Abstract
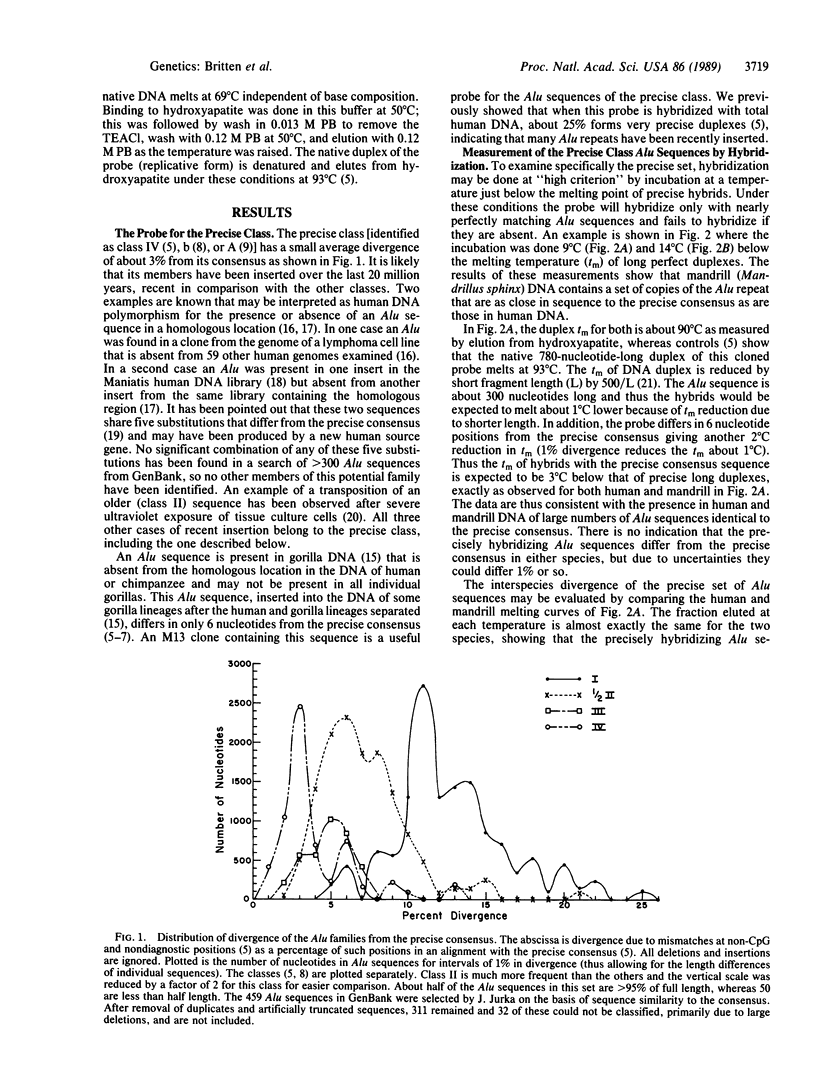
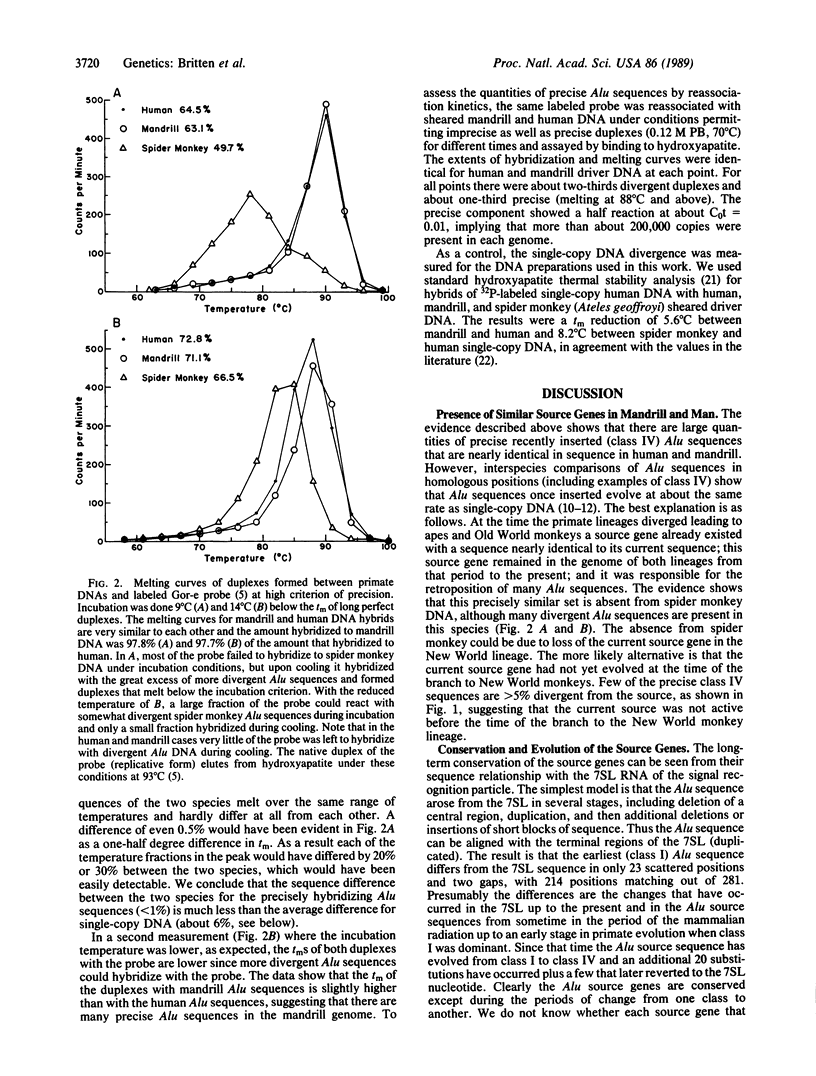
A significant fraction of human Alu repeated sequences are members of the precise, recently inserted class. A cloned member of this class has been used as a probe for interspecies hybridization and thermal stability determination. The probe was reassociated with human, mandrill, and spider monkey DNA under conditions such that only almost perfectly matching duplexes could form. Equally precise hybrids were formed with human and mandrill DNA (Old World monkey) but not with spider monkey DNA (New World). These measurements as well as reassociation kinetics show the presence in mandrill DNA of many precise class Alu sequences that are very similar or identical in quantity and sequence to those in human DNA. Human and mandrill are moderately distant species with a single-copy DNA divergence of about 6%. Nevertheless, their recently inserted Alu sequences arise by retroposition of transcripts of source genes with nearly identical sequences. Apparently a gene present in our common ancestor at the time of branching was inherited and highly conserved in sequence in both the lineage of Old World monkeys and the lineage of apes and man.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. S., Gibbs R. A., Ranier J. E., Nguyen P. N., Caskey C. T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988 Dec 9;16(23):11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Degen S. J., Rajput B., Reich E. The human tissue plasminogen activator gene. J Biol Chem. 1986 May 25;261(15):6972–6985. [PubMed] [Google Scholar]
- Deininger P. L., Slagel V. K. Recently amplified Alu family members share a common parental Alu sequence. Mol Cell Biol. 1988 Oct;8(10):4566–4569. doi: 10.1128/mcb.8.10.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou-Pachnis A., Tsichlis P. N. Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res. 1985 Dec 9;13(23):8379–8387. doi: 10.1093/nar/13.23.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. J., Grula J. W., Davidson E. H., Britten R. J. Evolution of sea urchin non-repetitive DNA. J Mol Evol. 1980 Dec;16(2):95–110. doi: 10.1007/BF01731580. [DOI] [PubMed] [Google Scholar]
- Hwu H. R., Roberts J. W., Davidson E. H., Britten R. J. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn R. M., Fritsch E. F., Parker R. C., Blake G., Maniatis T. The isolation and characterization of linked delta- and beta-globin genes from a cloned library of human DNA. Cell. 1978 Dec;15(4):1157–1174. doi: 10.1016/0092-8674(78)90043-0. [DOI] [PubMed] [Google Scholar]
- Lehrman M. A., Russell D. W., Goldstein J. L., Brown M. S. Alu-Alu recombination deletes splice acceptor sites and produces secreted low density lipoprotein receptor in a subject with familial hypercholesterolemia. J Biol Chem. 1987 Mar 5;262(7):3354–3361. [PubMed] [Google Scholar]
- Lin C. S., Goldthwait D. A., Samols D. Identification of Alu transposition in human lung carcinoma cells. Cell. 1988 Jul 15;54(2):153–159. doi: 10.1016/0092-8674(88)90547-8. [DOI] [PubMed] [Google Scholar]
- Maeda N., Bliska J. B., Smithies O. Recombination and balanced chromosome polymorphism suggested by DNA sequences 5' to the human delta-globin gene. Proc Natl Acad Sci U S A. 1983 Aug;80(16):5012–5016. doi: 10.1073/pnas.80.16.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M. M., Slightom J. L., Goodman M. Phylogenetic relations of humans and African apes from DNA sequences in the psi eta-globin region. Science. 1987 Oct 16;238(4825):369–373. doi: 10.1126/science.3116671. [DOI] [PubMed] [Google Scholar]
- Quentin Y. The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol. 1988;27(3):194–202. doi: 10.1007/BF02100074. [DOI] [PubMed] [Google Scholar]
- Sawada I., Willard C., Shen C. K., Chapman B., Wilson A. C., Schmid C. W. Evolution of Alu family repeats since the divergence of human and chimpanzee. J Mol Evol. 1985;22(4):316–322. doi: 10.1007/BF02115687. [DOI] [PubMed] [Google Scholar]
- Slagel V., Flemington E., Traina-Dorge V., Bradshaw H., Deininger P. Clustering and subfamily relationships of the Alu family in the human genome. Mol Biol Evol. 1987 Jan;4(1):19–29. doi: 10.1093/oxfordjournals.molbev.a040422. [DOI] [PubMed] [Google Scholar]
- Trabuchet G., Chebloune Y., Savatier P., Lachuer J., Faure C., Verdier G., Nigon V. M. Recent insertion of an Alu sequence in the beta-globin gene cluster of the gorilla. J Mol Evol. 1987;25(4):288–291. doi: 10.1007/BF02603112. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Waring M., Britten R. J. Nucleotide sequence repetition: a rapidly reassociating fraction of mouse DNA. Science. 1966 Nov 11;154(3750):791–794. doi: 10.1126/science.154.3750.791. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Willard C., Nguyen H. T., Schmid C. W. Existence of at least three distinct Alu subfamilies. J Mol Evol. 1987;26(3):180–186. doi: 10.1007/BF02099850. [DOI] [PubMed] [Google Scholar]



