset phenomenon is that UDP-(5F)-GlcNAc binds slowly to the open form of the enzyme. However, given the modest structural change versus UDP-GlcNAc, this seems unlikely. Instead, if UDP-(5F)-GlcNAc binds to a closed form of the apo enzyme, a possible physical mechanism for the slow binding may arise from a small loop (residues 16–22) that becomes ordered in the closed conformation. This loop is proposed to sequester the uridine moiety of UDP-GlcNAc from solvent. It is possible that the movement of this loop limits the rate at which UDP-(5F)-GlcNAc can bind to the apo enzyme resulting in the time-dependent inhibition.
While a number of time-dependent inhibitors of glycosidase enzymes have been reported14, we believe that this is the first reported glycosyltransferase inhibitor to act in a slow-onset manner. The potent inhibition of CgMshA by the fluoro-substituted compound is consistent with the oxo-carbenium transition-state structure proposed for retaining glycosyltransferase enzymes based on kinetic isotope effects.15,16 While structural studies indicate a large conformational change upon sugar nucleotide binding, the kinetic results suggest that more subtle structural changes are involved in the time-dependent inhibition of CgMshA by UDP-(5F)-GlcNAc. Taken together, these conclusions provide data on the electronic structure of the transition state of this retaining glycosyltransferase and the possibility of alternate conformations of the enzyme. This information will aid in the design of future inhibitors for the large family of GT-B glycosyltransferase enzymes.
Supplementary Material
Acknowledgement
We thank Matthew W. Vetting for providing the purified enzyme and Matthew Hartman for a sample of UDP-(5F)-GlcNAc. This work was supported by NIH Grant AI33696 (J.S.B.) and a postdoctoral fellowship from the Charles H. Revson Foundation (P.A.F.)
Footnotes
Supporting Information Available: Experimental procedures and steady-state inhibition plots are described. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lovering AL, Gretes M, Strynadka NC. Curr Opin Struct Biol. 2008;18:534–543. doi: 10.1016/j.sbi.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Sears P, Wong CH. Cell Mol Life Sci. 1998;54:223–252. doi: 10.1007/s000180050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slawson C, Housley MP, Hart GW. J Cell Biochem. 2006;97:71–83. doi: 10.1002/jcb.20676. [DOI] [PubMed] [Google Scholar]
- 4.Coutinho PM, Deleury E, Davies GJ, Henrissat B. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 5.Vetting MW, Frantom PA, Blanchard JS. J Biol Chem. 2008;283:15834–15844. doi: 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 7.Hartman MC, Coward JK. J Am Chem Soc. 2002;124:10036–10053. doi: 10.1021/ja0127234. [DOI] [PubMed] [Google Scholar]
- 8.Hartman MC, Jiang S, Rush JS, Waechter CJ, Coward JK. Biochemistry. 2007;46:11630–11638. doi: 10.1021/bi700863s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrison JF, Walsh CT. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 10.Buschiazzo A, Ugalde JE, Guerin ME, Shepard W, Ugalde RA, Alzari PM. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boix E, Zhang Y, Swaminathan GJ, Brew K, Acharya KR. J Biol Chem. 2002;277:28310–28318. doi: 10.1074/jbc.M202631200. [DOI] [PubMed] [Google Scholar]
- 12.Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Proc Natl Acad Sci U S A. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. Proc Natl Acad Sci U S A. 2003;100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gloster TM, Davies GJ. Org Biomol Chem. 2010;8:305–320. doi: 10.1039/b915870g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SC, Singh AN, Raushel FM. J Biol Chem. 1988;263:10151–10154. [PubMed] [Google Scholar]
- 16.Murray BW, Wittmann V, Burkart MD, Hung SC, Wong CH. Biochemistry. 1997;36:823–831. doi: 10.1021/bi962284z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.