Summary
Archaea, one of three major evolutionary lineages of life, encode proteasomes highly related to those of eukaryotes. In contrast, archaeal ubiquitin-like proteins are less conserved and not known to function in protein conjugation. This has complicated our understanding of the origins of ubiquitination and its connection to proteasomes. Here we report two small archaeal modifier proteins, SAMP1 and SAMP2, with a β-grasp fold and C-terminal diglycine motif similar to ubiquitin, that form protein-conjugates in the archaeon Haloferax volcanii. SAMP-conjugates were altered by nitrogen-limitation and proteasomal gene knockout and spanned various functions including components of the Urm1 pathway. LC-MS/MS-based collision-induced dissociation demonstrated isopeptide bonds between the C-terminal glycine of SAMP2 and the ε-amino group of lysines from a number of protein targets and Lys58 of SAMP2 itself, revealing poly-SAMP chains. The widespread distribution and diversity of pathways modified by SAMPylation suggest this type of protein-conjugation is central to the archaeal lineage.
Keywords: archaea, ubiquitin, proteasome, protease, AAA ATPases
Introduction
In eukaryotic cells, the conjugation of ubiquitin (Ub) and ubiquitin-like (Ubl) proteins to protein targets plays an integral role in a wide variety of processes including proteasome-mediated proteolysis, heterochromatin remodeling and protein trafficking1,2. Elaborate ATP-dependent systems mediate these covalent attachments including the use of E1 Ub-activating, E2 Ub-conjugating and E3 Ub-protein ligase enzymes1,2. Of these, E1 catalyzes the ATP-dependent adenylation of the Ub/Ubl C-terminal carboxylate and transfers this activated form of Ub/Ubl to a conserved cysteine on E1. This Ub/Ubl thioester intermediate is transferred to an E2 to form a second thioester linkage. The E2 Ub-conjugating enzyme then transfers the Ub/Ubl to an ε-amino group of a lysine residue either within a target protein or on a growing poly-Ub/Ubl chain2,3. Transfer to Nα-amino groups has also been observed4. Often Ub-transfer is with assistance from an E3 Ub-protein ligase either forming an E3-Ub/Ubl thioester intermediate or with E3 facilitating Ub/Ubl-transfer from E2 directly to the substrate protein.
Although universal in eukaryotes, the presence of Ub-like protein conjugation systems in prokaryotes is less clear. PUP, the first example of a protein covalently attached to target proteins in prokaryotes5,6, appears restricted to Actinobacteria and Nitrospriae and is distinct from ubiquitination in its use of deamidase and glutamine synthetase-like ligase6,7 reactions for conjugation and its disordered structure8,9. The β-grasp fold of Ub/Ubl proteins, however, are common to a growing superfamily of proteins involved in diverse functions that span all three domains of life10-12. Of these β-grasp functions, the enzymology and mechanism of sulphur activation for the biosynthesis of thiamine, tungsten and molybdenum cofactors bears striking resemblance to the activation of Ub/Ubl13. Jab1/MPN domain metalloenzyme (JAMM) motifs common to deubiquitinating enzymes used for the recycling of Ub and removal of Ubl modifiers are also conserved in many prokaryotes14-16. On the basis of these features, it is unclear (i) whether eukaryotic Ub/Ubl-systems were derived from a combination of various prokaryotic β-grasp fold pathways that function in related yet distinct chemistry or (ii) whether prokaryotes figured out how to conjugate Ub/Ubl-proteins to protein targets prior to the divergence of eukaryotes. Here we demonstrate two small archaeal modifier proteins (SAMPs) of the β-grasp superfamily are differentially conjugated to protein targets in the archaeon Haloferax volcanii, thus providing an evolutionary link in Ub/Ubl-protein conjugation systems.
SAMP1 and SAMP2 form protein conjugates
Small proteins with a β-grasp fold and C-terminal di-glycine motif similar to Ub are widespread among Archaea10-12. Although presumed to activate sulphur for the biosynthesis of cofactors such as thiamine, tungsten and molybdenum, the biological function of these proteins remains unknown. In this study, Ub-like β-grasp proteins were identified in the deduced proteome of H. volcanii (Fig. 1). The proteins were fused to an N-terminal FLAG-tag and synthesized in H. volcanii grown under various conditions including complex and minimal media, nitrogen-limitation and salt concentrations ranging from suboptimal to optimal (1.0 to 2.5 M NaCl). The FLAG-tagged proteins were analyzed for conjugate formation by anti-FLAG immunoblot (α-FLAG IB) of cell lysate separated by reducing SDS-PAGE.
Figure 1. Multiple amino acid sequence alignment of the C-termini of Ub, Urm1 and PUP to select di-glycine motif proteins of H. volcanii.
C-terminal di-glycine motifs are shaded in red. Identical and similar amino acids are shaded in black and grey, respectively. Amino acid length of protein and membership in the Ub/ThiS/MoaD β-grasp superfamily are indicated. HVO, Haloferax volcanii; Sc, Saccharomyces cerevisiae; Mt, Mycobacterium tuberculosis; Hvo2619, SAMP1; Hvo0202, SAMP2.
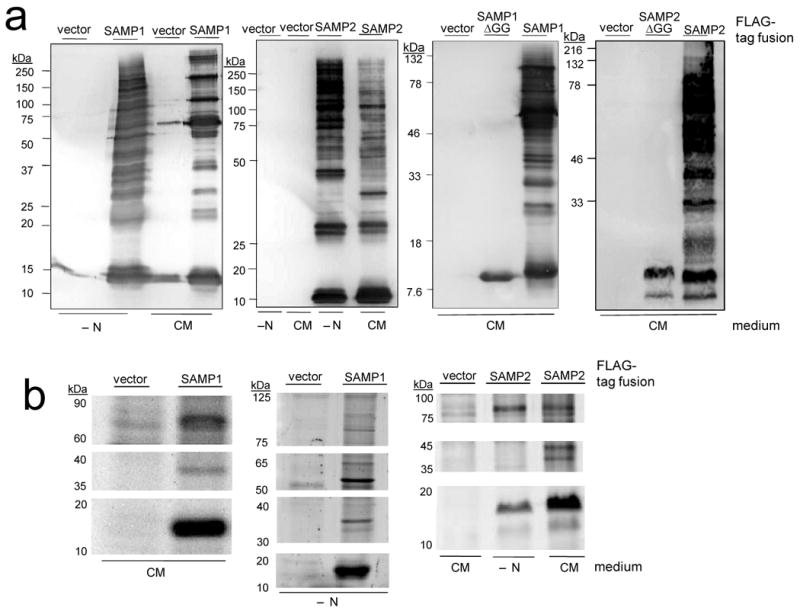
Using this approach, two Ubl-proteins, HVO_2619 (SAMP1) and HVO_0202 (SAMP2) that share only 21 % identity and 30 % similarity in amino acid sequence, were found to form differential protein conjugates that were modulated by growth condition (Fig. 2). Protein conjugates were not detected for the remaining proteins examined (HVO_2177, HVO_2178 and HVO_0383) (Supplemental Fig. 1). Although the number of SAMP-conjugates detected was minimal when cells were grown under standard conditions in complex medium with only two discrete protein bands detected for each SAMP (58- and 14-kDa for SAMP1 and 18- and 16-kDa for SAMP2) (Fig. 2a), a dramatic increase in the number of SAMP-conjugates was observed when cells were transferred to glycerol-alanine minimal medium (Fig. 2b). Systematic supplementation of media with glycerol, alanine and ammonium chloride revealed low nitrogen was the signal for this prominent increase (Fig. 2b). Each of the SAMPs was associated with distinct patterns of protein-conjugates suggesting the presence of a relatively complex regulatory network of SAMPylation that not only senses environmental cues, but also discriminates and differentially conjugates the two SAMP proteins to their protein targets. Interestingly, the predominant SAMP2-conjugates detected migrated in regular intervals of ∼ 11-12 kDa by SDS-PAGE, suggesting SAMP2 formed free SAMP2 polymers.
Figure 2. SAMP1 and SAMP2 are differentially conjugated to proteins and influenced by nitrogen-limitation.
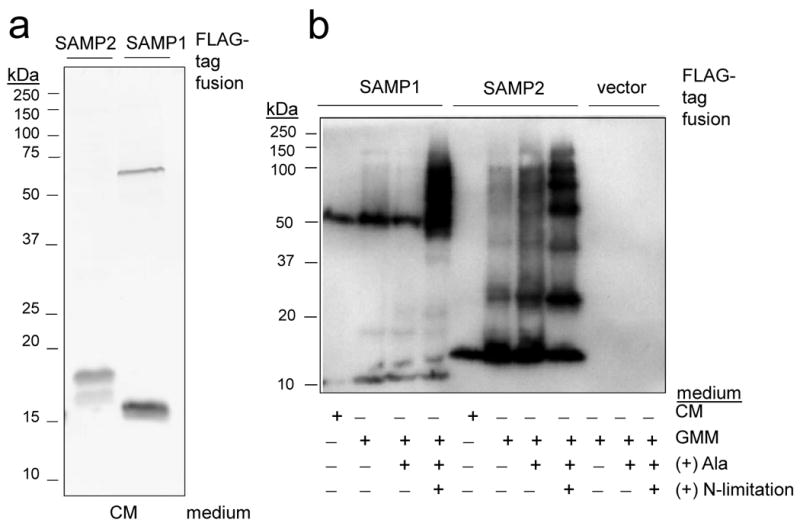
a) α-FLAG immunoblot of SAMP1 and SAMP2 expressed as N-terminal FLAG-tagged fusions in H. volcanii cells grown on complex medium (CM). b) FLAG-SAMP fusions similarly expressed and analyzed from cells grown on CM, glycerol minimal medium (GMM), GMM supplemented with alanine (+ Ala) and GMM + Ala devoid of NH4Cl (+ N-limitation). All details on experimental procedures and strains are available as supplemental data.
Proteasomes alter SAMP-conjugates
H. volcanii mutant strains with markerless deletions in proteasomal genes, including those encoding the subunits of the 20S proteasomal core particle (CP) and Rpt-like ATPase subtypes17, were used to examine the influence of proteasome function on the levels of SAMP-conjugate formation. Site-2-type metalloprotease (S2P) knockout strains were also included in this analysis. Unlike some archaea that synthesize a single CP of α- and β-type subunit composition and do not encode Rpt-like ATPases, H. volcanii synthesizes multiple proteasomal subtypes including: (i) CPs with a β-type subunit that associates with α1 and/or α2 subunits as well as (ii) PAN-A and PAN-B proteins that are closely related to eukaryotic 26S proteasomal Rpt subunits18,19. Of these, α1 and PAN-A are highly abundant during all phases of growth19, double knockout of the Rpt-like genes has little impact on standard growth and synthesis of CPs containing either α1 or α2 can be separately abolished17. However, conditional knockout of all CP subtypes renders cells inviable17.
Analysis of the FLAG-SAMP fusions in the various proteasomal mutants revealed significant differences in SAMP-conjugate levels compared to wild type. A substantial increase in SAMP1-conjugate and decrease in SAMP2-conjugate levels was observed during nitrogen-limitation in ΔpanA ΔpsmA mutant strains (deficient in synthesis of PAN-A and α1), while deletion of site-2-type metalloprotease genes had no effect (Fig. 3). Consistent with this, ΔpanA ΔpsmA single and double knockouts have the most pronounced phenotypes of the viable proteasomal mutant strains of H. volcanii, with enhanced sensitivity to nitrogen-limitation, hypo-osmotic shock and the amino acid analogue L-canavanine17. The enhanced levels of SAMP1-conjugates in the ΔpanA ΔpsmA mutant suggest SAMP1 targets proteins for destruction by proteasomes. Other functions of SAMPylation are also likely based on the decrease in SAMP2-conjugates observed in select proteasomal mutant strains.
Figure 3. SAMP-conjugates are altered by proteasomal gene knockout.
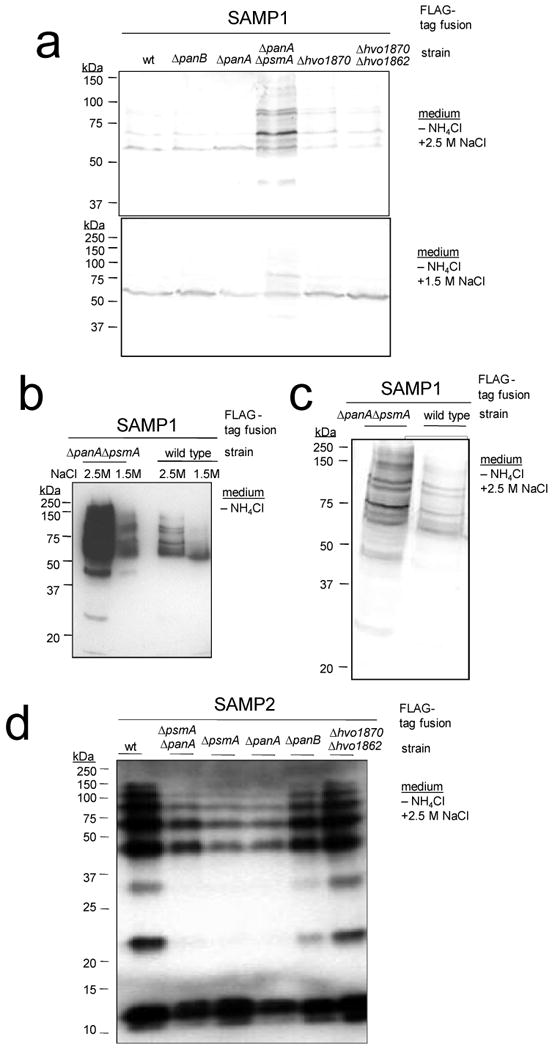
a-c) α-FLAG immunoblot of SAMP1 expressed as an N-terminal FLAG-tagged fusion in H. volcanii wild type and protease mutant strains grown under nitrogen-limiting conditions with 2.5 M NaCl or 1.5 M NaCl as indicated. d) SAMP2 was similarly expressed and analyzed in wild type and mutant strains. SAMP1-conjugate levels of ΔpsmA and ΔpanA ΔpanB mutant strains were similar to wild type, and SAMP-conjugates were not detected in strains with vector alone (data not shown). psmA (CP α1), panA and panB (Rpt-like AAA ATPases), HVO_1870 and HVO_1862 (site-2 type metalloprotease homologs).
Identification of SAMP-conjugates
SAMP-conjugates were purified from H. volcanii cells expressing the FLAG-SAMP fusions by α-FLAG immunoprecipitation (IP) compared to cells expressing the FLAG-SAMP fusions with deletions in their C-terminal diglycine motif (ΔGG) or vector alone (Fig. 4). Unlike most proteins, the vast majority of proteins from haloarchaea are highly acidic and require high salt (> 1 M) for stability and activity20. Non-covalent protein complexes from these ‘salt-loving’ organisms typically dissociate in the low salt and detergent conditions required for IP. Consistent with this, SAMP-conjugates were readily purified by IP from H. volcanii based on α-FLAG immunoblot and SYPRO Ruby stain of these fractions (Fig. 4). The purified SAMP-conjugates were resistant to boiling in the presence of SDS and reducing reagents (Fig. 4a). The results also demonstrated that the C-terminal diglycine motif of SAMP1 and SAMP2 was required for their conjugation to proteins and that IP enhanced the ability to detect a remarkable diversity of SAMP-conjugates present in cells grown under rich and nitrogen-limiting conditions. It should also be noted that the SAMP-conjugate banding patterns were not influenced by addition of reducing reagents. Thus, IP combined with boiling, separation by SDS-PAGE and staining with SYPRO Ruby proved ideal for the isolation of covalently-linked FLAG-SAMP-conjugates (Fig. 4b). Proteins specific for the FLAG-SAMP expressing strains were excised from the gels, digested with trypsin and identified by mass spectrometry (MS). Using this approach, thirty-four SAMP-protein conjugates were identified including those present in cells grown under nutrient rich and nitrogen-limiting conditions (Table 1). Of the proteins identified, all were unique to the strains expressing the FLAG-SAMP fusions compared to cells with vector alone, and two of the conjugates were common to both SAMP1 and SAMP2 (HVO_0558 and HVO_A0230; Table 1). Consistent with their role as small archaeal modifier proteins, SAMP1 and SAMP2 were the only proteins identified in SDS-PAGE gel slices that spanned a wide-range of molecular masses (5 – 125 kDa, Supplementary Table 3).
Figure 4. SAMP-conjugates are isolated by immunoprecipitation.
SAMP1 ± ΔGG and SAMP2 ± ΔGG were expressed as N-terminal FLAG-tagged fusions in H. volcanii grown in complex medium (CM) and nitrogen-limiting conditions (− N). Proteins were immunoprecipitated with α-FLAG, boiled and separated by either: a) reducing 12 % SDS-PAGE and analyzed by α-FLAG immunoblot or b) non-reducing 12 % SDS-PAGE and stained for total protein by SYPRO Ruby. Molecular mass standards and range of gel slices excised for MS-analysis are indicated on left. H. volcanii with vector alone served as a negative control in all experiments including MS-analysis of gel slices.
Table 1. H. volcanii SAMPs and SAMP-conjugates identified by MSa.
| Protein | Homolog/Description | CM | -N | CM | -N | Relation to Ub, Sulfur and Proteasomes |
|---|---|---|---|---|---|---|
| FLAG-SAMP1 | FLAG-SAMP2 | |||||
| Ubl-/S-chemistry: | ||||||
| HVO_2619 | SAMP1 | + | + | − | − | Ubl β-grasp |
| HVO_0202 | SAMP2 | − | − | + | + | Ubl β-grasp |
| HVO_0558 | UBA/E1/MoeB, Ub- and sulphur-activating enzymes | + | + | + | + | Homolog of the N-terminal domain of Uba4p, the E1-enzyme of the Urm1 pathway |
| HVO_1864 | N-terminal domain related to MobB P-loop NTPase; C-terminal domain related to MoaE sulphur-conjugating enzyme | + | + | − | − | S-conjugation |
| HVO_2305 | MoeA, functions with MoaB in metal insertion into molybdopterin | − | − | + | − | Mo/W-insertion |
| HVO_0025 | SseA/TssA, tandem RHD thiosulfate sulfurtransferase | − | + | − | + | Homolog of Urm1-associated Yor251cp21,25 |
| HVO_0861 | SufB/SufD, cysteine desulfurase activator subunit | − | − | − | + | Cysteine desulfurase activator; accumulates in HVO after cLβL treatment36 |
| HVO_0580 | N-type ATP PPases and ATP sulfurylases | − | − | + | + | Homolog of Urm1-associated Ncs6p, functions in tRNA adenylation21,25,32 |
| N-limitation/stress response: | ||||||
| HVO_A0230 | MsrA, methionine-S-sulfoxide reductase | − | + | + | + | |
| HVO_2402 | Glycine cleavage P-protein, catalyzes initial step of oxidative cleavage of glycine to NH4+, CO2 and methylene group (-CH2-) | − | + | − | − | |
| HVO_2900 | FumC, ROS-resistant fumarase C | − | + | − | − | |
| HVO_1289 | OsmC, osmotically inducible protein C peroxiredoxin | − | − | − | + | OsmC accumulates in HVO ΔpanA37, Ahp1p is a peroxiredoxin and the only known target of urmylation30 |
| HVO_1250 | Peroxiredoxin-/thioredoxin-like | − | − | + | − | |
| HVO_2682 | Dodecin-flavoprotein, may prevent riboflavin degradation and trap phototoxic lumichrome waste | − | − | − | + | |
| Metabolism: | ||||||
| HVO_2583 | HmgA, 3-hydroxy-3-methylglutaryl CoA reductase | − | − | + | − | |
| HVO_2328 | Isochroismatase | − | − | + | − | |
| HVO_1545 | DhaL, dihydroxyacetone kinase (DHAK) subunit | − | − | + | − | Components of DHAK-PTS system accumulate in HVO after cLβL treatment and ΔpanA36,37 |
| HVO_1496 | PtsI, PTS system EI | − | − | + | − | |
| HVO_0481 | GAPDH, glyceraldehyde-3-P DH | − | − | + | − | HVO_0480 (3-phospho-glycerate kinase) encoded within operon accumulates in HVO ΔpanA37 |
| HVO_1000 | Acetyl-CoA synthetase | − | − | + | − | |
| HVO_0887 | 2-oxoglutarate oxidoreductases, β | − | − | + | − | homolog HVO_1304, accumulates in HVO after cLβL treatment36 |
| HVO_A0379 | agaF, N-methyl-hydanoinase A | − | − | + | − | HVO_A0378 (oxoprolinase homolog) within operon accumulates in HVO after cLβL treatment36 |
| HVO_0980 | NdhG, NADH-quinone OR, chain c/d | − | − | + | − | |
| DNA replication, transcription, translation and RNA processing: | ||||||
| HVO_1727 | TATA-box binding protein E | − | − | − | + | |
| HVO_1478 | TFB, transcription initiation factor | − | − | + | − | |
| HVO_0359 | EF-1α, translation elongation factor | − | + | − | − | accumulates in HVO after cLβL treatment36, putative isopeptidase26,27 |
| HVO_0966 | aIF2ba, archaeal translation initiation factor | − | − | + | + | |
| HVO_1921 | SerS, seryl-tRNA synthetase | − | − | + | − | |
| HVO_0677 | AspS, aspartyl-tRNA synthetase | − | − | + | − | |
| HVO_1572 | GyrB, DNA gyrase B | − | − | + | − | |
| HVO_1344 | Shwachman-Bodian-Diamond syndrome protein, putative role in RNA metabolism | − | − | + | − | gene neighbor of archaeal α-type 20S proteasomal subunits29 |
| HVO_1577 | Putative winged-helix transcriptional regulator, C-terminal CBS domains | − | − | + | − | HVO 20S proteasome associated protein28 |
| Conserved: | ||||||
| HVO_0736 | DUF302 | − | − | − | + | |
| HVO_B0053 | C-terminal H-X3-H motif protein | − | − | − | + | |
−, undetectable. +, MS-identified protein-conjugate unique to IP fractions of H. volcanii strains expressing the FLAG-tagged β-grasp Ub-like protein SAMP1 or SAMP2 compared to vector alone. Cells were grown on complex medium (CM) or under nitrogen-limiting conditions (-N). Protein identities are reported according to the H. volcanii gene locus tag from the USCS Archaeal Genome Browser (April 2007 version). SAMP-conjugates were reproducibly purified by immunoprecipitation (IP) with α-FLAG, boiled in SDS buffer, separated by SDS-PAGE and analyzed by immunoblot with α-FLAG. Only α-FLAG reactive bands were further analyzed by MS for protein identity and covalent linkages. Proteins were identified using a hybrid quadrupole-TOF (ABI QSTAR XL) and hybrid quadrupole-linear ion trap (ABI 4000 QTRAP). All details on experimental procedures, MS-data and FASTA files of identified protein sequences are available as supplemental data.
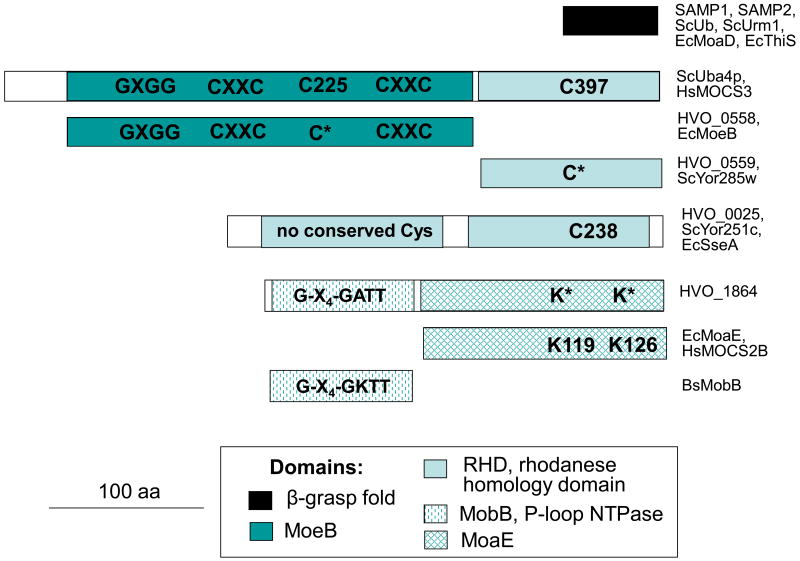
Many of the SAMPylated proteins were homologs of enzymes associated with Ubl-conjugation and/or sulphur-activation (Table 1). These included homologs of Uba4p, Yor251c and Ncs6p/Ncs2p associated with the Urm1 pathway involved in thiolation of tRNA and protein conjugation21,22 as well as MobB, MoaE, MoeA and SufB/D, all predicted to be involved in pathways associated with sulphur metabolism. Interestingly, homologs of the N- and C-terminal domains of Uba4p are encoded as separate proteins in H. volcanii and other archaea. HVO_0558, identified as a SAMP-conjugate, is similar to the Uba4p N-terminal domain and Cys225 active site required for adenylyltransferase activity21,23,24 (Fig. 5), while the divergently transcribed HVO_0559 is related to the Uba4p C-terminus including the rhodanese domain (RHD) and Cys397 needed for persulfide formation in sulphurtransferase reactions25. Whether HVO_0558 functions as an E1 and activates the SAMPs for protein-conjugation and/or sulphur transfer to tRNA or cofactors such as molybdopterin remains to be determined; however, its association with both SAMP proteins under all conditions examined and its relationship to the Urm1 pathway is consistent with this possibility.
Figure 5. SAMP and SAMP-conjugates are related to sulphur-activation and ubiquitination pathways.
SAMP1 and SAMP2 cluster to the β-grasp superfamily. HVO_0558 is related to Uba4p of the Urm1 pathway and molybdopterin (MPT) synthase sulphurases (e.g., MoeB, MOCS3). Although the RHD common to Uba4p is not conserved in HVO_0558 it is found in the gene neighbour HVO_0559. HVO_1864 is related to MoaE proteins that associate with β-grasp proteins to form active MPT synthases38,39 and MobB, a P-loop NTPase of MPT synthesis40. HVO_0025 is a dual RHD protein related to 3-mercaptopyruvate sulphurtransferases that form persulfide intermediates41,42 and ScYor251c of the Urm1 pathway21.
A wide-variety of proteins spanning functions from stress response to basic transcription, translation and DNA replication were also conjugated to the SAMPs (Table 1). Many of these proteins were previously found to accumulate in H. volcanii cells after chemical and/or genetic perturbation of proteasome function (as indicated in Table 1). Furthermore, many have been linked to Ubl/Ub-proteasome systems including the translation elongation factor EF-1α26,27, predicted transcriptional regulator HVO_1577 associated with H. volcanii 20S CPs28, Shwachman-Bodian-Diamond syndrome protein encoded in proteasomal operons in archaea29 and HVO_1250 and HVO_1289 of similar antioxidant function to the Urm1-target Ahp1p30.
Mapping sites of SAMPylation
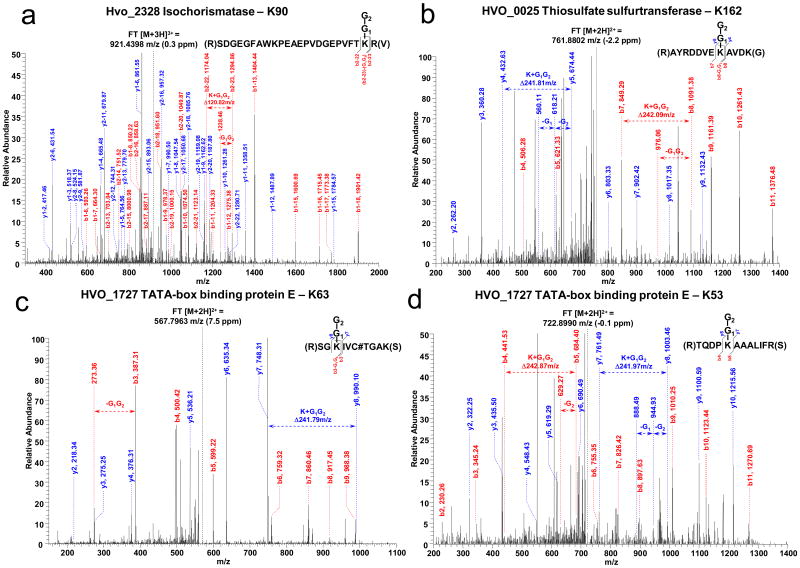
To enhance MS-coverage and map the sites of SAMPylation, FLAG-SAMP2-conjugates were purified by α-FLAG in liquid phase for analysis of trypsinized peptides by reversed phase liquid chromatography coupled with tandem mass spectrometry (RP-LC-MS/MS) using a data dependent MS/MS scan mode and parent mass list method. Unlike SAMP1, which has a limited number of C-terminal trypsin cleavage sites, SAMP2 has a lysine at position 64. Thus, if an isopeptide bond is formed between the C-terminal carboxylate of SAMP2 and an amino group of the substrate protein, SAMP2 will leave a ‘GG-footprint’ on the target site after trypsinization. Using this approach, eleven sites of SAMP2 modification were mapped by collision-induced dissociation (CID) based MS/MS (Table 2). The sites were based on the mass differences between the y and b ion series containing the SAMP2-derived GG-footprint on lysine residues (Fig. 6 and Supplementary Fig. 2). The SAMPylated peptides were detected from doubly- to quadruply-charged molecular ions and mapped by more than one peptide on the same protein.
Table 2. SAMP2-conjugate sites mapped by MS/MSa.
| No. | ORF No. | Protein Description | Z | Mass accuracy (ppm) | Xcorr | Sf | Residue modified | Peptides |
|---|---|---|---|---|---|---|---|---|
| 1 | HVO_0202 | SAMP2 | 2 | 1.2 | 2.01 | 0.71 | K58 | (R)VK@VLR(L) |
| 2 | HVO_0966 | eif2ba / aIF-2BII translation initiation factor | 4 | -0.1 | 4.90 | 0.94 | K210 | (R)YLNDVDHVLVGADAVAADGSVINK@IGTSGLAVNAR(E) |
| 3 | -3.3 | 7.61 | 0.99 | |||||
| 3 | HVO_1572 | GyrB, DNA gyrase B subunit | 2 | -13.2 | 0.97 | 0.30 | K624 | (R)K@QFIK(D) |
| 4 | HVO_2328 | Isochorismatase | 4 | 0.4 | 4.01 | 0.94 | K90 | (R)SDGEGFAWKPEAEPVDGEPVFTK@R(V) |
| 3 | 0.3 | 5.52 | 0.97 | |||||
| 2 | -0.2 | 3.59 | 0.92 | |||||
| 5 | HVO_0558 | MoeB, molybdopterin biosynthesis protein | 3 | 0.5 | 4.12 | 0.93 | K113 | (R)VDK@SNVHEVVAGSDVVVDASDNFPTR(Y) |
| 6 | HVO_0980 | NdhG, NADH-quinone oxidoreductase chain c/d | 2 | 28.5 | 0.73 | 0.47 | K517 | (R)FK@IR(S) |
| 7 | HVO_1289 | OsmC-like protein superfamily | 2 | 3.9 | 2.78 | 0.69 | K59 | (R)VGGQK@TGFDDLGK(V) |
| 8 | HVO_1727 | TATA-box binding protein E | 2 | 7.5 | 3.27 | 0.91 | K63 | (R)SGK@IVC#TGAK(S) |
| 2 | -0.1 | 2.65 | 0.88 | K53 | (R)TQDPK@AAALIFR(S) | |||
| 9 | HVO_0025 | SseA/TssA, tandem RHD thiosulfate sulfurtransferase | 2 | -2.2 | 3.14 | 0.81 | K162 | (R)AYRDDVEK@AVDK(G) |
| 2 | 0.6 | 2.00 | 0.52 | K166 | (K)AVDK@GLPLVDVR(S) |
Abbreviations: Z, Charge state; Xcorr, Cross correlation; Sf, Final score;
SAMP2-modification;
alkylated cysteine.
Figure 6. MS/MS spectra of SAMP2-conjugate sites.
SAMP2-modification of: (a) HVO_2328 K90 based on mass difference between b2-22 and b2-23 ions and loss of Gly1-Gly2 at 1238.46 m/z from the b2-23 ion derived from the triply charged precursor ion. (b) HVO_0025 K162 based on mass difference between both ion series derived from the doubly charged precursor ion: (i) y4 and y5 ions and loss of Gly1-Gly2 at 618.21 and 560.11 m/z, (ii) b7 and b8 ions and loss of Gly1-Gly2 at 976.06 m/z. (c - d) HVO_1727 K63 and K53. SAMP2 C-terminal diglycine (-Gly1-Gly2). Other MS/MS spectra, Supplementary Figure 2.
A number of fundamental insights were revealed by the CID-based MS/MS spectra concerning how SAMP2 modifies proteins. First and foremost, the C-terminal glycine of SAMP2 is covalently attached through an isopeptide bond to the ε-amino group of lysine residues of at least nine different substrate proteins. Secondly, SAMP2 can modify a single substrate protein at multiple sites based on the finding that TATA-binding protein E (HVO_1727) and SseA/Yor251cp (HVO_0025) homologs are modified at either of two lysine residues in close proximity. Thirdly, although a thioester bond was not detected by MS/MS between SAMP2 and any of the cysteine residues of HVO_0558, SAMP2 did modify this Uba4p homolog through an isopeptide bond at K113 suggesting SAMP2 may regulate the adenylation of either itself or SAMP1. Furthermore, the MS/MS data revealed that SAMP2 forms polymeric chains with itself at lysine 58 similar to Ub and other Ubl proteins (such as SUMO2/3 and NEDD8).31 Whether the SAMP2 polymeric chains are free or covalently attached to substrate proteins and the full diversity of these SAMP2 chains (i.e., homotypic, heterologous or mixed with SAMP1) remain to be determined. Likewise, it remains to be determined whether SAMP1 and SAMP2 compete for the same or different lysine residues on substrates and target these proteins for different fates, or whether they are mutually exclusive in their sites of protein targeting. Proteins with multiple SAMP-sites occupied remain to be identified. Our results do reveal, however, that the same protein can be modified by either SAMP1 or SAMP2 (i.e., Uba4p and MsrA homologs) and that the same protein can be modified on different lysine residues (i.e., TATA-binding protein E and SseA/Yor251cp homologs).
Widespread distribution of SAMP homologs
While SAMP1 and SAMP2, share limited primary sequence identity to each other, both proteins are members of a large superfamily that shares a common β-grasp fold and includes members from all archaea10-12. In addition to this common 3-D fold, SAMP1 and SAMP2 are related in primary amino acid sequence to small proteins from other archaea including species of haloarchaea, methanogens and Archaeoglobus (30 to 80 % identity)(Supplementary Fig. 3 and 4). SAMP1 also shares a close relationship with the N-termini of small proteins that have a C-terminal domain of unknown function (DUF1952) from thermophilic bacteria of the deep branching Thermus species (33 to 39 % identity) (Supplementary Fig. 3). Interestingly, a number of these SAMP homologs (5 from haloarchaea and 2 from Thermus) have 2 to 82 amino acid residues carboxyl to the diglycine motif and, thus, would likely require proteolytic cleavage prior to covalent attachment if functioning similar to the H. volcanii SAMPs.
The organization of the SAMP1 and SAMP2 genes on the H. volcanii genome is also revealing (Supplementary Fig. 5). Unlike eukaryotes that encode Ub as fusion proteins that are proteolytically processed to expose a functional C-terminal diglycine motif, SAMP1 and SAMP2 are encoded as single small proteins (of 87 and 66 amino acids, respectively) with the diglycines apparently exposed after translation. Comparison of the SAMP operons to other microbial genomes reveals a high conservation of immediate gene order between H. volcanii and other diverse haloarchaea. This includes the prediction that SAMP1 is co- and divergently transcribed with genes encoding proteins with regulatory of K+ conductance (RCK) domains likely to form K+ channels for cellular defense against osmotic stress. Likewise, haloarchaeal SAMP2 genes appear to be commonly co- and divergently transcribed with Gcn5-related N-acetyltransferase (GNAT) and AAA ATPase replication factor C small subunit homologs. This conservation in gene order suggests that SAMPylation is linked to osmotic stress, DNA replication and/or protein acetylation. Although SAMP-conjugates were not altered by low salt stress (Fig. 3a and data not shown), a strong and constitutive rRNA P2 promoter was used to drive expression of the FLAG-SAMP genes for this analysis. Interestingly, we did detect an increase in the levels and change in the types of SAMP-conjugates formed during nitrogen-limitation suggesting stress and/or reduced growth rate may be associated with SAMP function.
Discussion
H. volcanii forms a relatively elaborate network of protein conjugates including the covalent linkage to target proteins of at least two different Ubl-proteins, SAMP1 and SAMP2, that are conserved among diverse archaea. These data suggest ubiquitin-like protein conjugation system origins reside in archaea. H. volcanii forms these differential SAMP-conjugates in the presence of only a single E1 and in the absence of any apparent E2 or E3 homologs suggesting a streamlined Ubl-system for protein conjugation. In support of this possibility, (i) the related eukaryotic E1s can be relatively promiscuous and activate more than one type of structurally distinct Ubl protein32, (ii) E2-intermediates have yet to be identified for the ancient Urm1 pathway and (iii) ubiquitination can occur in the absence of E3 Ub-ligases33. Thus, an archaeal E2- and E3-independent Ubl-conjugation mechanism is feasible. Common to SAMP1 and SAMP2, was their conjugation to HVO_0558 under all conditions examined suggesting a close association of this E1 (Uba4p N-terminal domain) homolog with SAMPylation. The multiple RHD proteins that are related to the C-terminus of Uba4p and encoded as separate proteins in most archaea, including H. volcanii, may add functional flexibility to the SAMPylation system. Small Zn-finger proteins such as Brz34, prevalent in archaea and similar to the RING domains of E3 Ub ligases35, may also assist in discerning the various interactions required for SAMPylation. Although it has yet to be determined the full extent of poly- vs. mono-SAMPylation and whether the SAMPs are reused, SAMP2-polymeric chains were detected in this study and archaea encode proteins with JAMM motifs similar to eukaryotic deubiquitinating enzymes14-16 suggesting a SAMP-recycling mechanism is conserved.
Methods Summary
Small proteins were selected from the deduced proteome of H. volcanii based on the presence of a β-grasp fold and C-terminal diglycine motif. N-terminal FLAG-tagged fusions of these proteins were expressed in H. volcanii (± proteasomal gene mutations) grown under rich and nitrogen-limiting conditions. Formation of SAMP-conjugates was monitored by α-FLAG-immunoblot of cell lysate that was separated by reducing SDS-PAGE. SAMP-conjugates were enriched from cell lysate by α-FLAG-immunoprecipitation and further purified by SDS-PAGE prior to identification by MS (compared to cells with FLAG-SAMPΔGG or vector alone). Sites of SAMPylation were mapped by LC-MS/MS-based CID of FLAG-SAMP2-conjugates purified by α-FLAG chromatography.
Methods
Materials
Biochemicals were purchased from Sigma-Aldrich (St. Louis, MO). Other organic and inorganic analytical grade chemicals were from Fisher Scientific (Atlanta, GA) and Bio-Rad (Hercules, CA). Desalted oligonucleotides were from Integrated DNA Technologies (Coralville, IN). DNA modifying enzymes and polymerases were from New England Biolabs (Ipswich, MA).
Strains, media, and plasmids
H. volcanii and E. coli strains, oligonucleotide primers used for cloning, and plasmids are summarized in Supplementary Tables 1 and 2. E. coli DH5α was used for routine recombinant DNA experiments, and E. coli GM2163 was used for isolation of plasmid DNA for transformation of H. volcanii as previously described43. H. volcanii wild type and protease mutant strains expressing N-terminal FLAG-tagged fusions were grown to stationary phase (OD600 of 1.5 - 2.2) at 42 °C and 200 rpm. Media included: i) ATCC 974 composed of 2.14 M NaCl, 246 mM MgCl2·6H2O, 29 mM K2SO4, 0.91 mM CaCl2·2H2O, 0.5 % yeast extract (Difco) and 0.5% tryptone; ii) YPC composed of 0.5% yeast extract (Difco), 0.1% peptone (Oxoid), 0.1% casamino acids (Difco) with 18 % salt water (2.5 M NaCl, 88 mM MgCl2·6H2O, 85 mM MgSO4·7H2O, 56 mM KCl, 3 mM CaCl2) and 12 mM Tris-HCl buffer pH 7.5 according to Allers et al.43 and iii) GMM composed of 20 mM glycerol with 18 % salt water, 5 mM NH4Cl, trace minerals (1.8 μM MnCl2·4H2O, 1.5 μM ZnSO4·7H2O, 8.3 μM FeSO4·7H2O, 0.2 μM CuSO4·5H2O), cofactors (3 μM thiamine or vitamin B1 and 40 nM biotin or vitamin H) and buffers (42 mM Tris-HCl pH 7.5 and 1 mM KPO4 pH 7.5). Media was supplemented with alanine (25 mM) (+Ala), devoid of ammonium chloride (−N) or reduced to 1.5 M NaCl as indicated. Media was also supplemented with novobiocin (0.1 μg·ml-1) and uracil (50 μg·ml-1) as needed. Uracil was solubilized in 100 % DMSO at 50 mg·ml-1 prior to addition to media.
DNA purification and analysis
The H. volcanii genes encoding HVO_2619 (SAMP1), HVO_0202 (SAMP2), HVO_2177, HVO_2178 and HVO_0383 were isolated from genomic DNA by PCR using primers listed in Supplementary Table 1, H. volcanii genomic DNA as template, Phusion DNA polymerase and 3 % (v/v) DMSO according to supplier (New England Biolabs). PCR was performed with a thermal gradient for annealing at ± 5 °C primer Tm using an iCycler (BioRad Laboratories). PCR products were analyzed on 2 % (w/v) agarose gels in TAE buffer (40 mM Tris acetate, 2 mM EDTA, pH 8.5) using Hi-Lo DNA molecular weight marker (Minnesota Molecular, Minneapolis, MN.) and ethidium bromide staining at 0.5 μg ml−1. DNA fragments of appropriate molecular mass were purified by MinElute (Qiagen) or isolated from SeaKem GTG agarose (FMC Bioproducts, Rockland, ME) gels using the QIAquick gel extraction kit (Qiagen) as needed. DNA fragments were ligated into the NdeI and BlpI sites of pJAM202 or NdeI and KpnI sites of pJAM939 using appropriate restriction enzymes, Antarctic phosphatase and T4 ligase as indicated in Supplementary Tables 1 and 2. Plasmid DNA was isolated from E. coli strains using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). Fidelity of all PCR amplified products was confirmed by sequencing the DNA of plasmid inserts by Sanger automated DNA sequencing using an Applied Biosystems Model 3130 Genetic analyzer (UF ICBR Genomics Division).
Immunoblot
H. volcanii cells expressing N-terminal FLAG-tagged fusions were harvested by centrifugation (10,000 × g, 10 min, 25 °C), boiled 20 to 30 min in SDS-loading buffer with reducing reagents (2.5 % [v/v] β-mercaptoethanol or 10 mM dithiothreitol) and separated by SDS-PAGE (10 and 12 %) at 0.065 OD600 units per lane. Equivalent protein loading was confirmed by staining with Coomassie Blue. Proteins were electroblotted onto Hybond-P polyvinylidene fluoride (PVDF) membranes (Amersham) (14.5 h at 20 V or 2.5 h at 90 V; 4°C). FLAG-tagged fusions were detected by immunoblot using: i) anti-FLAG M2 antibody (Stratagene) and anti-mouse IgG-alkaline phosphatase (AP) antibody raised in goat (Sigma) and ii) AP-linked anti-FLAG M2 monoclonal antibody (Sigma). AP activity was detected colormetrically using nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and by chemiluminescence using CDP-Star according to supplier's protocol (Applied Biosystems, Foster City, CA) with X-ray film (Hyperfilm; Amersham Biosciences).
Preparation of cell lysate for IP and FLAG column elution
H. volcanii cells expressing FLAG-SAMP fusions and vector alone (100 ml cultures) were harvested by centrifugation (6,000 × g, 20 min, 25 °C) and resuspended in 1 ml of lysis buffer (50 mM Tris-Cl buffer at pH 7.4 with 1 % [v/v] Triton-X-100, 5 mM EDTA, 0.02 % [w/v] sodium azide, 10 mM iodoacetamide, 1 mM PMSF, 300 mM NaCl, 1 U·ml-1 DNase I). Debris was removed by centrifugation (14,000 × g, 20 min, 25 °C).
Immunoprecipitation
α-FLAG M2 agarose (Sigma; product number A2220) was prepared for immunoprecipitation (IP) by washing 2 × with PBS and 2 × with wash buffer (50 mM Tris-Cl buffer at pH 7.4 with 0.1 % [v/v] Triton-X-100, 300 mM NaCl, 5 mM EDTA, 0.02 % [w/v] sodium azide, 0.1% [w/v] SDS, 0.1% [w/v] deoxycholine). Clarified cell lysate (1 ml) was added to washed-agarose beads (100 μl) and incubated by rocking at 4 °C for 12 – 16 h. Protein-bound-beads were washed 10 × with wash buffer (1 ml per wash) and eluted with either SDS-PAGE or glycine buffer as described below.
For SDS-PAGE, proteins were eluted from beads by boiling for 10 min in 40 μl SDS-PAGE buffer (100 mM Tris-Cl buffer at pH 6.8 with 2 % [w/v] SDS, 10 % [v/v] glycerol, 0.6 mg·ml-1 bromophenol blue). Sample (20 μl) was separated by 12 % SDS-PAGE at 200 V for 40 to 50 min. Gels were stained with SYPRO Ruby according to manufacturer's protocol (BioRad) or developed with AP-linked α-FLAG M2 monoclonal antibody as described above. Gels were imaged on a BioRad XR imager and gel pieces were cut manually for mass spectrometry analysis by QSTAR and QTRAP (see below for details).
For glycine elution, 100 μl of 0.1 M glycine-HCl buffer at pH 2.5 was added to the protein-bound agarose beads and gently rocked (5 min at room temperature). The agarose beads were centrifuged (8,500 × g, 30 sec at room temperature), and the supernatant was added to a sterile 1.5 ml microcentrifuge tube that contained 20 μl of 1M Tris-HCl buffer at pH 8.0 supplemented with 1M NaCl. The addition of 0.1 M glycine-HCl buffer at pH 2.5 was repeated twice to maximize elution from the beads, and eluted proteins were collected in the same 1.5 ml microcentrifuge tube.
FLAG column elution
A polypropylene column (0.5 × 5 cm2; BioRad) was packed with α-FLAG M2 agarose to a total bed volume of 0.5 ml, as directed by the manufacturer (Sigma). After preparation of the resin, the column was equilibrated with 10 column volumes of TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.4). Clarified lysate (1 ml) (prepared as described above) was applied to the column (4 ×) and washed with 20 column volumes of TBS. Bound proteins were eluted with five column volumes of TBS containing 1× FLAG peptide (Sigma) at 100 μg/ml. Eluted proteins were collected in nine fractions (∼300 μl each). The column was regenerated immediately after use with three column volumes of 0.1 M glycine-HCl, pH 3.5, re-equilibrated with 13 column volumes of TBS, and stored in TBS with 50% [v/v] glycerol and 0.02% [w/v] sodium azide, as directed by the manufacturer (Sigma). All buffers were filtered with a 0.45 μm surfactant-free cellulose acetate (SFCA) filter (Nalgene Nunc) prior to use.
Mass spectrometry
SAMP-conjugates were identified from SYPRO-Ruby stained SDS-PAGE gels by mass spectrometry using a QTRAP triple quadrupole ion-trap mass spectrometer and a QSTAR quadrupole time-of-flight mass spectrometer with an inline capillary reverse-phase high-performance liquid chromatography (HPLC) separation of protein digests (UF ICBR Proteomics Division). A PepMap™ C18 column (75-μm inside diameter, 15-cm length; LC Packings, San Francisco, CA) was used for reverse-phase separation in combination with an Ultimate capillary HPLC system (LC Packings) operated at a flow rate of 200 nl·min-1 with a 60-min gradient from 5 to 50 % (v/v) acetonitrile in 0.1% (v/v) acetic acid. In-gel proteins were extracted by successive washes of gel slices in acetonitrile to a final volume of 100 μl. Extracted proteins were dried under vacuum centrifugation. The resulting desiccant was suspended in 100 μl of 50 mM NH4HCO3 (pH 7.5). Samples were reduced by the addition of 5 μl of 200 mM dithiothreitol (DTT solution) for 1 h at 25 °C. Samples were alkylated by the addition of 4 μl of 1 M iodoacetamide for 1 h at 25 °C. Alkylation was stopped by the addition of 20 μl of DTT solution. Samples were digested with a 1:20 mg ratio of trypsin or AspN to protein for 18-24 h at 37 °C. Digested peptides were purified using 300 μl C18 spin columns and dried under vacuum centrifugation. The resulting dessicant was resuspended in 5 – 10 μl of 5 % ACN (loading buffer for HPLC).
Mapping of SAMPylation sites was performed as follows. H. volcanii (pJAM949, FLAG-SAMP2) and (pJAM202c, vector alone) cells grown on complex medium (ATCC 974) to stationary phase were used for generation of cell lysate. Clarified lysate (1 ml) was bound to the α-FLAG agarose beads and eluted by glycine buffer or 1× FLAG peptide as described above. Eluted protein samples were diluted into 40 mM ammonium bicarbonate (NH4HCO3), reduced with 10 mM DTT for 1 hr at 56 °C, carboxy-amidomethylated with 55 mM iodoacetamide for 45 min in the dark, and digested with 3 μg of trypsin (Promega) in 40 mM NH4HCO3 overnight at 37 °C. After digestion, the peptides were acidified with trifluoroacetic acid (TFA) at a final concentration of 0.1 % TFA. Desalting was performed with C18 spin columns (Vydac Silica C18, The Nest Group, Inc.) and the resulting peptides were dried down in a Speed Vac and stored at -20 °C until analyzed. The peptides were resuspended with 19.5 μL of mobile phase A (0.1% formic acid, FA, in water) and 0.5 μL of mobile phase B (80% acetonitrile, ACN, and 0.1% formic acid in water) and filtered with 0.2 μm filters (Nanosep, PALL). The sample was loaded off-line onto a nanospray tapered capillary column/emitter (360 × 75 × 15 μm, PicoFrit, New Objective) self-packed with C18 reverse-phase (RP) resin (10.5 cm, Waters) in a nitrogen pressure injection cell for 10 min at 1000 psi (∼5 μL load) and separated using a 160 min linear gradient of increasing mobile phase B at a flow rate of ∼200 nL/min directly into the mass spectrometer. LC-MS/MS analysis was performed on a LTQ Orbitrap XL ETD mass spectrometer (ThermoFisher, San Jose, CA) equipped with a nanospray ion source. A full FTMS (Fourier transform mass spectrometry) spectrum at 30,000 resolution was collected at 250-2000 m/z followed by 6 data dependent MS/MS spectra in ITMS (Ion trap mass spectrometry) of the most intense ion peaks following CID (36 % normalized collision energy). For the parent mass list method, 5 data dependent MS/MS spectra from the full FTMS were activated in the most intense ion peaks from parent mass list following 36% CID. The parent mass width was set up ± 20.0 ppm. To obtain the parent mass list, the identified protein sequences were theoretically digested by trypsin allowing to one internal miss cleavage. The masses of theoretical tryptic peptides were allowed for dynamic modifications with the masses of oxidized methionine, alkylated cysteine, and two glycines on lysine (15.9949, 57.0215 and 114.0429 Da) respectively and then calculated with up to quintuply charge states. The masses were selected between 250-2000 m/z at each charge state for the parent mass list.
MS Data Analysis
SAMP-conjugate peptides were identified from the MS data using MASCOT algorithms44 that searched a custom, non-redundant database based on the hypothetical proteome of translated open-reading frames from the H. volcanii genome (April 2007 version, http://archaea.ucsc.edu/). Probability-based MOWSE scores were estimated by comparison of search results against estimated random match population and are reported as ∼10 × log10(p), where p is the absolute probability. Individual ion scores greater than 22 indicates identity or extensive homology (p<0.05). Carbamidomethylation was used as a fixed modification due to sample preparation. Variable modifications that were searched included deamidation of asparagines and glutamine, oxidation (single and double) of methionine, glycine-glycine addition on lysine, thiocarboxylation of C-termini, and pyro-glutamine of N-terminal glutamine.
Data generated for site mapping of SAMP2-protein conjugates was searched against the H. volcanii sequence database containing the common contaminants database using the TurboSequest algorithm (BioWorks 3.3.1 SP1, Thermo Fisher Scientific, Inc.). Spectra with a threshold of 15 ions, a TIC of 1 × 103, and a mass range of [MH]+ = 500-5000 m/z were searched. The SEQUEST parameters were set to allow 30.0 ppm of precursor ion mass tolerance and 0.5 Da of fragment ion tolerance with monoisotopic mass. Only fully tryptic peptides were allowed with up to three missed internal cleavage sites. Dynamic mass increases of 15.9949, 57.0215, and 114.0429 Da were allowed for oxidized methionine, alkylated cysteine, and two glycines on lysine residue respectively. Proteins identified by more than two peptides were only considered to be statistically significant at ≥ 1% false discovery rate (FDR) using the ProteoIQ software package (BIOINQUIRE, GA). The fragmentations of all peptides containing an internal Gly-Gly modified lysine residue were subjected to manual validation.
Protein sequences
All H. volcanii protein sequences described in this study are included with gene locus tag numbers as supplemental information. The following protein sequences were also described: ScUb (P61864); ScUrm1 (P40554); EcMoaD (CAA49864); EcThiS (O32583); ScUba4p (P38820); HsMOCS3 (O95396); EcMoeB (P12282); ScYor285W (Q12305); ScYor251c (Q08686); EcSseA (P31142); EcMoaE; (P30749); HsMOCS2B (O96007); BsMobB (O31704) (GenBank or Swiss-Prot accession numbers in parenthesis; Sc, Saccharomyces cerevisiae; Ec, E. coli; Bs, Bacillus subtilis; Hs, Homo sapiens).
Supplementary Material
Acknowledgments
We thank the staff at UF ICBR including C. Diaz and R. Zheng for MS and S. Shanker for DNA sequencing. Thanks to M. Terns and F. Aydemir for helpful advice on purification of SAMP-conjugates from α-FLAG agarose, N. Furlow for plasmid DNA preparations and J. Foster for other helpful advice. We thank also T. Allers, M. Mevarech, M. Dyall-Smith and M. Danson labs for H. volcanii strains and plasmids. This work was funded in part by NIH 1S10 RR025418-01 to SC, Integrated Technology Resource for Biomedical Glycomics at UGA (supported by NIH/NCRR P41 RR018502, LW senior investigator) and NIH R01 GM057498 and DOE DE-FG02-05ER15650 to JMF.
Footnotes
Author Contributions: JMF, MAH, DK, JP and HM performed cloning and immunoblot experiments. MAH and HM purified SAMP-conjugates by α-FLAG IP and chromatography. GZ transformed H. volcanii and prepared media. SC directed the identification of SAMP-conjugates by MS. J-M L and LW mapped the SAMP-conjugate sites by CID-based MS/MS. JMF, LW, MAH and J-M L interpreted the data. JMF planned the studies and wrote the manuscript. All authors commented on the manuscript.
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., III Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Striebel F, et al. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1597. in press. [DOI] [PubMed] [Google Scholar]
- 8.Liao S, et al. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, et al. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392:208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like β-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burroughs AM, Balaji S, Iyer LM, Aravind L. A novel superfamily containing the β-grasp fold involved in binding diverse soluble ligands. Biol Direct. 2007;2:4. doi: 10.1186/1745-6150-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: Implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2008;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol Rev. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 14.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 15.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 16.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 17.Zhou GY, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaczowka SJ, Maupin-Furlow JA. Subunit topology of two 20S proteasomes from Haloferax volcanii. J Bacteriol. 2003;185:165–174. doi: 10.1128/JB.185.1.165-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leidel S, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 22.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008;105:18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz J, et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 25.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonen H, et al. Protein synthesis elongation factor EF-1α is essential for ubiquitin-dependent degradation of certain Nα-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci U S A. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonen H, Dickman D, Schwartz AL, Ciechanover A. Protein synthesis elongation factor EF-1α is an isopeptidase essential for ubiquitin-dependent degradation of certain proteolytic substrates. Adv Exp Med Biol. 1996;389:209–219. doi: 10.1007/978-1-4613-0335-0_26. [DOI] [PubMed] [Google Scholar]
- 28.Humbard MA, Stevens SM, Jr, Maupin-Furlow JA. Posttranslational modification of the 20S proteasomal proteins of the archaeon Haloferax volcanii. J Bacteriol. 2006;188:7521–7530. doi: 10.1128/JB.00943-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maupin-Furlow JA, Wilson HL, Kaczowka SJ, Ou MS. Proteasomes in the archaea: from structure to function. Front Biosci. 2000;5:d837–d865. doi: 10.2741/furlow. [DOI] [PubMed] [Google Scholar]
- 30.Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2:930–936. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeller D, et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Tarasov VY, et al. A small protein from the bop-brp intergenic region of Halobacterium salinarum contains a zinc finger motif and regulates bop and crtB1 transcription. Mol Microbiol. 2008;67:772–780. doi: 10.1111/j.1365-2958.2007.06081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 36.Kirkland PA, Reuter CJ, Maupin-Furlow JA. Effect of proteasome inhibitor clasto-lactacystin-β-lactone on the proteome of the haloarchaeon Haloferax volcanii. Microbiology. 2007;153:2271–2280. doi: 10.1099/mic.0.2007/005769-0. [DOI] [PubMed] [Google Scholar]
- 37.Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase a mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leimkuhler S, Freuer A, Araujo JA, Rajagopalan KV, Mendel RR. Mechanistic studies of human molybdopterin synthase reaction and characterization of mutants identified in group B patients of molybdenum cofactor deficiency. J Biol Chem. 2003;278:26127–26134. doi: 10.1074/jbc.M303092200. [DOI] [PubMed] [Google Scholar]
- 39.Matthies A, Rajagopalan KV, Mendel RR, Leimkuhler S. Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc Natl Acad Sci U S A. 2004;101:5946–5951. doi: 10.1073/pnas.0308191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLuskey K, Harrison JA, Schuttelkopf AW, Boxer DH, Hunter WN. Insight into the role of Escherichia coli MobB in molybdenum cofactor biosynthesis based on the high resolution crystal structure. J Biol Chem. 2003;278:23706–23713. doi: 10.1074/jbc.M301485200. [DOI] [PubMed] [Google Scholar]
- 41.Colnaghi R, Cassinelli G, Drummond M, Forlani F, Pagani S. Properties of the Escherichia coli rhodanese-like protein SseA: contribution of the active-site residue Ser240 to sulfur donor recognition. FEBS Lett. 2001;500:153–156. doi: 10.1016/s0014-5793(01)02610-2. [DOI] [PubMed] [Google Scholar]
- 42.Spallarossa A, et al. The “rhodanese” fold and catalytic mechanism of 3-mercaptopyruvate sulfurtransferases: crystal structure of SseA from Escherichia coli. J Mol Biol. 2004;335:583–593. doi: 10.1016/j.jmb.2003.10.072. [DOI] [PubMed] [Google Scholar]
- 43.Dyall-Smith M. The Halohandbook: Protocols for Halobacterial Genetics. 2008. [Google Scholar]
- 44.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.