Abstract
Cortical development depends upon tightly controlled cell fate and cell survival decisions that generate a functional neuronal population, but the coordination of these two processes is poorly understood. Here we show that conditional removal of a key apical complex protein, Pals1, causes premature withdrawal from the cell cycle, inducing excessive generation of early-born postmitotic neurons followed by surprisingly massive and rapid cell death, leading to the abrogation of virtually the entire cortical structure. Pals1 loss shows exquisite dosage sensitivity, so that heterozygote mutants show an intermediate phenotype on cell fate and cell death. Loss of Pals1 blocks essential cell survival signals, including the mammalian target of rapamycin (mTOR) pathway, while mTORC1 activation partially rescues Pals1 deficiency. These data highlight unexpected roles of the apical complex protein Pals1 in cell survival through interactions with mTOR signaling.
Keywords: apical complex, Pals1, mTOR, neural cell fate, cell polarity, cell death
INTRODUCTION
The cerebral cortex is formed by the orderly generation of postmitotic neurons through symmetric and asymmetric cell division of neural progenitors during neurogenesis (Farkas and Huttner, 2008; Gotz and Huttner, 2005; Huttner and Kosodo, 2005; Miyata et al., 2001). The proliferation of cortical progenitor cells, followed by their irreversible depletion near the time of birth, is tightly orchestrated by cellular and molecular events that balance the generation of early-born neurons with the maintenance of progenitors for later born neurons (Caviness et al., 1995; Gotz and Huttner, 2005; Takahashi et al., 1996). Two neural progenitor cell types are identified in the developing cortex. One is a radial neuroepithelial cell, with a cell body in the ventricular zone and an apical process that inserts into the ventricular lining, and a long, thin basal processes that reaches the pial surface at the outside of the brain (Anthony et al., 2004; Fishell and Kriegstein, 2003; Noctor et al., 2001; Noctor et al., 2002; Tamamaki et al., 2001). These radial cells serve both as progenitors and migratory guides for newly born neurons. The other progenitor cell type is a more recently characterized basal progenitor, which localizes primarily to the subventricular zone (SVZ) and undergoes one or more cell divisions, typically symmetrically, to generate neurons in the cerebral cortex (Haubensak et al., 2004; Kowalczyk et al., 2009; Miyata et al., 2004; Noctor et al., 2004; Noctor et al., 2008). Radial neuroepithelial progenitors form an epithelial structure with their apical, ventricular processes connected to adjoining cells by adherens junctions. Although considerable progress has been made in understanding the cellular events of radial glial progenitor cell division, the molecular control of cell fate decisions remains poorly understood.
Recent work suggests that similar functions of apical complex proteins in cell fate determination may unite flies and mammals (Doe, 2008; Wodarz, 2005). The apical polarity complex of flies, which includes the proteins Bazooka (Par3), Par6, and aPKC, restricts cell fate determinants to one side of neural stem cells, termed neuroblasts, such that these determinants are inherited preferentially by one daughter cell that retains the neuroblast cell fate. On the other hand, the basal complex proteins, Lgl/Dlg/Scb and Numb, are segregated to the other daughter cell, termed the ganglion mother cell (Roegiers and Jan, 2004; Wodarz, 2005), which is an intermediate progenitor that generates two neurons or two glial cells. Genetic studies of the vertebrate Numb orthologues have uncovered a number of roles in neurogenesis (Li et al., 2003; Petersen et al., 2002; Petersen et al., 2004; Rasin et al., 2007). However, in contrast to its role in invertebrates, deletion of aPKCλ in mice at E15 (midway through neurogenesis) did not clearly affect cell fate decisions in a fashion comparable to its role in invertebrates (Imai et al., 2006). On the other hand, mouse Lgl1 mutants showed hyperproliferation of progenitors in the brain, in conjunction with Numb mislocalization (Klezovitch et al., 2004). Recent work suggests that Par3 is segregated asymmetrically in cortical neuroblasts, and also serves as a cell autonomous regulator of Notch signaling (Bultje et al., 2009). In addition, knockdown and overexpression studies of Par3 and Par6 (Costa et al., 2008) and conditional knockout of their upstream regulator Cdc42 (Cappello et al., 2006) suggest that the apical complex proteins are essential for self-renewal of neural progenitors in the developing mammalian cortex. While it seems likely that evolutionarily conserved polarity complex proteins have roles in proliferation control, whether they also play a role in maintaining neural progenitor cell fate has not been shown genetically.
Here, we study a functionally related apical complex protein Pals1 (Protein associated with Lin7), and extend the role of the apical complex to control cell survival as well. Pals belongs to the MAGUK (Membrane Associated Guanylate Kinase) family of proteins, which are scaffolding factors containing both postsynaptic density 95/discs large/zona occludens-1 (PDZ) and Src homology 3 (SH3) domains (Kamberov et al., 2000). Pals1 also contains two L27 domains that bind PatJ and Lin7, both critical components of tight junction formation and maintenance. The PDZ domain of Pals1 links it to Crb1, 2, and 3, Crumbs vertebrate homologues, which encode transmembrane proteins with EGF-like repeats, thus forming a tripartite PatJ-Pals1-Crbs complex (Roh et al., 2002a). In addition, an evolutionarily conserved domain, whose structure is not yet understood, mediates binding of Pals1 to the Par6-Par3-aPKC apical complex. Thus, Pals1 links these two evolutionary conserved apical complex signaling pathways (Hurd et al., 2003). The critical function of Pals1 (nagie oko in zebrafish) is well established in epithelial polarity and adherens junction assembly in mammalian cells, in zebrafish embryos and in invertebrates (Bachmann et al., 2001; Hong et al., 2001; Straight et al., 2004; Wei and Malicki, 2002), but a role for Pals1 has not been studied in cell fate decisions.
In this study, we show that Pals1 loss causes defects not only in cell fate decisions, but also surprisingly in cell survival, and use genetics to elucidate key downstream effectors of these roles. We find that absence of Pals1 leads to the depletion of progenitor cells by premature withdrawal from the cell cycle, generating excessive early-born postmitotic neurons. Unexpectedly however, Pals1-deficient cells undergo massive and rapid cell death, which leads to the total abrogation of almost the entire cortical structure. Since accumulating evidence has implicated mammalian target of rapamycin (mTOR) pathway components and polarity proteins (Massey-Harroche et al., 2007; Pinal et al., 2006; von Stein et al., 2005), we investigated the genetic relationship between Pals1 and mTOR signaling. Activation of the mTOR signaling pathway by elimination of a negative regulator, Tuberous sclerosis complex subunit 2 (Tsc2), partially restores the medial cortex in Pals1 mutants, suggesting genetic interaction between the two signaling pathways. Taken together, our findings identify a new connection between the apical complex and mTOR signaling that couples cell fate and cell survival during cortical development.
RESULTS
Pals1 is essential for histogenesis of the mammalian cortex
Several aspects of Pals1 expression suggested that Pals1 plays an important role during mammalian neurogenesis. First, Pals1 expression in cortical progenitors was very high during the period of neurogenesis, and rapidly downregulated over the course of neurogenesis so that Pals1 mRNA and protein were significantly reduced by P0, when neurogenesis nears completion (Supplementary Figure S1A)(Ishiuchi et al., 2009). In wild type mice, Pals1 localized apically in the cortical neuroepithelium along the ventricular surface, and its expression overlapped extensively with members of both apical polarity complexes, including Crb2, aPKCζ, and Patj (Supplementary Figure S2A). In addition, several apical complex proteins co-immunoprecipitated with Pals1 from E13 forebrain lysates (Supplementary Figure S2A), suggesting that murine apical complex proteins physically interact as in other species (Hurd et al., 2003). Apical proteins localized adjacent to adherens junctions marked by β-catenin (Supplementary Figure S2A), reflecting an intimate link between the apical complex and adherens junctions in the developing brain.
We removed Pals1 in mouse embryos using a conditional mutation created by inserting LoxP sites into introns 2 and 3 of the mouse Pals1 gene (Supplementary Figure S1B), since complete loss of Pals1 was lethal at early embryonic ages (data not shown). Pals1 floxed homozygote neonates and adults showed no identifiable phenotype and had normal lifespan and breeding. Cre-mediated recombination removed exon 3, resulting in a nonsense mutation with early truncation of the 867-amino acid Pals1 protein at amino acid 122 (Supplementary Figure S1C), removing most of Pals1’s known functional domains (Roh et al., 2002b). Deleting Pals1 using Emx1-Cre (Pals1loxp/loxp: Cre+ (CKO) animals), which drives Cre-mediated recombination in cortical progenitors of medial cortex and hippocampus (Gorski et al., 2002), resulted in undetectable Pals1 expression by E11 (Supplementary Figure S1D) in progenitor cells of these structures, confirmed by immunostaining with three distinct antisera (Supplementary Figure S1D and data not shown)(Chae et al., 2004; Roh et al., 2002b).
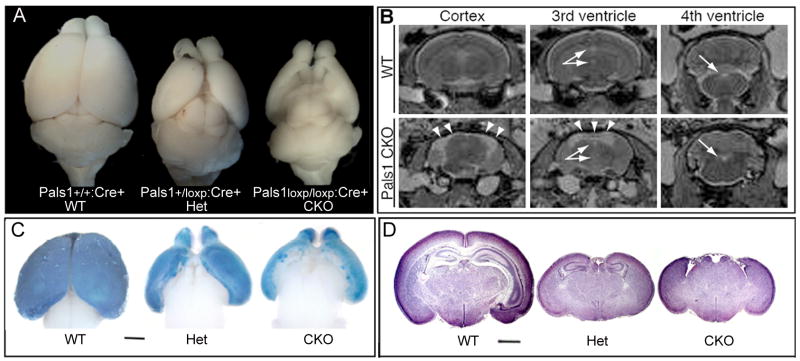
Surprisingly, given the expected role of apical complex proteins in cell fate determination, Emx1-Cre mediated removal of Pals1 (Pals1 CKO) produced not merely a smaller cortex, but a cortex that was essentially completely absent, lacking virtually all cortical neurons. Extreme thinning was seen in lateral cortex, where some Pals1 expression was retained due to weaker or delayed Cre expression (Figure 1A-D)(Gorski et al., 2002). Heterozygote Pals1loxp/+:Cre+ animals (Het) also showed an extremely small cortex, with small residual medial cortical structures, including the hippocampus. Magnetic resonance imaging (MRI) revealed that the space typically occupied by the neocortex in the wild type mouse was replaced by a fluid-filled cystic space contiguous with the lateral ventricles in the Pals1 CKO mouse (Figure 1B). There was no apparent change in the size of the 3rd and 4th ventricles, where Cre was not expressed (data not shown). Therefore, in contrast to Drosophila Stardust, which is not required for neuroblast cell fate determination (Bachmann et al., 2001; Hong et al., 2001), Pals1 is required for normal histogenesis of the cerebral cortex, and interestingly this requirement is dose-dependent with milder heterozygote phenotypes.
Figure 1. Cerebral cortex-specific ablation of Pals1 causes severe cortical malformation.
(A) Complete loss of medial cortex is seen after cortical-specific deletion of Pals1, using Emx1Cre (Figure S3). The heterozygote cortex is smaller, though lateral cortex is relatively spared. (B) Representative rostral, medial, and caudal 2T weighted MRI sections of wild type (WT) and Pals1 CKO adult mice. Arrows point to intact 3rd and 4th ventricles. Arrowheads point to a fluid-filled cystic space in Pals1 CKO mice that would normally be occupied by the cerebral cortex in WT mice. (C) X-gal labeling at E16 show that areas with strong Cre expression in normal brain are absent in Pals1 CKO; Rosa26 brain. (D) Histology of the whole brain at P4 shows a reduced brain size in both Pals1 Het and Pals1 CKO animals. The medial structures are completely absent in Pals1 CKO animals. The size bar shows 1mm. See also Figure S1.
Preserved and defective behaviors in Pals1 CKO mice
Remarkably, Pals1 CKO mice survive into adulthood, representing an adult mouse with virtually no cerebral cortex. We found that Pals1 CKO mice survived, fed, and bred, in the laboratory environment, and in general appeared surprisingly healthy, though their overall body weight was lower than age-matched wild type (WT) controls (Figure 2A), and their fur appeared less kempt in adulthood (data not shown). Basic neurological reflexes, such as the righting reflex, appeared intact (data not shown). Pals1 CKO mice responded to tail pinch, and there were no detectable differences in their speed to tail withdrawal in the hot water tail flick test when compared to Het or WT controls (data not shown). Preservation of the Preyer reflex indicated that the Pals1 CKO and Het mice registered auditory cues (data not shown). Perhaps most remarkably, considering the ablation of the entire frontal motor cortex, no differences were detected between groups of mice in a motor coordination test using the accelerating rotarod, in either acquiring the task (data not shown) or in test day performance (Figure 2B). Hence, Pals1 CKO mice uncover a series of behaviors that are governed predominantly by cortex-independent circuitry.
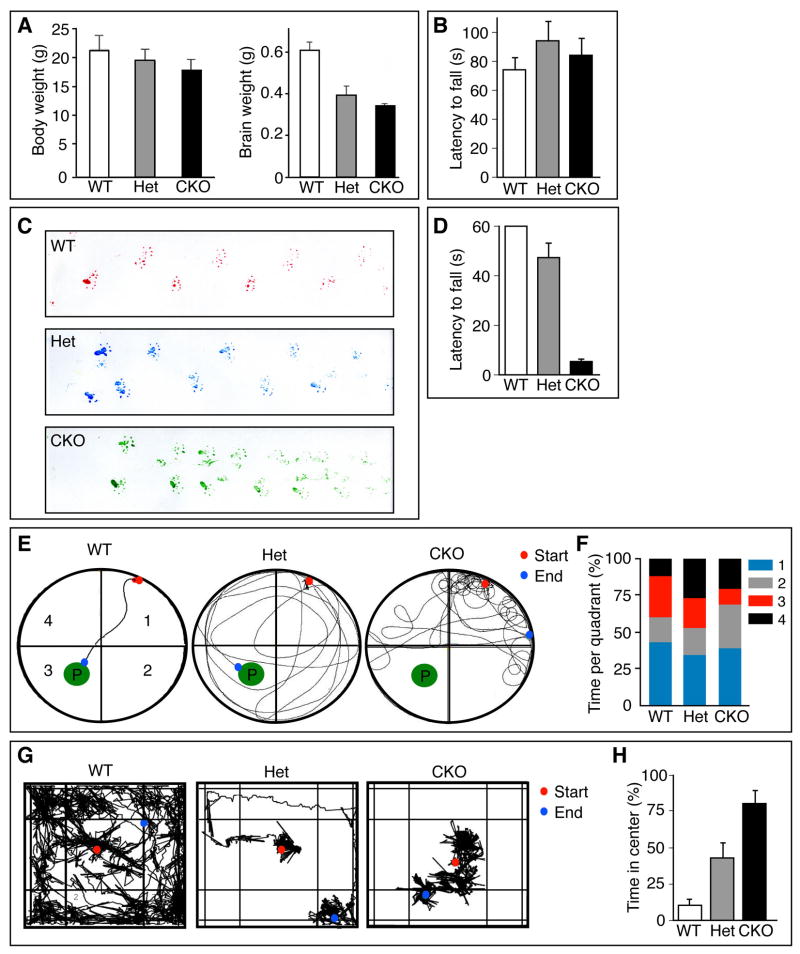
Figure 2. Pals1 loss reveals dorsal cortex-dependent and -independent behaviors.
(A) Body and brain weight of littermates at 4 weeks of age. The brain mass of Pals1 CKO and Pals1 Het mice are smaller than WT (t-test, p<0.001; n=3–5). (B) Mice tested on the accelerating rotarod showed no significant differences between Pals1 CKO, Pals1 Het, or control mice. (C) Similar footprint patterns of wild type (top), Pals1 Het (middle), and Pals1 CKO (bottom) mice. (D) Pals1 CKO mice had reduced latency to fall in the wire hang test versus controls and Pals1 Het mice (t-test: p<0.0005 and p<0.05, respectively, n=10). (E) Representative traces of swimming patterns of WT (left), Pals1 Het (middle), and Pals1 CKO (right) mice in a Morris water maze where mice swam towards a visible platform (P) located in quadrant 3. (F) Quantification of time spent per quadrant during swimming trial in (E). Pals1 CKO mice spent the least time in the quadrant with the platform versus wild type and Pals1 Het mice (t-test: p<0.005; n=10). (G) Representative traces of exploratory behavior of WT (left), Pals1 Het (middle), and Pals1 CKO (right) mice during a 10 minute open field trial. (H) Quantification of time spent in center quadrant during open field test (G). Pals1 Het and Pals1 CKO mice spent less time exploring the open field versus WT controls (t-test: p<0.05 and p<0.0005, respectively; n=10).
Not surprisingly, Pals1 CKO mice exhibited severely compromised behaviors in several tests previously shown to depend upon intact cortical circuitry. For example, their movements were irregular and jumpy, disrupting their stride and gait (Figure 2C). Pals1 CKO performed poorly in the wire hang (Figure 2D). Pals1 CKO mice swam; however while there was no significant difference between the groups of mice in swimming speed, Pals1 CKO mice did so in a characteristic circling pattern (Figure 2E). They typically failed to locate a visible platform in the Morris water maze (success rates: +/+ = 10/10; +/− = 5/10; −/− = 4/10; Figure 2E), as if they were blind to it, and spent less time searching for the visible platform in the appropriate quadrant than Het or WT controls (Figure 2F). Pals1 CKO mice also failed in the visual forepaw reach test (data not shown). Finally, Pals1 mutant mice had reduced exploratory initiative and locomotor behavior in the open field test (Figure 2G). After 3 minutes in the open field, 6/10 of Het mice, and 9/10 of Pals1 CKO mice remained in the center quadrant while 10/10 WT control mice had by this time started exploring the novel environment. Neither Pals1 CKO nor Het mice were noticeably distressed in the center of the field. During the course of the full 10 minute trial, both Pals1 CKO and Het mice preferred the center quadrant over any other quadrant (Figure 2H). In fact, 4/10 of the Pals1 CKO mice remained in the center quadrant for the entire duration of the experiment. Taken together, these data demonstrate a broad range of behavioral deficits arising from the absence of appropriate cortical feed-forward and feedback circuitry in the Pals1 CKO mouse. In addition, the Het displays an intermediate behavioral phenotype, underscoring the importance of appropriate Pals1 gene dosage during cortical development.
Pals1 is necessary and sufficient for cortical progenitor self renewal
In order to investigate the reason for the absence of almost the entire cortex, we examined Pals1 CKO animals and noted striking changes in cortical size and morphology at E12 and E14 (Figure 3A, B). Consistent with the proposed role of the apical complex in regulating cell fate (Bultje et al., 2009; Costa et al., 2008), Pals1 CKO animals did show dramatic changes in progenitor cell renewal and cell fate (Figure 3C-G). S-phase cells, marked by BrdU incorporation, were diminished almost two-fold compared to the WT. M-phase cells, marked by phospho-Histone H3 (PH3) immunoreactivity (Figure 3D, E, F), were similarly reduced at E12.5 and E14.5, with Hets showing intermediate numbers in both cases. To determine the cause of progenitor cell loss, we labeled progenitor cells with BrdU at E10-11, and labeled those progenitors still cycling 18–24 hours later by immunostaining with Ki67, as previously described (Chenn and Walsh, 2002). Of the progenitors labeled at E10, 15.7% of Pals1 CKO progenitors exited the cell cycle on E11 (and were hence not double-labeled with Ki67), compared to 8.5% in wild type and 9.7% in Hets, suggesting that Pals1-deficient progenitors prematurely withdraw from the cell cycle. Mutant progenitors labeled at E11 exited the cell cycle by E12 at 2.3-fold the rate of normal progenitors (Figure 3G and H). Furthermore, postmitotic neurons (marked by Reelin, Dcx, Map2 and Tbr1) were increased at E11 and E12 transiently in CKO brains (Supplementary Figure S3A-F), suggesting that some progenitor cells exit the cell cycle to become neurons in the absence of Pals1. Basal progenitors, i.e., intermediate progenitors located in subventricular zone and labeled by Tbr2 immunoreactivity, were also significantly reduced in number at E12 and later stages (Supplementary Figure S3G-J).
Figure 3. Pals1 deficiency depletes the neural progenitor pool by premature cell cycle exit.
(A) The Pals1 CKO cortex is smaller than littermate controls at E12. (B) At E14, both Pals1 Het and Pals1 CKO cortices are extremely small compared to controls. (C) A schematic of interkinetic movement of neural progenitors. (D) S-Phase cells transiently labeled by BrdU, and M-phase cells labeled with anti-P-H3 are substantially reduced in the Pals1 CKO brain. Intermediate reduction is seen in Pals1 Het embryo, compared to WT at E12 and E14 (D, E, F, respectively)(t-test; BrdU at E12, p<0.001; BrdU at E14, p<0.005; P-H3 at E12, p<0.01; P-H3 at E14, p<0.01; n=3 and n=4). (G, H) Neural progenitors exiting the cell cycle after 24 hours are double-labeled by BrdU and Ki67. Cells exiting the cell cycle are positive for BrdU, but negative for Ki67, and are increased in the Pals1 CKO versus controls. See also Figure S3.
To determine if these defects in self-renewal are cell-autonomous, we employed shRNA technology to acutely knockdown Pals1. Pals1 shRNA effectively reduced the expression of Pals1 in cultures of NIH 3T3 cells and in electroporated cells in vivo (Supplementary Figure S4A, B and data not shown)(van Rossum et al., 2006). Pals1 shRNA effects were rescued by co-electroporating a shRNA resistant human PALS1 expression vector, indicating that the defects were specific to Pals1 (Supplementary Figure S4C). Two days following shRNA administration, endogenous Pals1 was undetectable by Pals1 immunostaining in the electroporated cortical area (Supplementary Figure S4A). Progenitors electroporated with Pals1 shRNA also showed defects in self-renewal as revealed by PH3 or transient BrdU labeling compared to control (Figure 4A-J, arrow). Furthermore, greater numbers of shRNA treated cells were positive for the neuronal marker Tuj1 compared to control shRNA treated cells, indicating a cell autonomous generation of neurons accompanying reduction of Pals1 (Figure 4K-N).
Figure 4. Pals1 is necessary and sufficient for self-renewal of neural progenitors.
(A-E) Progenitors in mitosis (P-H3-positive staining cells) are decreased in the Pals1 silenced (Pals1 shRNA) cortical region, and dispersed from the apical margin. (A) Non-target shRNA vectors were used as controls. (B-E) The proportion of P-H3-positive cells in the Pals1 shRNA treated cells (GFP-positive) reduced to 17% as compared to control 21% (t-test; p<0.005; n=5). (G-J) Reduction and scattering of S-phase proliferative cells in Pals1 shRNA tissue was analyzed 1-hr after a BrdU pulse. Arrows (panels C, H) separate electroporated regions from adjacent nonelectroporated regions. (J) Quantification of BrdU-positive cells among shRNA treated cells reveals fewer S-phase proliferating cells upon Pals1 shRNA versus control (t-test; p<0.0001; n=5). (L-N) Pals1 downregulation increases Tuj1-positive staining neurons compared to control, indicating a premature cell-cycle exit of cortical progenitors. GFP and Tuj1 immunoreactivity largely overlap, indicating a substantial cell-autonomous impact of Pals1 silencing in promoting neuronal differentiation. (O, S) E13.5 embryos were electroporated with GFP (O, S) or pCAGGS-Pals1 (P, Q, R, T), and cortical tissue was analyzed at P3. (O, S) GFP electroporated neurons are clustered in the cortical plate. (P, T) Conversely, Pals1 overexpressing cells are scattered more widely in the cortical and subcortical regions, suggesting abnormalities in exiting the VZ, and potentially in neuronal migration. A small fraction of cells were still localized in the SVZ, suggesting failure to leave the proliferative regions (arrows). (Q) High magnification images show Pals1 overexpression. (R) Example of Pals1 overexpressing cells confined to the SVZ. These cells continue to express Ki67, indicating their persistent proliferative state. (S, T) Higher magnification images of the boxed areas in O and P respectively. (U, V) Two days after pCAGGS-Pals1 electroporation, Pals1 is highly expressed in the cytoplasm and in the apical junction in overexpressing cells. (W, X) High magnification pictures of boxed areas of V. See also Figure S4.
We next tested if overexpression of Pals1 in neural progenitors is sufficient to maintain progenitors in the proliferating state. Cortical progenitor cells electroporated at E13 and examined later at postnatal day 3 (P3) revealed that the majority of Pals1-overexpressing cells were located in the cortical plate (CP). However, some Pals1-overexpressing cells persisted in proliferative regions such as the SVZ (Figure 4P, arrows). By contrast, no Pals1 expressing cells were observed in the SVZ in control experiments, and all control cells were tightly localized in the cerebral cortex by P3 (Figure 4O). The persistent SVZ cells overexpressing Pals1 were frequently immunoreactive for Ki67 (64% ± 8 %) while no Ki67-positive cells were found in the CP (Figure 4Q, R). The cells in the CP appeared to be postmitotic neurons based on their typical neuronal morphology and the fact that they were Ki67 negative. Pals1 overexpressing cells were also more widely dispersed in the cortical wall than in control electroporated cells (Figure 4O, P, S, T), suggesting that Pals1 overexpression may delay or disrupt cell cycle exit and/or normal cortical migration. When overexpressed, Pals1 localized to apical junctions as well as the cytoplasm (Figure 4U-X), so that additional signaling roles of Pals1 beyond the maintenance of the polarized structure of progenitors cannot be completely ruled out. Taken together, these findings indicate that Pals1, like Par3 (Bultje et al., 2009; Costa et al., 2008) is both necessary and sufficient to maintain progenitors in the cycling state.
Pals1 removal causes widespread apoptotic cell death
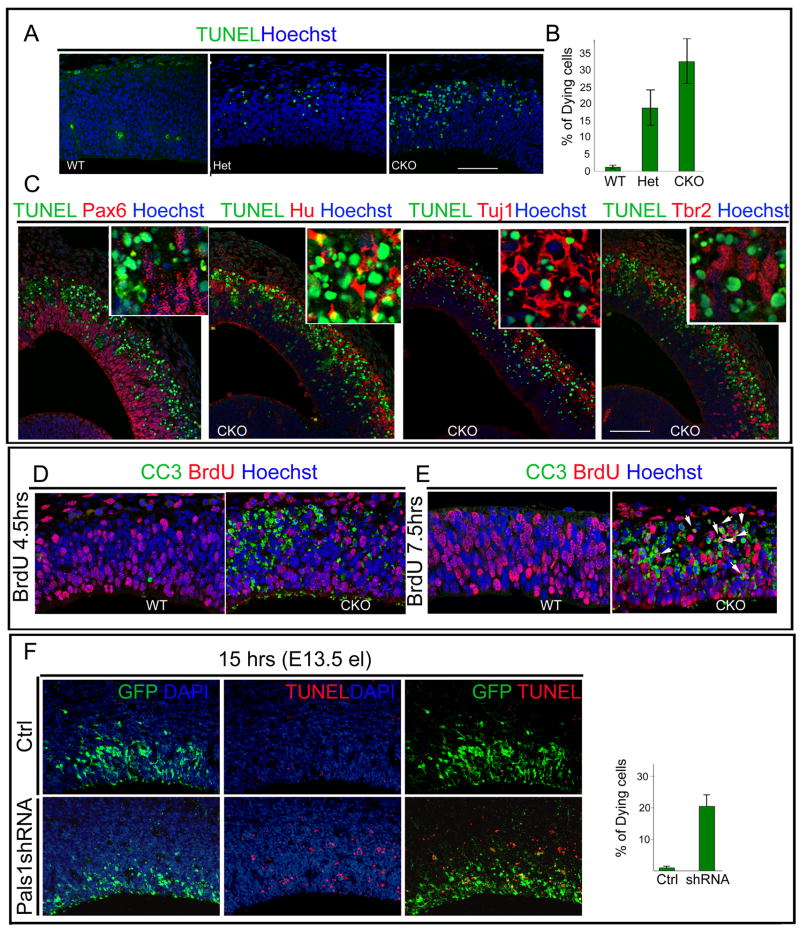
Though Pals1 regulates cell fate, the profound loss of medial cortex in the Pals1 CKO suggested that additional mechanisms beyond merely a cell fate change must contribute, and we found that massive apoptotic cell death took place in the developing mutant medial cortex. Cell death could be observed in Pals1 CKO mice as early as E10.5 (Figure 5, data not shown). In order to determine the types of cells dying, we first visualized dying cells by TUNEL staining. TUNEL-positive cells primarily localized dorsally in the intermediate zone and cortical plate in Pals1 CKO mice, where extensive cell death peaked at E11-12 (Figure 5A, B and data not shown). Double staining with the nuclear neuronal marker Hu revealed many TUNEL-Hu double-positive cells, suggesting that cell death involved many postmitotic neurons. The neuronal identity of TUNEL-positive cells was also consistent with their location outside of the VZ where they overlapped with cells staining with the neuronal marker Tuj1 (Figure 5C). In contrast, dying cells could be rarely, if ever, double-labeled with progenitor markers such as Pax6 and Tbr2 (Figure 5C).
Figure 5. Pals1 deficiency causes massive and rapid cell death of abnormally generated postmitotic neurons.
(A, B) More cells in the Pals1 CKO undergo apoptotic cell death than in Pals1 Het or WT littermates, as seen by TUNEL staining at E12. Dying cells are observed by E10 and peak at E11-E12 (ANOVA; WT vs. Het p<0.05; WT vs. CKO p<0.0005; Het vs. CKO p<0.05; n=3 and n=5). (C) The cells undergoing apoptotic cell death are primarily neuronal, as shown by double-labeling with progenitor markers, (apical progenitors: Pax6; basal progenitors: Tbr2) and the nuclear neuronal markers Hu, Tuj1 together with TUNEL staining. Hu and Tuj1 extensively overlap with TUNEL-stained dying cells while progenitor markers rarely overlap with TUNEL-positive staining cells. The size markers represent 75μm. (D) The cells in M phase or G2 phase, which are labeled by a 4.5 hour BrdU pulse, rarely overlap with cleaved caspase 3 (CC3)-positive staining cells. (E) Cells labeled by a 7.5 hour BrdU pulse are mainly in G1 or exiting the cell cycle. In the Pals1 CKO brain, most BrdU-positive cells double label with CC3 (E, arrow). (F) Pals1 knockdown cells acutely undergo apoptotic cell death since over 20% of Pals1 shRNA-treated cells are TUNEL-positive as early as 15 hrs after electroporation. See also Figure S5.
Cleaved Caspase 3 (CC3), an earlier marker of apoptosis, also identified dying cells predominantly outside of the VZ. However in contrast to TUNEL staining, CC3-positive cells were also found in the VZ even at the apical surface as early as E11. Pax6 and CC3 immunoreactivity co-localized only very rarely (Supplementary Figure S5), further suggesting that most dying cells are recently born neurons. However, we cannot rule out a contribution of progenitor cells to the apoptotic population since 9 hours after Pals1 knockdown, most GFP-positive cells, which were apoptotic, were weakly Pax6-positive (data not shown). Double-labeling studies using CC3 and BrdU also suggested that dying cells had mainly withdrawn from the cell cycle. Following a BrdU pulse at E11, we examined dying cells either after 4–5 hours, at which time BrdU labeled cells are primarily in M-phase and G2-phase (Takahashi et al, 1995) or after 7.5 hours. CC3-positive cells rarely overlapped with BrdU 4–5 hours after labeling (Figure 5). By contrast, 7.5 hours following the BrdU pulse, we observed BrdU and CC3 double-labeled cells (Figure 5D, E, arrows), suggesting that the majority of dying cells were exiting the cell cycle. Similar to genetic ablation, some neurons generated by shRNA-mediated Pals1 knockdown underwent apoptotic cell death, and could be double-labeled with TUNEL (Figure 5F). Already 15 hours following Pals1 knockdown, over 20% of Pals1 knockdown cells were TUNEL-positive, with the proportions of dying cells reducing slightly with time (Supplementary Figure S5). Pals1 may be essential in the transition from progenitor to neuron, or at the very earliest stages of neuronal differentiation, since removal of Pals1 in postmitotic neurons using synapsin Cre, or RNAi-mediated knockdown of Pals1 in cortical neurons in vitro, did not result in massive neuronal death (data not shown). Taken together, these data highlight a new and unexpected role for Pals1 in neurons and neural progenitors in cell survival, implicating potential interactions between the apical complex and cell survival pathways.
Pals1 removal disrupts other apical and basolateral components
Removal of Pals1 disrupted the in vivo expression and localization of all apical complex proteins examined, suggesting that Pals1 is a central hub for apical complex proteins. Crb2 and aPKCλ immunoreactivity were already diminished in E11-12 CKO mice, the earliest date we observed reduction of Pals1 expression (Figure 6A, B). In time-course experiments using shRNA-electroporated mouse embryonic cortices, Crb2 mislocalization occurred 9 hours after electroporation (Figure 6C), whereas aPKC localization was normal at 9 hours after RNAi, but became abnormal at later time points, such as 48 hours post-electroporation (Supplementary Figure S2). These findings suggest that, while Pals1 links both Par3/Par6/aPKC and Pals1/Crb/Patj apical complexes, one of the earliest defects associated with Pals1 loss is a disruption of the Pals1/Crb/Patj apical complex (Figure 6C).
Figure 6. Pals1 loss disrupts apical complex proteins and adherens junctions.
(A) The expression of Pals1 in the neuroepithelium at E12 is diminished in a medial to lateral gradient in Pals1 CKO mutant mice versus WT. (B) At E12, the apical complex proteins aPKCλ and Crb2 were reduced in an analogous manner to the Pals1 loss in the Pals1 CKO. (C) Defects of Crb2 protein localization are observed as early as 9 hrs after Pals1 shRNA electroporation. The Crb2 protein diffuses in the cytoplasm in the shRNA treated cells, whereas the cells treated with control shRNA show apical localization. (D) Adherens junctions are relatively intact at E12 in Pals1 CKO mutant mice. At E13, adherens junctions are either displaced basally, or absent in Pals1 CKO mice. At E14, adherens junctions are completely absent in medial cortex of mutant mice. (E) Top panels: Scanning EM reveals fewer primary cilia in the Pals1 CKO neuroepithelium compared to wild type (arrowheads). Middle panels: Pericentrin staining was reduced in Pals1 CKO mice, indicating a reduction in basal bodies (the base of cilia). Lower panels: Pals1 CKO mice have shorter cilia than WT controls. In the Pals1 CKO, the apical membrane shows large and irregularly shaped particles compared to controls, which have smaller and relatively uniform particles at E12 (arrows). See also Figure S2.
Pals1 has also been implicated in adherens junction formation in cultured cells (Wang et al., 2006), and adherens junction components such as β-catenin (Chenn and Walsh, 2002; Zechner et al., 2003) and α(E)-cadherin (Lien et al., 2006) regulate neural progenitor renewal. We first detected adherens junction defects in Pals1 CKO animals at E13, at which point adherens junctions were either displaced basally or absent altogether (Figure 6D). Adherens junction loss appeared to follow the loss of apical protein immunoreactivity by about 24 hours, as adherens junctions appeared relatively intact at E12 (Figure 6D). Adherens junction defects were also observed in Pals1 knockdown cells with a time course mirroring that observed in Pals1 CKO mice (data not shown).
Disruption of the apical complex also led to other apical defects. Basal bodies marking the base of cilia identified by pericentrin staining were reduced (Figure 6E), and scanning EM revealed that primary cilia were reduced in number and size in Pals1 CKO compared to wild type controls (Figure 6E). Together, these data suggest that loss of apical complex leads to impaired adherens junctions, as well as apical structures.
Loss of mTOR activity is in part responsible for postmitotic neuron apoptosis
In order to understand Pals1’s role in cell survival, we examined alterations in signaling pathways that regulate cell survival. Several recent studies suggest that cell polarity and the mammalian target of rapamycin (mTOR) pathway, a major cell growth and survival pathway, have physical and functional interactions (Feng et al., 2008; Pinal et al., 2006). We used the level of phosphorylated ribosomal S6 protein (pS6), which promotes the synthesis of protein upon phosphorylation by activated mTORC1, as a measure of mTORC1 activity (Sarbassov et al., 2005). Pals1 CKO animals showed striking down-regulation of pS6 levels in neurons marked by Tuj1 (Figure 7A). To quantify the postmitotic cells that were pS6-positive, we employed the double staining of nuclear staining of cell cycle inhibitor p27 (Kip1) immunostaining (Nguyen et al., 2006), which extensively overlapped with early neuron markers, Tbr1 at E11 (data not shown) and pS6 (Figure 7B). While 71% of p27 (Kip)-positive cells were pS6-positive in wild type controls, only 30% of p27 (Kip1)-positive cells were pS6-positive in Pals1 CKOs and 48% of p27 (Kip1)-positive cells were pS6-positive in Pals1 Het animals, (Figure 7C), suggesting that Pals1 loss impairs mTOR signaling in neurons in a dose-dependent manner. Thus, Pals1 function appears essential, presumably at the transition from progenitor to neuron, to form neurons capable of normal mTOR signaling.
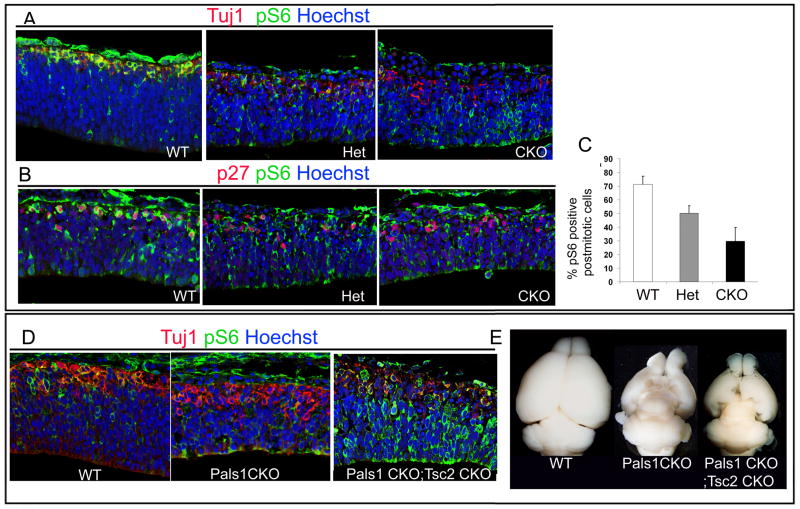
Figure 7. mTOR activity is attenuated in Pals1 CKO neurons.
(A-C) Abnormally generated neurons marked by Tuj1 (A) and p27 (B, C) in the Pals1 CKO mice show reduced or absent pS6 expression, indicating attenuation of mTOR activity compared to WT littermates at E11. In control animals, 72% of p27-positive cells express pS6 while in the Pals1 Heterozygote and Pals1 CKO mice, 48% and 30% of p27-postive cells overlap with pS6 respectively (t-test, p<0.05; n= 3 for WT n=3 for Het and n=5 for CKO). (D, E) The activation of mTOR by elimination of a negative regulator, Tsc2, genetically restores pS6 staining in postmitotic neurons marked by Tuj1 in the double mutants (Pals1 CKO; Tsc2 CKO; Emx1Cre) at E12 compared to single mutant (Pals1 CKO). (E) At P21, the medial cortex of the Pals1; Tsc2 double mutant is partially restored compared to the Pals1 CKO single mutant in which the medial cortex is almost absent.
To test whether activation of mTOR could rescue the dying cells in the Pals1 CKO animals, we created a double mutant of Pals1 and a Tsc2 floxed allele, both under the control of Emx1Cre (Gorski et al., 2002). Tsc2 negatively regulates mTOR through inhibition of Rheb, a small GTPase protein, by its GAP (GTPase activating protein) activity, so that the removal of Tsc2 activates mTORC1 by Rheb activation (Figure 7D). At E12, the expression of pS6 was increased in the Tsc2; Pals1 double mutant and remarkably, the expression of pS6 in the postmitotic neurons was restored in the double mutant. At postnatal day 21, unlike the Pals1 CKO animals which do not have any medial cortex, double mutants partially restored the medial cortex of Pals1 mutants, in which both early-born neurons and late-born neurons were found (Figure 7E, data not shown). These data suggest that activation of mTOR signaling by Tsc2 mutation can partially rescue the neuronal death caused by Pals1 removal.
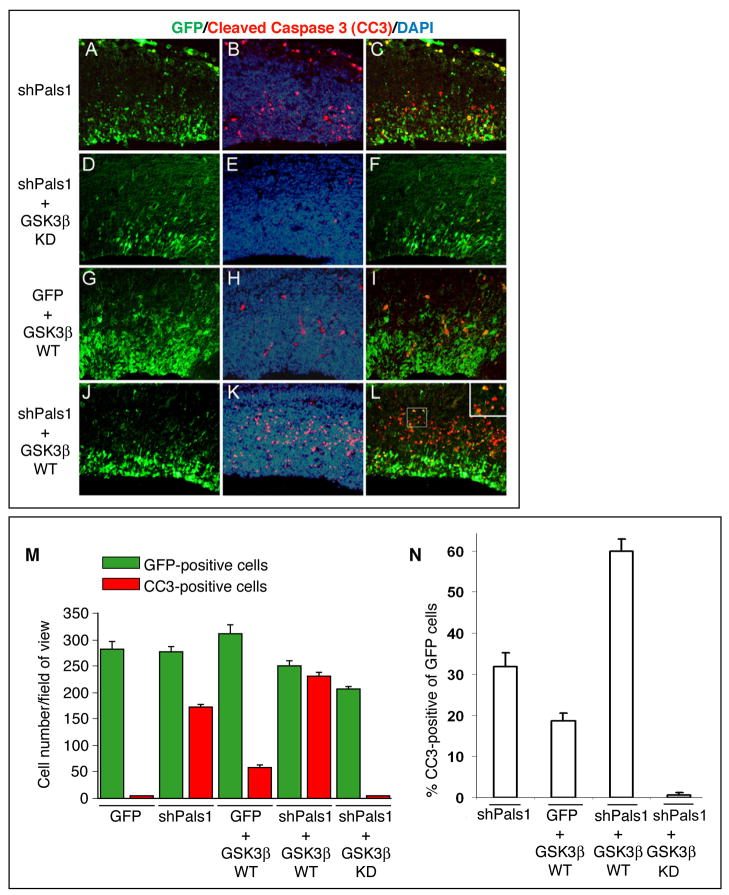
To further examine mTOR function in Pals1-deficient neurons, we down-regulated GSK-3β in order to activate downstream mTOR signaling. Since GSK-3ß can inhibit mTOR signaling via phosphorylation of Tsc2 (Buller et al., 2008; Inoki et al., 2006), we co-electroporated the Pals1shRNA together with a kinase-dead GSK-3β construct, which inhibits GSK-3β signaling (Stambolic and Woodgett, 1994), into developing cortices at E13.5 to test whether the down-regulation of GSK-3β activity would rescue the apoptotic cell death caused by Pals1 knockdown. Interestingly, 31 % of GFP positive electroporated cells were CC3-positive in the Pals1 shRNA targeted cortical domain, while only a minority (~1%) of cells were CC3-positive upon simultaneous electroporation of the kinase-dead GSK-3β (Figures 8A-8F, and 8M, 8N), suggesting that GSK-3β knockdown efficiently rescues the cell death induced by loss of Pals1 activity. Conversely, the proportions of CC3-positive cells were increased two-fold when wild type GSK-3β was electroporated together with the Pals1 shRNA (Figure 8G-L, 8M, N). Taken together, these data strongly suggest that the reduction of mTOR activity in the abnormally generated neurons likely contributes to the massive cell death in Pals1 CKO animals.
Figure 8. GSK-3β activity has a synergistic effect on neuronal cell death together with Pals1 down-regulation.
(A-F) Neuronal cell death induced by Pals1 down regulation (A-C) is efficiently rescued by simultaneous downregulation of GSK-3β activity obtained by expression of a kinase-dead GSK-3β expression vector (D-F). (G-L) GSK-3β overexpression elicits only a minor effect on cell death in control cortical tissue, while when combined with Pals1 down-regulation exerts a dramatic increase in neuronal cell death (J-L). DNA constructs are electroporated in E13.5 mouse cortices and results analyzed 48hr later. C, F, I, L show merged images of CC3-positive (red, apoptotic cells) and GFP electroporated cells (green). (M) Quantification of the electroporated (GFP+, green) and apoptotic (CC3+, red) cells per field for each different condition analyzed in (A-L)(ANOVA; p<0.0001; n=3). (N) Percentage of CC3-positive apoptotic figures within the total electroporated GFP-positive each different condition analyzed (ANOVA; p<0.0001; n=3).
DISCUSSION
Our genetic study demonstrates a near-complete abrogation of cortical development in the absence of the apical complex protein Pals1. Surprisingly, despite the absence of most cortical structures, Pals1 CKO mice survive and display relatively normal health, thus providing a unique animal model that distinguishes essential functions of the cerebral cortex. Our data also suggest that the role of Pals1 in cell fate determination, in which mutation of Pals1 results in a cell fate shift to generate neurons instead of progenitor cells, likely relates to the proper localization and maintenance of apical complex proteins in the apical membrane, and the establishment of normal apical membrane structures, such as primary cilia. Finally, we have uncovered an unexpected role of Pals1 in cell survival, which is closely coupled to the cell fate defects. Virtually all of the abnormally generated neurons die, and their death is associated with defects in mTOR signaling. This unexpected connection between cell fate and survival signaling by the apical complex protein Pals1 sheds light on how Pals1 integrates signaling events during neurogenesis to regulate neuronal number.
Pals1 in cell fate and cell survival
Our study uncovers a previously unknown role for Pals1 in regulating progenitor cell fate and cell survival. Pals1-deficiency produces premature exit from the cell cycle that is tightly coupled to defects in survival of recently differentiated neurons (Figures 3–5). The rapid clearance of dying cells does not allow us to definitively exclude roles for Pals1 in regulating the survival of progenitors as well, though at most few double-labeled dying progenitors are seen. Since the expression of Pals1 decreases in postmitotic neurons (Supplementary Figure S1A), and since neither Pals1 knockdown in postmitotic neurons nor neuron-specific Pals1 deletion promotes neuronal death (data not shown), it appears that loss of Pals1 in progenitor cells results in cells that express a neuronal fate, but which are deficient in mTOR signaling, and hence rapidly, and virtually quantitatively, die. Thus, Pals1 appears to play a key role not only in the regulation of asymmetric cell division (Bultje et al., 2009 ; Costa et al., 2008; Srinivasan et al., 2008), but also is essential for the subsequent transition from progenitor to neuron, and the survival of the neuron.
Defects in progenitor and neuron survival are also a feature of Notch 1 and Notch 3 conditional knockouts (Mason et al., 2006). The shared cell death phenotype between Notch and Pals CKOs raises the possibility that either Pals1 acts through Notch, or that Pals1 and Notch signaling converge on common targets to govern cell survival. Indeed similar to Drosophila neuroblasts (Wodarz, 2005), Par3 modulates Notch signaling through an interaction with Numb to regulate asymmetric division during mammalian cortical neurogenesis (Butje et al., 2009). Since Pals1 regulates localization of Par3 and other apical complex proteins in neural progenitors (data not shown and Figure 6B), Pals1 loss may disrupt Notch signaling by improper localization of Par3. Consistent with this model, up regulation of mTOR signaling through inactivation of Tsc1 or overexpression of Rheb orthologues in Drosophila activate Notch signaling, regulating cell fate choices of daughter cells of sensory organ progenitors (Karbowniczek et al. 2009).
Thus, the apical complex and mTOR signaling connect two major biochemical pathways involved in proliferation and survival. While the failure to appropriately specify neural progenitor cell fate has been associated with cell death (Ohtsuka et al., 1999; Petersen et al., 2002), the molecular pathways coupling these processes in cortical progenitor cells have remained incompletely understood. mTOR signaling is a well established regulator of cell survival through interactions with pro-apoptotic proteins including BAD and P53 (Feng et al., 2005; Freilinger et al., 2006). Here, we found mTOR signaling to mediate apical complex signaling events governing cell fate determination and survival. Both Tsc1 and Tsc2 localize to the cytoplasm and the apical membrane (Massey-Harroche et al., 2007), and Tsc2 interacts with Patj, providing a biochemical link between the apical complex to mTOR signaling pathway (Massey-Harroche et al., 2007). The brain specific Tsc2 mouse model has an enlarged brain and a significant reduction in the number of early-born neurons (Way et al., 2009), a phenotype in some ways complementary to the Pals1 CKO phenotype.
The expression level of Pals1 and other apical complex proteins is closely linked to the normal progression of neurogenesis (Supplemental Figure S1A, Costa et al., 2008). The ability of Pals1 overexpression to delay cell cycle exit of progenitors and the prominent defects in Pals1 Het mice support the conclusion that Pals1 regulates progenitor maintenance and cell survival in a dose-dependent manner. Interestingly, Pals1 Hets also displayed intermediate levels of attenuation of mTORC1 signaling (Figure 7A–C). The rescue of the Pals1 CKO cortex phenotype in the Tsc2 CKO; Pals1 CKO; Emx1Cre double knockout mice was only partial. The efficacy of rescue corresponds to the gradient of Emx1Cre expression, which is highest medially, where no rescue takes place, and lower laterally, where lower levels of Emx1Cre mediate less complete Pals1 recombination (Figure 1C, Figure 7). Thus, the dose-dependent function of Pals1 as a scaffolding protein may be essential for recruiting appropriate signaling molecules, such as components of the mTOR signaling pathway.
While most of the Tsc2 forebrain-specific mutant mice die before postnatal day 15 (data not shown), some rescue of both their viability and body size was observed in the Tsc2 CKO; Pals1 CKO; Emx1Cre double knockout mice (data not shown), further supporting a genetic interaction. Taken together, our data suggest that the interaction between Pals1 and mTOR signaling plays a key role in mediating neuronal survival, and more broadly, in ensuring appropriate brain development on the whole.
Because both apical complex proteins and the mTOR signaling cascade are highly conserved, the interactions between apical complex proteins and mTOR signaling may generalize beyond the cortex to other organ systems and species. Abnormal mTOR signaling was recently implicated as a potential common downstream target of several mouse models of Retinitis pigmentosa (Punzo et al., 2009), a degenerative condition in which retinal photoreceptors, whose highly specialized apical membranes form light-sensitive elements, are lost. Moreover, mutations in human Crumbs homolog 1 (CRB1), a Pals1 interacting protein, lead to Retinitis pigmentosa 12/Leber’s Congenital Amaurosis 8 (den Hollander et al., 1999). Thus together with our findings, these data suggest that in addition to mediating survival signaling during normal development, the apical complex, when dysregulated, may contribute to degenerative disease by regulating mTOR signaling.
Finally, the apical membrane gives rise to primary cilia, which are significantly reduced in length in Pals1-deficient progenitors (Figure 6E). Primary cilia play important roles in relaying extracellular signals including Sonic hedgehog and Wnt to cells (Caspary et al., 2007; Eggenschwiler and Anderson, 2007; Rohatgi et al., 2007). Interestingly, cortical progenitor cells extend their primary cilia directly into the cerebrospinal fluid (Figure 6), raising the possibility that progenitor cells might obtain instructive cues from the embryonic cerebrospinal fluid. In this manner, impaired ciliary development may result in severely disrupted signaling for a host of developmental cues.
Distinguishing cortex-dependent from cortex-independent behaviors in Pals1 CKO mice
Despite a nearly complete loss of the cortex, Pals1 CKO mice survive, and are relatively healthy, unlike several other known mutants associated with severe defects in cortical structure (De Pietri Tonelli et al., 2008; Depaepe et al., 2005; Hatakeyama et al., 2004). Using a battery of behavioral paradigms, we found that, as expected, Pals1 CKO mice show severe impairment in tasks that are known to be cortex-dependent, including performance in the open field, the wire hang, and locating a visible platform in the Morris water maze (Figure 2)(Crawley, 2008). Consistent with the absence of visual and association cortices, we were not able to detect any evidence for intact vision (Figure 2E and data not shown). Several tasks known to depend principally on subcortical areas are spared in Pals1 CKO mice, despite deficits in cortical feedback projections to these areas. For example, the Pals1 CKO mice performed as well as control mice on the accelerating rotarod, despite the lack of a motor cortex and a pyramidal tract. In addition, the mice are able to swim (Figure 2E), an activity that typically requires bilateral coordination.
The ability of Pals1 CKO mice to retain some but not other behaviors presumably depends on compensatory pathways, and an intriguing candidate is the superior colliculus, a subcortical structure that appears intact and even somewhat enlarged in Pals1 CKO mice compared to wild type controls (data not shown), despite the loss of its sizable innervation from many cortical regions. The superior colliculus, an ancient structure that antedates the cerebral cortex evolutionarily, tends to be relatively larger in mammals with less developed, smaller cortices (Finlay et al., 2001). The superior colliculus is thought to underlie retained visual functions in decerebrate cats, such as luminance discrimination (Fischman and Meikle, 1965), as well as the phenomenon of “blindsight”, in which some patients with occipital lobe damage can accurately point to stimuli that they deny visual awareness of (Weiskrantz et al., 1974). The superior colliculus is also known to coordinate sensorimotor functions, consistent with our observations of intact rotarod and swimming behaviors. On a practical level, the long-term viability of the Pals1 CKO mouse, in the absence of virtually all cortical mantle and hippocampus, should facilitate in vivo studies of development and function of the superior colliculus and other subcortical structures, by allowing direct access for imaging and electrophysiology studies.
EXPERIMENTAL PROCEDURES
Mice and generation of targeted animals
Mice were housed and handled in accordance with protocols approved by the IACUC of Harvard Medical School, Stem Cell Research Institute, Dibit, H. San Raffaele, and University of Texas, Health Science Center at Houston. The targeted allele was generated by inserting LoxP sites flanking exon 3 of the Pals1 gene with a neomycin resistance (neor) cassette using homologous recombination in ES cells (Feng et al., 2006). The targeted allele created a truncated N-terminal Pals1 protein (122aa). The targeting construct was transfected into ES cells. Targeted clones were confirmed by Southern blot using a 5’ and 3’ probe with EcoRV and EcoR1 digestion. The targeted allele was detected by PCR analysis of DNA from tail biopsies or embryonic tissues. Genotyping was carried out using two primers: 5’ TTTTTCCCACTTCTCATTACAGTG; 3’ GCCCTCCGTTTCCTCTTATC. The conditional allele contained a LoxP site and flanking sequence, thereby generating a 236bp PCR product as compared to wild type sequence of 191bp. The conditional allele of Tsc2 mice were genotyped as previously described (Hernandez et al., 2007). Pals1 f/+, Tsc2f/+ Emx1Cre and Pals1 f/+, Tsc2f/+ Emx1Cre mice were bred for double conditional mutant mice.
Magnetic Resonance Imaging
Perfused adult mouse brains were imaged with an 8.5T magnet at the BIDMC Center for Basic MR Research. 2T weighted axial images were analyzed using ImageJ.
Behavior analysis
N = 10 for each group tested in all behavioral paradigms. No statistically significant differences were observed between male and female behavior in any group tested (data not shown). The righting reflex, wire hang, stride and gait, and visual reach were tested using standard procedures (Crawley, 2008). The hot water tail flick test was used to calculate the latency to withdraw tail from water maintained at 55°C (Fairbanks and Wilcox, 1997). The Preyer reflex was observed as mice responded with an ear flick to a 96 dBSL click generated by a Clickbox (MRC Institute of Hearing Research) held at a 30cm distance.
The accelerating rotarod apparatus (TSE RotaRod System V4.0.2) was used to measure motor coordination. Mice were trained for 2 consecutive days at constant speed (4.0 rpm) for a maximum duration of 300 sec. per trial. A single training session consisted of 3 trials with 5–10 minute intertrial intervals. During testing on day 3, mice were placed on an accelerating rotarod from 4.0 – 40.0 rpm for 300 sec. The testing day consisted of 3 trials with 10 minute intertrial intervals, and the latency to fall was measured. Rotarod data were analyzed by ANOVA followed by Fisher’s PLSD posthoc test. Data are represented as mean ± SEM.
The open field test was conducted in a brightly illuminated square arena (485mm3). Mice were placed in the center of the arena and allowed to explore the area for 10 min. Behavior was tracked using a ceiling-mounted camera (Panasonic WV BP332). Data were analyzed by ANOVA followed by Fisher’s PLSD posthoc test. Data are represented as mean ± SEM.
Swimming was assessed in a Morris water maze during a visible platform test. Training consisted of placing a mouse on a visible platform with a flag for 20 sec with 20 sec intertrial intervals for 3 trials in a pool (1.2 m in diameter) filled with water rendered white with non-toxic paint. The mice were then placed in water and guided to swim to the platform for 3 separate trials. Unassisted swimming was subsequently monitored and analyzed by ceiling-mounted video tracking as above.
In situ hybridization and histology
Tissue was fixed overnight in 4% paraformaldehyde/PBS at 4°C, transferred to 30% sucrose, embedded in OCT, and cryosectioned for in situ hybridization with the Pals1 (892-1600) probe. Alternatively, for histological analysis, tissue was embedded in paraffin and sectioned at 5μm. H&E staining was carried out by standard procedures.
X-gal labeling
Whole heads or brains were placed in a solution of 100mM sodium phosphate (pH7.3), 2mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 for 30 min. For the color reaction, 5mM potassium ferricyanide, 5mM potassium ferrocyanide, and 1mg/ml X-gal were added to this solution.
Quantification of S phase and M phase cells
BrdU labeled cells were counted after a 30 min. pulse. E12 and E14 Paraffin sections were stained with BrdU (Harlan Laboratories) and P-H3 (Upstate) antibodies. Quantification was performed on 3–5 WT, Pals1 Het and Pals1 CKO embryos for each time point. Cells were manually counted from four different areas of each embryo on a digital image generated either by confocal microscope (Leica) or inverted epiflouresence microscope (Zeiss).
Quantification of dying cells
Dying cells were labeled in 3–5 embryos at E12 with an In Situ Cell Death Detection kit (Roche). Four sections from each embryo were stained and total numbers of cells was determined by manually counting cells with positive Hoechst 33258 staining nuclei in the unit area on a digital image generated as described above.
Quit fraction using BrdU labeling and Ki67 staining
BrdU was injected into the timed pregnant females at E10 and E11. 3–5 pups of each genotype and age were harvested after 18 or 24 hours, and 4 sections of each embryo for each genotype were stained with BrdU and Ki67. The cells that are BrdU-positive but Ki67-negative were manually counted and compared on a digital image generated as described above.
Immunohistochemistry
5μm sections were prepared as described above, deparaffinized, and rehydrated through an ethanol series into distilled H2O. After antigen retrieval using Antigen Unmasking Solution (Vector Laboratories), sections were rinsed in PBS and incubated with antisera to ß-catenin (BD Biosciences), Nestin (Developmental Studies Hybridoma Bank), Glast (Chemicon), BrdU (Harlan), Ki67 (Nova Castra), Pals1 (Chae et al., 2004) (Upstate, gift from Dr. B. Margolis (Roh et al., 2002b), P-H3 (Upstate), aPKCζ, aPKC λ (Transduction Laboratories), Dcx (Gleeson et al., 1999), Reelin (G10, Chemicon), or Pericentrin-1 (Covance) in PBS containing 5% blocking sera for 2 hours at room temperature or overnight at 4C. Sections were washed 4 times in PBS for 10 minutes and incubated with secondary antibodies (goat anti-mouse IgG antibody)(Jackson Immuno Research Laboratories) or Alexa 488 goat anti-rabbit antibody (Molecular Probes) at 1:500, followed by Hoechst 33258 staining.
In utero electroporation
In utero electroporation was performed as described previously (Saito, 2006). Briefly, uterine horns of anesthetized pregnant dams were exposed through an incision in the peritoneum. DNA (3–4ug/ul) was injected manually through the uterine wall into the telencephalic vesicle using pulled micropipettes. Four pulses of 40–55 V (50ms duration) were applied across the uterine wall using a BTX ECM830 pulse generator. The uterus was then replaced in the abdominal cavity and embryonic development allowed to proceed normally.
Pals1 shRNA knockdown
Pals1 knockdown experiments were performed with previously reported and verified Pals1 shRNA constructs (van Rossum et al., 2006) or GFP or unrelated shRNAs (Mpp4, scrambled) that were injected and electroporated into developing cortices (E13 or E14). At least three different sections from three different embryos were analyzed with BrdU, P-H3, and Tuj1.
Pals1 overexpression
The Pals1 coding sequence (2027bp) was amplified from human cDNAs and cloned into a pCAGGS expression vector upstream of pIRES-GFP. pCAGGS-Pals1 or its pCAGGS empty vector control was injected into E13 cortices and analyzed at P3. Ki67-positive and GFP-positive cells that localized in the ventricular layer were counted. Three different sections for each of two Pals1 overexpressing brains together with three control GFP electroporated embryos that do not have any GFP-positive staining cells in the proliferative regions were used in the analyses.
Supplementary Material
Acknowledgments
We are grateful to members of the Walsh laboratory for critical reading of the manuscript. We thank K. Jones for Emx1Cre mice, the Developmental Studies Hybridoma Bank for Nestin and Pax6 antisera, B. Margolis for Pals1 antisera, A. Le Bivic for Crb2, Crb3, and Patj antibodies, T. Thomson for blastocyst injections and ES cell work, S. White for histology, the Harvard EM facility for scanning and transmission EM, D. Burstein and R. Akhavan for MRI scans, and A. Hafner, D. Sinclair, and L. Goodrich for use of their mouse behavioral equipment. This work is supported by a Helen Hay Whitney Postdoctoral Fellowship and institutional funding from the Department of Pediatrics, University of Texas, Health Science Center at Houston to S.K, a Sigrid Jusélius Foundation Fellowship to M.K.L., EC (QLG3-CT-2002-01266) and Telethon grant (GGP07181) to V.B., a IDDRC (P30HD018655) from the Children’s Hospital Boston and R01NS32457 and P01 NS40043 grants from the NINDS to C.A.W. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Bachmann A, Schneider M, Theilenberg E, Grawe F, Knust E. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature. 2001;414:638–643. doi: 10.1038/414638a. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- Chae TH, Kim S, Marz KE, Hanson PI, Walsh CA. The hyh mutation uncovers roles for alpha Snap in apical protein localization and control of neural cell fate. Nat Genet. 2004;36:264–270. doi: 10.1038/ng1302. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, van de Pol DJ, Payne AM, Bhattacharya SS, Kellner U, et al. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12) Nat Genet. 1999;23:217–221. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- Depaepe V, Suarez-Gonzalez N, Dufour A, Passante L, Gorski JA, Jones KR, Ledent C, Vanderhaeghen P. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435:1244–1250. doi: 10.1038/nature03651. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008;283:23440–23449. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci U S A. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB, Nicastro N. Developmental structure in brain evolution. Behav Brain Sci. 2001;24:263–278. discussion 278–308. [PubMed] [Google Scholar]
- Fischman MW, Meikle TH., Jr Visual Intensity Discrimination in Cats after Serial Tectal and Cortical Lesions. J Comp Physiol Psychol. 1965;59:193–201. doi: 10.1037/h0021811. [DOI] [PubMed] [Google Scholar]
- Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Curr Opin Neurobiol. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Freilinger A, Rosner M, Krupitza G, Nishino M, Lubec G, Korsmeyer SJ, Hengstschlager M. Tuberin activates the proapoptotic molecule BAD. Oncogene. 2006;25:6467–6479. doi: 10.1038/sj.onc.1209660. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez O, Way S, McKenna J, 3rd, Gambello MJ. Generation of a conditional disruption of the Tsc2 gene. Genesis. 2007;45:101–106. doi: 10.1002/dvg.20271. [DOI] [PubMed] [Google Scholar]
- Hong Y, Stronach B, Perrimon N, Jan LY, Jan YN. Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature. 2001;414:634–638. doi: 10.1038/414634a. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr Opin Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Imai F, Hirai S, Akimoto K, Koyama H, Miyata T, Ogawa M, Noguchi S, Sasaoka T, Noda T, Ohno S. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133:1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T. Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol. 2009;185:959–967. doi: 10.1083/jcb.200811030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamberov E, Makarova O, Roh M, Liu A, Karnak D, Straight S, Margolis B. Molecular cloning and characterization of Pals, proteins associated with mLin-7. J Biol Chem. 2000;275:11425–11431. doi: 10.1074/jbc.275.15.11425. [DOI] [PubMed] [Google Scholar]
- Karbowniczek M, Zitserman D, Khabibullin D, Hartman T, Yu J, Morrison T, Nicolas E, Squillace R, Roegiers F, Henske EP. The evolutionarily conserved TSC/Rheb pathway activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development. J Clin Invest. 2009;120:93–102. doi: 10.1172/JCI40221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311:1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HA, Rakowiecki SM, Gridley T, Fishell G. Loss of notch activity in the developing central nervous system leads to increased cell death. Dev Neurosci. 2006;28:49–57. doi: 10.1159/000090752. [DOI] [PubMed] [Google Scholar]
- Massey-Harroche D, Delgrossi MH, Lane-Guermonprez L, Arsanto JP, Borg JP, Billaud M, Le Bivic A. Evidence for a molecular link between the tuberous sclerosis complex and the Crumbs complex. Hum Mol Genet. 2007;16:529–536. doi: 10.1093/hmg/ddl485. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. Embo J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- Pinal N, Goberdhan DC, Collinson L, Fujita Y, Cox IM, Wilson C, Pichaud F. Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr Biol. 2006;16:140–149. doi: 10.1016/j.cub.2005.11.068. [DOI] [PubMed] [Google Scholar]
- Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12:44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Roh MH, Liu CJ, Laurinec S, Margolis B. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J Biol Chem. 2002a;277:27501–27509. doi: 10.1074/jbc.M201177200. [DOI] [PubMed] [Google Scholar]
- Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol. 2002b;157:161–172. doi: 10.1083/jcb.200109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Roosa J, Olsen O, Lee SH, Bredt DS, McConnell SK. MALS-3 regulates polarity and early neurogenesis in the developing cerebral cortex. Development. 2008;135:1781–1790. doi: 10.1242/dev.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, Woodgett JR. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J. 1994;303(Pt 3):701–704. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight SW, Shin K, Fogg VC, Fan S, Liu CJ, Roh M, Margolis B. Loss of PALS1 expression leads to tight junction and polarity defects. Mol Biol Cell. 2004;15:1981–1990. doi: 10.1091/mbc.E03-08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res. 2001;41:51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I, Arsanto JP, Le Bivic A, Wijnholds J. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Muller glia cells. Hum Mol Genet. 2006;15:2659–2672. doi: 10.1093/hmg/ddl194. [DOI] [PubMed] [Google Scholar]
- von Stein W, Ramrath A, Grimm A, Muller-Borg M, Wodarz A. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 2005;132:1675–1686. doi: 10.1242/dev.01720. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chen XW, Margolis B. PALS1 Regulates E-Cadherin Trafficking in Mammalian Epithelial Cells. Mol Biol Cell. 2006 doi: 10.1091/mbc.E06-07-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way SW, McKenna J, 3rd, Mietzsch U, Reith RM, Wu HC, Gambello MJ. Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse. Hum Mol Genet. 2009;18:1252–1265. doi: 10.1093/hmg/ddp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- Weiskrantz L, Warrington EK, Sanders MD, Marshall J. Visual capacity in the hemianopic field following a restricted occipital ablation. Brain. 1974;97:709–728. doi: 10.1093/brain/97.1.709. [DOI] [PubMed] [Google Scholar]
- Wodarz A. Molecular control of cell polarity and asymmetric cell division in Drosophila neuroblasts. Curr Opin Cell Biol. 2005;17:475–481. doi: 10.1016/j.ceb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Zechner D, Fujita Y, Hulsken J, Muller T, Walther I, Taketo MM, Crenshaw EB, 3rd, Birchmeier W, Birchmeier C. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.