Abstract
Loss-of-function mutations in the filaggrin gene (FLG), cause the semi-dominant keratinizing disorder, ichthyosis vulgaris1, and convey major genetic risk to atopic dermatitis/eczema, eczema-associated asthma2,3 and other allergic phenotypes5. Several low frequency FLG null alleles occur in Europeans and Asians, with a cumulative frequency of ~9% in Europe4. Here we report a 1-bp deletion mutation, 5303delA, highly analogous to common human FLG mutations, within the murine flg gene in the spontaneous mouse mutant flaky tail (ft). Importantly, we demonstrate that topical application of allergen to mice homozygous for this mutation results in cutaneous inflammatory infiltrates and enhanced cutaneous allergen priming with development of allergen-specific antibody responses. These data validate ft as a useful model of filaggrin deficiency and provide experimental evidence for the hypothesis that antigen transfer through a defective epidermal barrier is a key mechanism underlying elevated IgE sensitization and initiation of cutaneous inflammation in humans with filaggrin-related atopic disease.
Eczema is a common disease of childhood that affects 15–30% of children and 2–10 % of adults in developed countries6. There is a strong genetic association with atopic disorders including asthma, hay fever and food allergy7. Elevated total IgE is observed in 60–80% of eczema patients attending secondary care. While IgE sensitization increases in the first few years of life in children with eczema8, both the timing and the pathogenic role of IgE sensitization in development of atopic dermatitis remains uncertain9. Recent evidence strongly suggests that children with early onset (<3 months) and more severe eczema have the greatest risk of IgE sensitization. While debate remains as to the relative causal contributions of inherited barrier defects versus immunological aberrations, the major genetic risk identified to date is associated with prevalent loss-of-function mutations in the FLG gene, encoding the skin barrier protein profilaggrin/filaggrin2,4.
The spontaneous recessive mouse mutant flaky tail (ft) originally arose on the background of an existing recessive hair phenotype, matted (ma) in 1958 and since then has been maintained on a mixed strain10. Flaky tail mice have a dry, flaky skin and annular tail and paw constrictions in the neonatal period11. Western blotting analysis shows that ft/ft mice express a truncated profilaggrin (~215 kDa) instead of the normal high-molecular-weight profilaggrin (>500 kDa)11. Although processed filaggrin in these mice has been shown to be virtually absent at the protein level11, the underlying genetic mechanism remains unknown, however, recent studies of human filaggrin mutations4 suggest that this might be a nonsense or frameshift mutation in exon 3 of flg. The immunological and skin barrier characteristics of this mouse phenotype remain unknown10.
Given the probable genetic similarity between ft mice and the human conditions ichthyosis vulgaris and atopic eczema, we sought to identify the underlying genetic mechanism underlying ft and study in detail the function of the skin barrier in ft/ft mice with respect to ease of allergen priming.
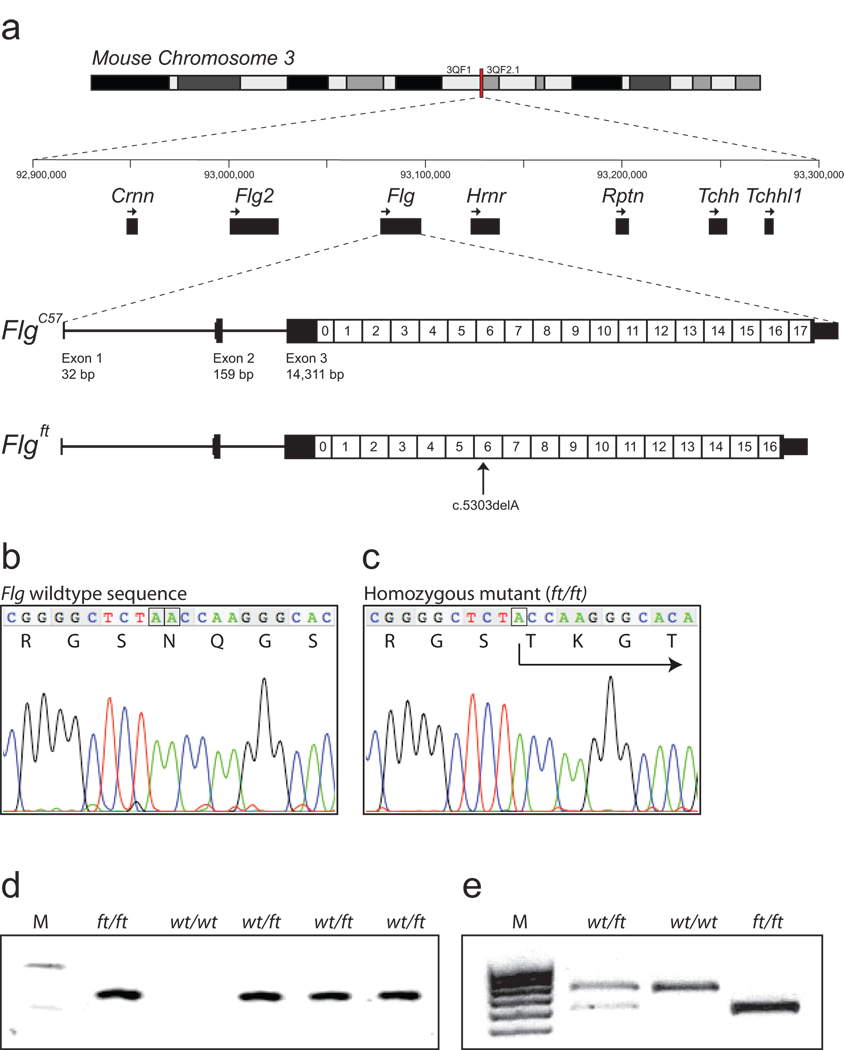
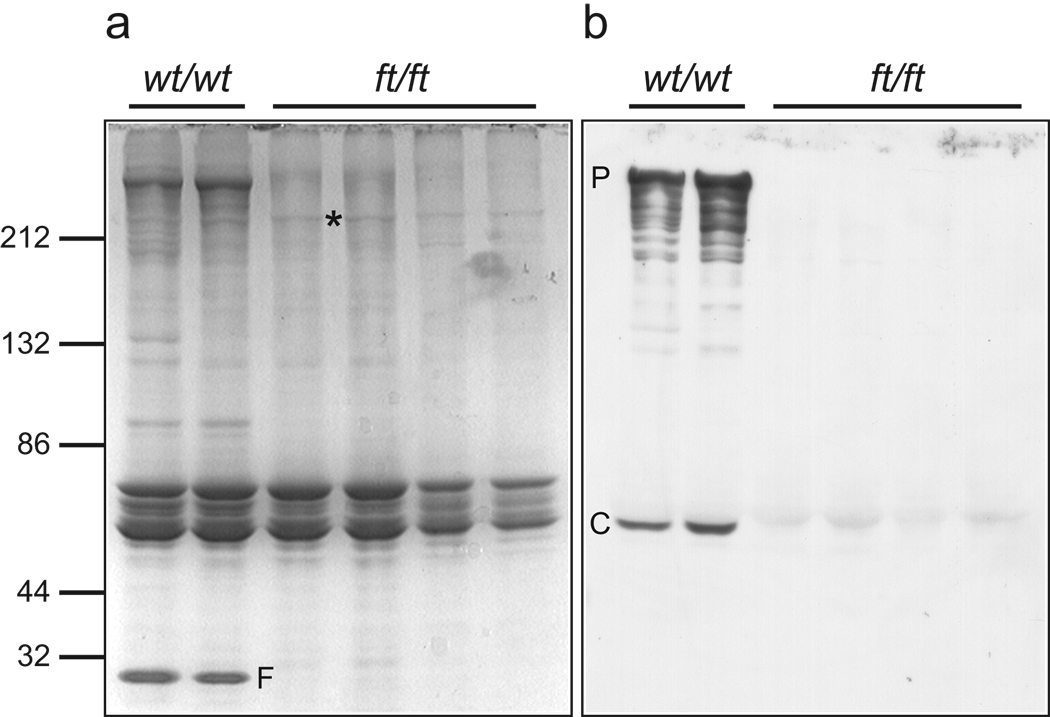
The murine filaggrin gene, flg, is located on mouse chromosome band 3QF2.1 within a cluster of seven genes encoding the fused S100 family of filaggrin-like proteins (Fig. 1a). The organization of this gene cluster is identical to the orthologous human cluster. The intron-exon organization of the 20,150 bp flg gene was manually annotated from the current build of the C57BL/6 mouse genome (Fig.1a, Supplementary Table S1). The flg genomic sequence derived from BAC clone RP23-227A12 contains small assembly errors due to incomplete resolution of the tandem repeats within the filaggrin gene (accession number AC158361). This issue was resolved by aligning the individual filaggrin repeats and correcting the 5 detected assembly errors (Supplementary Table S2), so that the complete ORF was in frame with the reported short partial 5’ and 3’ cDNA sequences for murine filaggrin12,13 (accession numbers AF510860 and J03458, respectively). The predicted 496 kDa murine profilaggrin consists of 4654 amino acids, including 16 full near-identical repeats of a 250 amino acid filaggrin consensus sequence (repeats 1–16), flanked by two incomplete repeats (repeats 0 and 17; Fig.1a). Most repeats are less than 1% different in protein sequence from each another. The amino-terminus consists of a conserved S100 calcium binding domain14 and a B-domain postulated to have a signalling function12,15. The protein domain organisation is identical to that of human profilaggrin4 except that the murine protein has 16 repeats of a smaller 250 amino acid sequence versus 10–12 repeats of 324 amino acids in humans4. The number of repeats has been shown to vary between mouse strains16, as it does in the human population4,17. Downstream of repeat 17 is a unique C-terminal domain. A polyclonal antibody raised against a synthetic peptide derived from the C-terminal tail domain recognized full-length ~500 kDa profilaggrin in wild type mice, as well as intermediates from profilaggrin-to-filaggrin processing (Fig. 2), therefore validating the predicted amino acid sequence. Although traces of truncated profilaggrin (~215 kDa) can be seen in ft/ft homozygotes by total protein staining (Fig. 2a) and in western blots using antibodies against a filaggrin repeat epitope, as previously reported11, this is not recognised by the C-terminal antibody, predicting a truncating mutation (Fig. 2b).
Figure 1. Murine filaggrin gene structure and mutation identification.
(a) The mouse filaggrin gene (flg) resides within a cluster of 7 genes encoding closely related fused-S100 proteins, spanning ~350 kb on murine chromosomal band 3QF21. Like the human ortholog, flg has a 3 exon gene structure. Exon 1 consists of 5’UTR sequences only; exon 2 contains the ATG and encodes part of the N-terminal S100 calcium binding domain of profilaggrin; and exon 3 encodes the remainder of the S100 domain, the B-domain and the filaggrin polyprotein repeat domain. In the July 2007 mouse genome sequence, annotated here (FlgC57), the latter consists of 16 near-perfect repeats encoding a 250 amino acid filaggrin protein (1–16) flanked by two imperfect repeats (0 and 17). Repeat 17 is followed by a short tail domain. The 485 bp 3’UTR contains a consensus polyadenylation signal. In ft mice, the exon 3 sequence determined here (Flgft) lacks one repeat due to a 750-bp in-frame deletion, as well as the frameshift mutation c.5303delA. In addition, there are several additional SNPs and small in-frame insertions/deletions in flg that differ between the two strains, so that their DNA sequences overall are 99.3% identical, excluding the repeat copy number variation.
(b) Wild-type flg sequence, corresponding to codons 1765–1771, with amino acid translation. The A dinucleotide involved in the mutation is boxed (compare panel c).
(c) Analogous flg sequence as shown in (b), derived from a homozygous ft/ft mouse, showing homozygous frameshift mutation designated c.5303delA, predicting truncation mutation p.Asn1768ThrfsX154. The single A nucleotide remaining at the site of the mutation is boxed (compare panel b).
(d) Allele-specific genotyping of the parents and F1 offspring of a cross between an ft/ft homozygote and a wild-type mouse. M, molecular weight markers, upper band = 118 bp; lower band = 72 bp. The allele specific band is only amplified from mice carrying the ft mutation.
(e) Genotyping for an Acc I restriction site polymorphism in filaggrin repeat 1, approximately 4 kb upstream of the 5303delA mutation. This assay is more convenient for scoring of heterozygotes. The 678 bp C57BL/6 allele does not digest; the ft allele cuts to yield fragments of 559 bp and 134 bp. M, molecular weight markers.
Figure 2. A truncated profilaggrin is expressed in flaky tail mouse skin, which lacks the C-terminus.
Urea/Tris protein extracts from control wild type and ft/ft mice were fractionated by SDS/PAGE and immunoblotted with the peptide antibody developed to the C-terminus of mouse profilaggrin. (a) Coomassie brilliant blue-stained protein gel to control for loading, and (b), corresponding immunoblot probed with the C-terminal antibody. The antibody detects both full-length profilaggrin (P) and a putative C-terminal processing product (C) in wild type mouse extracts, however, these immunoreactive products are not detected in ft/ft mice (b). Note the lack of filaggrin (F) in ft/ft mice (a), observed previously11. The mutant profilaggrin protein in ft/ft mice is faintly visible in (a) at an apparent molecular weight of ~215 kDa (*). This is readily detectable with a mouse filaggrin antibody as reported previously11. Molecular weight marker sizes are shown at left.
The long-range PCR methods used to identify human FLG mutations1,4 proved unable to amplify the larger exon 3 of the murine gene and so a shotgun sequencing method whereby six species of short PCR fragments were generated and cloned using primers designed to amplify any of the repeat units, as recently employed to detect human filaggrin mutations18. From the sequencing of >900 clones, a 1-bp deletion was detected in clones resulting from the four different PCR fragments (Fig. 1b&c). Allele-specific PCR genotyping showed that this deletion completely co-segregated with the ft phenotype in many mouse crosses (Fig. 1d). An Acc I restriction fragment length polymorphism was also detected in filaggrin repeat 1 that is in tight linkage with the mutation. This marker proved to be a convenient method for genotyping ft mice, since heterozygotes are more easily scored than by allele-specific PCR (Fig. 1e). Using these genotyping assays, the ft mutation was backcrossed and maintained in our laboratories on the C57BL/6 background.
Given the nature of the shotgun method initially used, it was not possible to fully assemble the complete ft sequence for flg exon 3, however, we constructed a BAC library from an ft/ft homozygous animal and screened out a clone containing the entire flg locus. Exon 3 was subcloned by recombineering and the resultant plasmid was ~9-fold over-sequenced by random transposon insertion methodology. Exons 1 and 2 were sequenced from ft/ft mice by standard PCR methods, allowing full assembly of the flg sequence from the mutant strain (Genbank accession 1148514). By this means, the mutation could be unambiguously designated as c.5303delA in filaggrin repeat 6 leading to a premature termination codon 154 codons downstream (designated p.Asn1768ThrfsX154). The flg sequence from ft mice was one filaggrin repeat shorter than the C57BL/6 sequence, consistent with earlier studies reporting variation in filaggrin copy number both in mouse and humans4,17. Otherwise, the coding sequence showed 99.3% identity between C57BL/6 and ft with no other loss-of-function variants detected. Further partial sequencing of the gene in different mouse strains suggested that the flg locus in ft was originally derived from the C3H strain (data not shown).
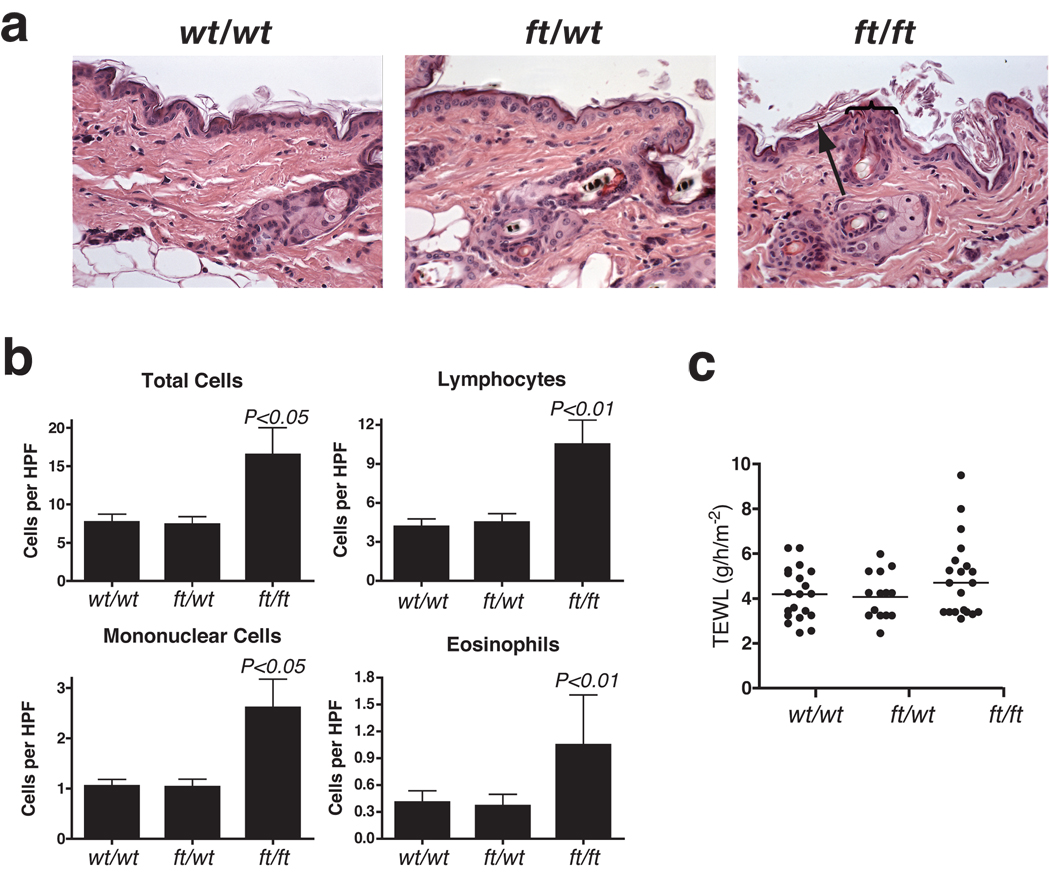
Skin biopsies for histopathological analysis were taken from adult 6–8 week old age-and sex-matched wt/wt (wild type), wt/ft (heterozygous) and ft/ft (homozygous) mice for routine histological analysis. No overt differences were noted between between wt/wt and wt/ft (Fig. 3a). In contrast, ft/ft mice had a mild diffuse orthokeratotic hyperkeratosis. (a hypertrophy of the stratum corneum of the skin characterized by the presence of non-nucleated cells). Occasional foci of acanthosis (abnormal thickening of the stratum spinosum layer of the skin) was observed, which varied in magnitude between individual ft/ft mice (Fig. 3a). In our hands, the ft/ft mice have a phenotype consistent with the original description of the ft/ft mice11, but did not have significant acanthosis or alterations to the stratum granulosum previously as reported11, possibly due to histopathological analysis being performed at a younger age. A broad spectrum of severity of skin pathologies was observed between individual ft/ft mice including some with sporadic superficial dermal cellular infiltrates. We further characterized the cutaneous cellular infiltrate and, based on cells detected per field (x1,000), significantly more total cells (P<0.05), lymphocytes (P<0.01), eosinophils (P<0.05) and mononuclear cells (P<0.01) were observed in ft/ft mice relative to wt/wt and wt/ft mice (Fig. 3b). To specifically address the baseline integrity of the skin barrier of flaky tail animals, transepidermal water loss (TEWL) was measured on the skin of adult age-matched wt/wt, wt/ft and ft/ft mice. Although there were higher individual TEWL readings in ft/ft mice, possibly reflecting the spectrum of severity in skin inflammation within this group, there was no statistically significant difference in baseline TEWL between the three groups of mice (Fig. 3c).
Figure 3. Cutaneous inflammation in untreated ft/ft mice but not wt/wt or wt/ft animals.
(a) Representative photomicrographs of skin sections from age- and sex-matched wt/wt, wt/ft and ft/ft mice. Ft/ft mice had orthokeratotic hyperkeratosis (arrow) with occasional foci of acanthosis (bracket) compared to wt/wt and ft/wt mice. (Original magnification x40.)
(b). Numbers of skin infiltrating cells, lymphocytes, eosinophils and mononuclear cells were detected per high-power fields (HPF). Cells were counted on 15–20 HPF (x1,000) on hematoxylin and eosin stained sections of 5–6 wt/wt, wt/ft and ft/ft mice. Data represent the mean; error bars represent standard error of the mean. Student’s t-test, or corrected Welch corrected t-test, was used to determine statistical differences between groups. NS = Non-significant.
(c) TEWL analysis of untreated wt/wt, wt/ft and ft/ft mice. Values are individual mice and Mean bars are shown.
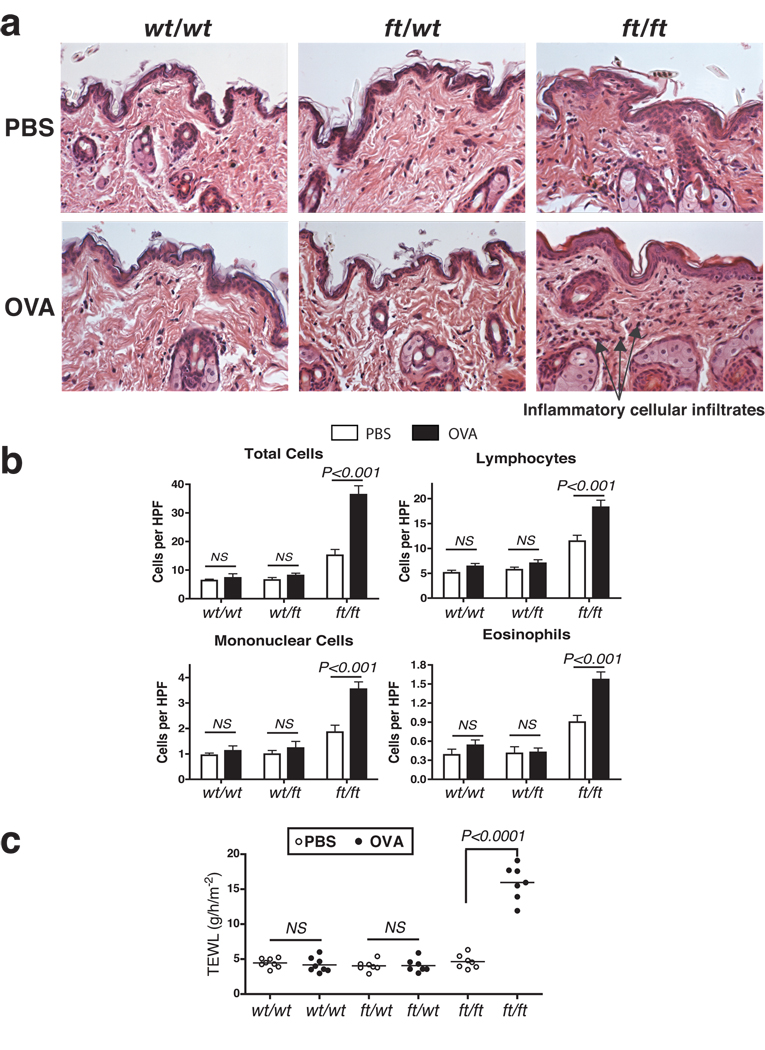
To test allergen priming of the skin in these mice, we used the widely studied and clinically relevant allergen ovalbumin (OVA). OVA or PBS, as a vehicle control, was applied to the shaved abdominal skin of wt/wt, wt/ft and ft/ft mice. (Full protocol in Supplementary Fig. S1a). Cutaneous exposure to OVA did not lead to gross skin lesions in any mice (data not shown). Skin removed from the site of allergen challenge revealed evidence of some edema and non-significant increase in cellular infiltrates but no gross damage or inflammation in either wt/wt or ft/wt mice.(Fig. 4a&b). In contrast, exposure of OVA to the skin of ft/ft mice induced diffuse mild acanthosis, with significant infiltrates of mixed, predominantly lymphocytic inflammatory cells, in addition to eosinophils and mononuclear cells (Fig. 4a&b). Measurement of TEWL at the site of allergen challenge 24 hours after OVA application revealed a significant (P<0.0001) elevation in TEWL of OVA-treated ft/ft mice relative to PBS-treated ft/ft mice, and wt/wt and wt/ft mice (Fig. 4c). Application of OVA to wt/wt or wt/ft caused no alteration in TEWL (Fig. 4c).
Figure 4. Allergen exposure exacerbates skin inflammation ft/ft mice but not wt/wt or wt/ft animals.
(a) Representative photomicrographs of skin sections from age- and sex-matched wt/wt, wt/ft and ft/ft mice exposed to OVA or PBS as a vehicle control. Ft/ft mice had diffuse mild acanthosis and marked increases in dermal cell infiltration when cutaneously challenged with OVA, compared to wt/wt and wt/ft mice. (Original magnification x40.)
(b). Quantification of numbers of skin infiltrating cells, lymphocytes, eosinophils and mononuclear cells detected per high-power fields (HPF). Cells were counted at on 15–20 HPF (x1,000) on hematoxylin and eosin stained sections of 11–14 wt/wt, wt/ft and ft/ft mice. Data represent the mean; error bars represent standard error of the mean. Student’s t-test, or corrected Welch corrected t-test, was used to determine statistical differences between groups. NS = Non-significant.
(c) TEWL analysis of skin of OVA exposed wt/wt, wt/ft and ft/ft mice. Values relate to individual mice and mean bars are shown.
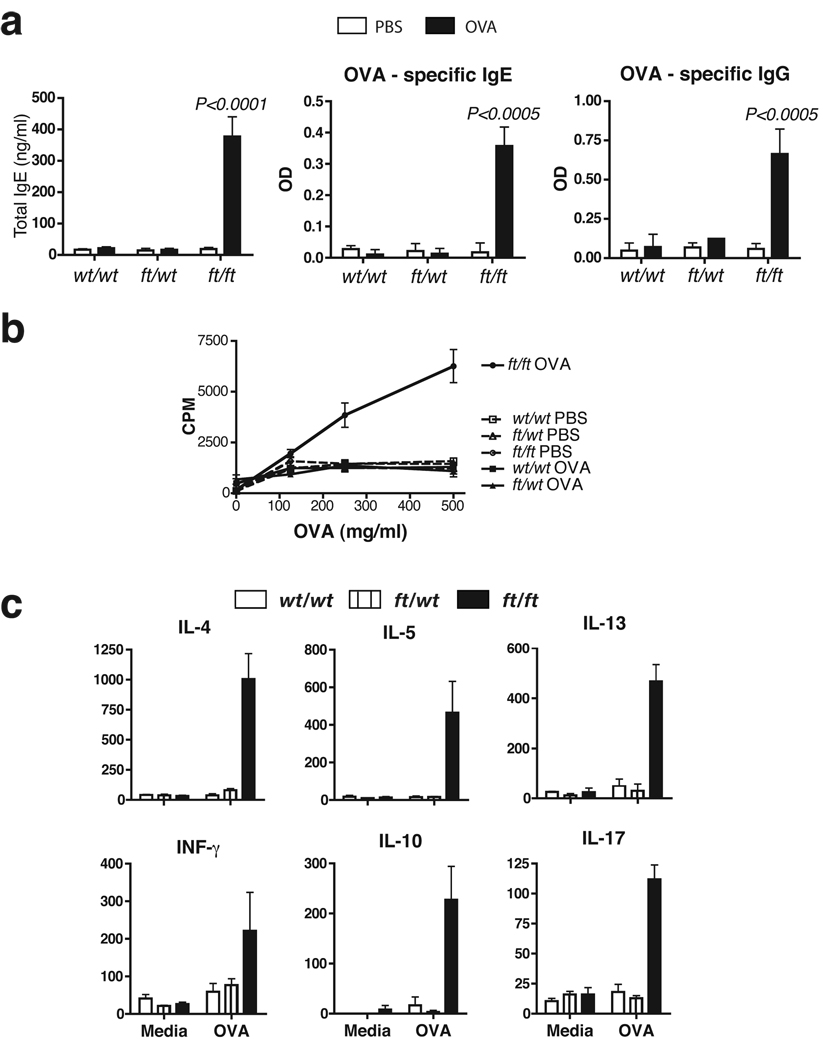
We then analysed the systemic immune response to OVA. Following cutaneous allergen exposure wt/wt mice did not have elevated levels of total serum IgE nor did they generate an antibody (IgG or IgE) response to OVA (Fig 5a; Supplementary Fig. S2), and spleen cells from wt/wt mice did not proliferate or produce cytokines when exposed to OVA in vitro (Fig. 5b&c).. In marked contrast, percutaneous allergen exposure in ft/ft mice generated OVA-specific IgG and OVA-specific IgE that was significantly (P>0.005) elevated relative to other PBS- or OVA-treated groups (Fig. 5a). The absence of allergen-specific responses in wt/wt mice upon allergen-challenge to intact skin has been reported previously in the background strain of the ft/ft mice used here, C57BL/6 mice19, however, an allergic response can be generated when OVA is applied after the skin barrier has been disrupted by tape-stripping20Spleen cells from OVA-exposed ft/ft mice dose-dependently proliferated to OVA, while cells from other test groups did not proliferate (Fig. 5b). None of the PBS-treated groups developed OVA-specific cytokine responses in splenic cells (data not shown). Spleen cells from OVA-treated wt/wt and wt/ft mice did not secrete cytokines in response to OVA (Fig. 5c). In contrast, ft/ft mice produced OVA-specific Th2 (IL-4, IL-5, IL-13), Th1 (IFN-γ), regulatory (IL-10) and Th17 (IL-17) cytokines (Fig. 5c), indicating a generalized allergen-specific cytokine response that was not solely Th2 skewed. Consistent with a mixed T helper cytokine response in ft/ft mice treated with OVA cutaneously, there was elevation of serum levels of both OVA-specific IgG2a and IgG1, antibody isotypes that are induced by Th1 and Th2 cytokines, respectively21 (Supplementary Fig. S2b). In these studies wt/ft animals were comparable to wt/wt mice with respect to all parameters analyzed (Fig. 3–Fig 5).
Figure 5. Elevated OVA-specific immune response in allergen exposed ft/ft mice but not wt/wt or wt/ft animals.
(a) ELISA detection of levels of total IgE, OVA-specific IgE and IgG in serum from wt/wt, wt/ft and ft/ft mice treated with PBS or OVA on skin. Data are Mean plus Standard Error of the Mean (SEM) from 7–12 mice.
(b) Cell proliferation to OVA of spleen cells from wt/wt, wt/ft and ft/ft mice treated with PBS or OVA on skin. Data represent the mean of spleens from 3–4 individual mice per group; error bars represent standard error of the mean.
(c) Cytokine production by spleen cells from OVA exposed wt/wt, wt/ft and ft/ft mice treated with PBS or OVA. Cells were cultured in media or OVA (500 µg/ml) and cytokines detected by ELISA. Data represent the mean of spleens from 3–4 individual mice per group; error bars represent standard error of the mean. Student’s t-test, or corrected Welch corrected t-test, was used to determine statistical differences between groups. NS = Non-significant.
These data show that ft/ft mice generate allergen-specific IgEand cytokine responses following cutaneous allergen challenge to intact skin. Several large studies have now shown that FLG loss-of-function mutations predispose to asthma in the context of AD5,22–24. To experimentally address this association using the mouse model, ft/ft and wt/wt animals that had been percutaneously treated with OVA were treated with OVA aerosol to evoke pulmonary sensitization and airway hyper responsiveness (AHR), and pulmonary responses were analyzed (Supplementary Fig. S1b). AHR, determined as airway resistance (RL), was not induced in OVA exposed ft/ft or wt/wt mice, with both OVA-exposed groups having similar RL values as PBS-exposed mice (Supplementary Fig. S4). No airway inflammation was seen on lung histology sections from wt/wt or ft/ft mice exposed to OVA via the skin (Supplementary Fig. S5a). and was subsequently confirmed and quantified by scoring of histology sections (data not shown). Furthermore, there was no significant elevation in the numbers of total cells and eosinophils recovered in bronchoalveolar lavage (BAL) fluidbetween OVA-exposed or PBS-treated ft/ft and wt/wt mice (Supplementary Fig. S5c & d). OVA application to barrier disrupted skin of wt/wt mice, as used to induce AHR on the more Th2 immunoreactive strain of BALB/c mice by Spergel and colleagues25, also failed to induce AHR in the tape-stripped C57BL/6 mice (Supplementary Fig. S4a). The inability to induce AHR after cutaneous OVA challenge may therefore reflect the use of C57BL/6 mice, which are less susceptible than BALB/c mice to allergen-induced Th2 inflammation.
To confirm that the OVA sensitization demonstrated here in ft/ft is due to a specific defect in the skin barrier rather than a generalized altered immunity, ft/ft and wt/wt mice received intraperitoneal injectionof OVA and adjuvant (Alum).Allergen-specific responses were then evaluated (Supplementary Fig. S3b). The OVA and adjuvant intraperitoneally immunized ft/ft mice developed comparable allergen-specific immune responses to wt/wt animals, with both groups developing a Th2-biased OVA-specific cytokine and antibody isotype response (Supplementary Fig. S6). To formally address if the lack of pulmonary inflammation in ft/ft mice following allergen challenge was due to to altered lung immunity and development of AHR in ft/ft mice these animals and wt/wt mice were challenged intraperitoneal with OVA and adjuvant followed by pulmonary allergen sensitization (Supplementary Fig. S3a), a conventional protocol for this model. There was comparable induction of AHR, determined as lung resistance (RL), in wt/wt and ft/ft mice, with no difference in RL between both groups (Supplementary Fig. S7). Furthermore, intraperitoneal challenge with OVA and adjuvant resulted in airway inflammation in both wt/wt and ft/ft mice (Supplementary Fig. S5b), with no differences between groups in the magnitude of lung inflammation detected (data not shown). Therefore ft/ft mice develop allergic lung inflammation after intraperitoneal priming with OVA and adjuvant but not when sensitized via the skin to OVA in the absence of adjuvant. Further work is needed to dissect if this is due to the type of OVA-specific response generated in the absence of using Alum, a Th2-inducing adjuvant, as opposed to the response generated following skin exposure to allergen in the absence of adjuvant.
Here we have identified the ft mutation as a frameshift mutation 5303delA, predicted to occur in repeat 6 of the flg gene (Fig. 1). The mutation is fully consistent with the loss of a C-terminal profilaggrin epitope and appearance of a ~215 kDa truncated profilaggrin molecule in the skin of mutant mice (Fig. 2). Several truncation mutations have been reported in the human FLG gene4, including three in filaggrin repeat 6. Importantly, the truncated profilaggrin species resulting from mutations in repeats 7 and 10 of FLG have been shown to be highly unstable and incapable of being processed to functional filaggrin4, exactly as shown in the ft mouse here (Fig. 2), and previously11. Thus, the 5303delA mutation is a functional null allele, highly analogous to many of the human filaggrin mutations now emerging and is perhaps a better model for the human mutations than a knockout mouse, which has proven difficult to generate due to the repetitive nature of this locus (R.B.P. and W.H.I.M., unpublished data). Availability of specific genotyping assays for the mutant allele allows generation of a range of congenic strains and double mutant mice to further dissect the immunological aspects of the biological effects of filaggrin mutations.
Although ft/ft mice have been shown to closely model the phenotypic, histological and ultrastructural characteristics of ichthyosis vulgaris in humans11, we show here that these filaggrin-deficient mice are additionally predisposed to develop sensitization following percutaneous exposure to a model allergen and that these homozygous mice develop cutaneous inflammatory infiltrates and allergen-specific immune responses following sensitization to allergen (Fig. 5). Following sensitization a further defect in skin barrier as measured by elevated TEWL is seen, suggesting that the mild initial heritable barrier defect is exacerbated by allergic sensitization. These results suggest that sensitization may also be an early event in filaggrin-deficient humans. A number of known modifiers, for example Th2 cytokines, including IL-13 and IL-4 have been shown to down-regulate expression of filaggrin26 and other key epidermal structural proteins27, and unknown genetic factors may also influence the functionality of ft/ft mice; these areas require further experimentation. It is noteworthy that the PBS-treated ft/ft mice showed no elevation of total IgE up to 12–13 weeks of age (Fig. 5). Indeed, in 8 month old untreated ft/ft mice, we have observed no increase in non-specific IgE or elevated Th2 cytokine (IL-4, IL-5 and IL-13) production, and there are no previous reports of spontaneous atopy or eczema in ft/ft mice10,11. Thus ft/ft mice maintained under Specific Pathogen-Free conditions do not appear to develop spontaneous allergic responses, in contrast to NC/Nga mice28.
As noted above the role of elevated IgE in human atopic dermatitis remains unresolved. Elevated IgE may contribute to disease pathogenesis or may be an epiphenomenon reflective of disease severity, or both. Given a recent report on the development of a mouse model of intrinsic atopic eczema29, highlighting the potential for eczema in mice in the absence of evaluated IgE, caution is needed in assigning an obligate pathogenic role for elevated IgE in murine atopic dermatitis. While clarification of this relationship will require more experimentation on mouse models in addition to human studies, we have shown enhanced percutaneous priming in a murine analogue of human filaggrin deficiency that hints at possible mechanisms in the human disease.
We have demonstrated that the causative mutation in the ft mouse is the 5303delA frameshift mutation in the flg gene, analogous to many human loss of function filaggrin mutations.. Furthermore, we show that homozygous, but not heterozygous, ft mice have elevated susceptibility to cutaneous exposure to a model allergen, indicative of altered skin barrier immunity in these mice. The identification of this murine flg mutation and demonstration of altered allergen sensitization in ft sets the scene for the use and further refinement of this animal model to further dissect the role of filaggrin in skin barrier function and the genesis of atopic eczema.
METHODS
Sequence analysis
The genomic sequence of the murine filaggrin locus derived from BAC clone RP23-227A12 (accession number AC158361.3) was annotated using the ClustalW tools within MacVector version 9.0 (MacVector Inc., Cambridge, UK). To identify the causative mutation in filaggrin in ft/ft mice, we applied the shotgun method recently developed to identify filaggrin mutations in human AD patients18. We used six pairs of PCR primers based on flg exon 3 sequence on BAC clone RP23-227A12, which were designed to amplify the each filaggrin homologous sequence unit (Supplementary Table S3). The PCR products were cloned into the TA-cloning vector pGEM-tEasy (Promega, Tokyo, Japan). Plasmid DNA template was generated using the TempliPhi system (GE Healthcare Biosciences, Tokyo, Japan) and directly sequenced. An identical one base deletion of A was identified in shotgun clones derived from four independent PCR fragments. To confirm the mutation and determine the full sequence of flg from the mutant strain, a BAC library was constructed from homozygous ft/ft mice, which was screened by hybridization and the resultant hits confirmed by PCR (Amplicon Express Inc., Pullman, WA). From a BAC containing the entire locus, exon 3 was subcloned into a plasmid vector by recombineering (Red/ET system, Gene Bridges GmbH, Heidelberg, Germany). The subclone was subjected to random transposon insertion using the EZ-Tn5 <KAN-2> insertion kit (EPICENTRE Biotechnologies Inc., Madison, WI) and 96 transposon insertion clones were sequenced bidirectionally from the Tn5 insertion site, allowing full assembly of exon 3 with ~9-fold over-sequencing.
Genotyping
For allele-specific PCR detection for the ft mutation, genomic DNA (2 ng) was amplified with primers mFlgdelA-F and mFlgdelA-R primers (Supplementary Table 3) using x2 KOD FX PCR buffer containing 1.5 nmol MgCl2, 0.2 U of KOD FX DNA polymerase (Toyobo Biochemicals, Osaka, Japan). PCR amplification conditions were follows: (94°C, 2 min) x1; (94°C 15 sec, 70°C 30 sec, 72°C 2 min) x35; (72°C 5 min) x1. PCR products were analyzed on 12% polyacrylamide gels. This PCR primer set is positive for ft/ft and wt/ft but is negative for wt/wt (Fig. 2d). To more conveniently genotype homozygotes, heterozygotes and wild-type mice, a PCR assay was developed to detect a 15 bp insertion polymorphism at the 5’ end of exon 3 in linkage disequilibrium with the ft mutation. This insertion creates an Acc I restriction site which was exploited for rapid genotyping. Primers FTins15.F and FTins15.R (Supplementary Table 3) were used in standard PCR reactions using the following amplification conditions: (94°C 5 min) x1; (94°C 30 sec, 58°C 30 sec, 72°C 1 min) x35; (72°C 5 min) x1. 5 µl of PCR product was digested overnight with 2.5 U Acc I and resolved on 1.5% agarose gels. The 678 bp C57BL/6 allele does not digest; the ft allele cuts to yield fragments of 559 bp and 134 bp (Fig. 2e).
C-terminal antibody
The C-terminal antibody specific for mouse profilaggrin was generated against the amino acid sequence ESIFTAKHLDFNQSHS. The peptide was conjugated to KLH via a N-terminal cysteine residue prior to injection into New Zealand white rabbits (Genemed Synthesis, Inc., South San Francisco, CA). The serum was affinity purified prior to use. Urea/Tris extracts were prepared from newborn homozygous ft/ft mice and control C57BL/6 mice as described previously11. Protein samples were separated on 5–12% SDS/polyacrylamide gels and transferred to PVDF membrane (Millipore, Billerica, MA). Blots were probed with C-terminal antibody and immunoreactive proteins visualized with ECL (Amersham/GE Healthcare, Pittsburgh, PA). A duplicate gel was stained with Coomassie Brilliant Blue R-250.
Mice
Breeding pairs of flaky tail (ft/ft) mice were provided by J.S. The ft/ft mice were backcrossed four generations on a C57BL/6J strain background and C57BL/6J mice were used as wild-type (wt/wt) controls. Female mice were used in all experiments unless otherwise indicated. Flaky tail (ft/ft) mice were crossed with wild-type C57BL/6J mice to generate flaky tail heterozygous (wt/ft) mice. The OVA cutaneous sensitization experiments were repeated with F7 backcrossed mice, with littermates from wt/ft to wt/ft matings genotyped as wt/wt, wt/ft and ft/ft and used in experiments, giving identical results as F4 backcross (data not shown). The mice were housed in SPF conditions, with irradiated diet and bedding provided. All animal experiments were performed in compliance with Irish Department of Health and Children regulations and approved by the Trinity College Dublin’s BioResources ethical review board.
Cutaneous challenge with OVA
The intact skin of wt/wt, wt/ft and ft/ft mice was challenged with OVA (Sigma Chemicals, Poole, UK) by cutaneous exposure. OVA was applied in a regimen involving three cycles of daily exposure to OVA for 5 consecutive days (see Supplementary Fig. S1a). The abdominal hair was carefully shaved 24 hours before application of OVA; mice with any visual damage to the skin were not used. OVA (fraction V; Sigma) was prepared in PBS (Dulbecco’s PBS; Sigma) at a concentration of 1 mg/ml. Mice were restrained and 50 µl of OVA solution, or 50 µl of PBS, was applied to the shaved skin and allowed to air dry. To address systemic allergen treatment, wt/wt and ft/ft mice were challenged with OVA in alum as an adjuvant by intra-peritoneal injection, as described30, see Supplementary Fig. S3b.
Measurement of trans-epidermal water loss (TEWL)
A Courage and Khazaka Tewameter TM210 (Enviroderm, Evesham, UK) was used for measurement of TEWL. TEWL analysis of the abdomen of male and female adult (6–10 weeks of age) wt/wt and ft/ft mice occurred 24 hours after shaving. Any mice with visual damage to the skin following shaving were not included in the analysis. TEWL was recorded at ambient temperature 19–21°C and humidity 50% ± 5. In mice sensitized to OVA, TEWL was measured in PBS- and OVA-treated mice after OVA application.
Spleen cell preparation
Single cell suspensions were prepared from spleens. Spleen cells were cultured in RPMI 1640 (Gibco-Invitrogen, Dun Laoghaire, Ireland) supplemented with 10% (v/v) heat-inactivated fetal calf serum (FCS; Labtech, Ringmer, UK), 2 m M L-glutamine (Gibco), 50 U/ml penicillin and 50 µg/ml streptomycin (Gibco). Spleen cells were plated at 5 × 106 cells/ml. OVA was subjected to endotoxin removal for use in cell assays as described30. Cells were unstimulated (media) or stimulated with OVA (250 and 500 µg/ml), in a 24 well plate (Greiner Bio-One, Frickenhausen, Germany) at 37°C for 72 hours. Supernatants were harvested after 72 hours and cytokine levels (IL-4, IL-5, IL-13, IFN-γ, IL-10 and IL-17) were measured by ELISA. For cell proliferation analysis cells were exposed to a range of concentrations of OVA, in triplicate wells on 96 well plates. Cultures were pulsed with 1 µCi per well [3H]thymidine (Amersham, Little Chalfont, UK) for the last 14 hours of culture. Cells were harvested with a cell harvester (Tomec Inc., Hamden, CT, USA) and [3H]thymidine incorporation was measured by a Wallac β-counter (Perkin-Elmer Inc., Waltham, MA, USA).
Ab and cytokine ELISA
Serum OVA-specific IgG, IgG1 and IgG2a levels were detected by direct ELISA using serial dilutions of sera. OVA-specific IgE was measured using biotinylated-OVA with 1:10 sera dilutions. Total serum IgE was measured using PharMingen antibodies (BD Biosciences, Oxford, UK), according to the manufacturers instructions. Total serum IgG was detected using commercial capture antibodies and IgG standard (Zymed Laboratories, Ca, USA; Sigma). Sandwich ELISAs were performed to quantify levels of specific cytokines in the supernatants cell cultures (IL-4, IL-5 and IL-13, PharMingen; IL-10, IL-17 and IFN-γ, R&D Systems, Abingdon, UK).
Histology
Abdominal skin of age- and sex-matched mice (wt/wt; wt/ft and ft/ft) that were untreated or exposed to OVA was removed and fixed in 10% formaldehyde saline. Paraffin sections were stained with hematoxylin and eosin. Tissue sections were analysed by JJC with slides blinded to murine genotype and exposure to allergen. To quantify the individual inflammatory cells infiltrating the skin a previous described scoring system was used20. The total number of cells, lymphocytes, eosinophils and mononuclear cells were counted per x1,000 high-power fields (HPF) of view, with 15–20 HPFs scored per mouse. Lungs were processed for histopathological analysis as described previously30.
OVA challenge regimen for Airway hyper responsiveness (AHR)
wt/wt and ft/ft mice were exposed to OVA cutaneously or intraperitoneally and then exposed to 20 minutes of continuous aerosol of 1% OVA each day for five consecutive days, as shown in Supplementary Fig. S1b & S3a, respectively. Epicutaneous exposure to OVA was achived by repeated tape-stripping of shaved skin25, prior to OVA application using the regime in Supplementary Fig S1b. AHR was analyzed the day following the final aerosol challenge. Mice were anesthetized, tracheostomized and ventilated. AHR was measured using a whole-body plethysmograph with a pneumotachograph linked to a transducer (EMMS, Hants, UK). The average lung resistence (RL) and the area under the resistence curve (RL AUC) parameters were quantified for 3 minute periods at baseline and after exposure to nebulized endotoxin-free PBS (Sigma) and acetyl-β-methycholine chloride (Sigma) at 10, 30, 60, 120 mg/ml. Bronchoalveolar lavage (BAL) fluids were collected by canulating the trachea and lavaging the lungs with a 1.0 ml of ice cold PBS. The BAL cells were pelleted, washed, and counted. The numbers of eosinophils in BAL were analysed by flow cytometry, as described30. Lungs were removed from mice for histology, see above.
Statistics
All animal experiments were repeated at least twice. GraphPad Prism and GraphPad Instat software were used for data analysis. Difference between groups was determined by Student’s t-test or, if non-parametric, Welch corrected t-test. Differences were considered significant when P<0.05.
Supplementary Material
REFERENCES
- 1.Smith FJD, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 2.Palmer CNA, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 3.Baurecht H, et al. Toward a major risk factor for atopic eczema: meta-analysis of filaggrin polymorphism data. J Allergy Clin Immunol. 2007;120:1406–1412. doi: 10.1016/j.jaci.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 4.Sandilands A, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 5.Henderson J, et al. The burden of disease associated with filaggrin mutations: a population-based, longitudinal birth cohort study. J Allergy Clin Immunol. 2008;121:872–877. doi: 10.1016/j.jaci.2008.01.026. e9. [DOI] [PubMed] [Google Scholar]
- 6.Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immunol. 2006;118:209–213. doi: 10.1016/j.jaci.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 7.Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol. 2003;112:S118–S127. doi: 10.1016/j.jaci.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 8.Illi S, et al. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J Allergy Clin Immunol. 2004;113:925–931. doi: 10.1016/j.jaci.2004.01.778. [DOI] [PubMed] [Google Scholar]
- 9.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 10.Sundberg JP. The flaky tail (ft) mutation. in Handbook of mouse mutations with skin and hair abnormalities. In: Sundberg JP, editor. Animal models and biochemical tools. Vol. 2. Ann Arbor: CRC Press; 1984. pp. 269–273. [Google Scholar]
- 11.Presland RB, et al. Loss of normal profilaggrin and filaggrin in flaky tail (ft/ft) mice: an animal model for the filaggrin-deficient skin disease ichthyosis vulgaris. J Invest Dermatol. 2000;115:1072–1081. doi: 10.1046/j.1523-1747.2000.00178.x. [DOI] [PubMed] [Google Scholar]
- 12.Pearton DJ, Dale BA, Presland RB. Functional analysis of the profilaggrin N-terminal peptide: identification of domains that regulate nuclear and cytoplasmic distribution. J Invest Dermatol. 2002;119:661–669. doi: 10.1046/j.1523-1747.2002.01831.x. [DOI] [PubMed] [Google Scholar]
- 13.Rothnagel JA, Mehrel T, Idler WW, Roop DR, Steinert PM. The gene for mouse epidermal filaggrin precursor. Its partial characterization, expression, and sequence of a repeating filaggrin unit. J Biol Chem. 1987;262:15643–15648. [PubMed] [Google Scholar]
- 14.Presland RB, Bassuk JA, Kimball JR, Dale BA. Characterization of two distinct calcium-binding sites in the amino-terminus of human profilaggrin. J Invest Dermatol. 1995;104:218–223. doi: 10.1111/1523-1747.ep12612770. [DOI] [PubMed] [Google Scholar]
- 15.Zhang D, Karunaratne S, Kessler M, Mahony D, Rothnagel JA. Characterization of mouse profilaggrin: evidence for nuclear engulfment and translocation of the profilaggrin B-domain during epidermal differentiation. J Invest Dermatol. 2002;119:905–912. doi: 10.1046/j.1523-1747.2002.00133.x. [DOI] [PubMed] [Google Scholar]
- 16.Rothnagel JA, Steinert PM. The structure of the gene for mouse filaggrin and a comparison of the repeating units. J Biol Chem. 1990;265:1862–1865. [PubMed] [Google Scholar]
- 17.Gan SQ, McBride OW, Idler WW, Markova N, Steinert PM. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry. 1990;29:9432–9440. doi: 10.1021/bi00492a018. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki T, et al. Sequence analysis of filaggrin gene by novel shotgun method in Japanese atopic dermatitis. J Dermatol Sci. 2008;51:113–120. doi: 10.1016/j.jdermsci.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Terada M, et al. Contribution of IL-18 to atopic-dermatitis-like skin inflammation induced by Staphylococcus aureus product in mice. Proc Natl Acad Sci U S A. 2006;103:8816–8821. doi: 10.1073/pnas.0602900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spergel JM, Mizoguchi E, Oettgen H, Bhan AK, Geha RS. Roles of TH1 and TH2 cytokines in a murine model of allergic dermatitis. J Clin Invest. 1999;103:1103–1111. doi: 10.1172/JCI5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snapper CM, Paul WE. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 22.Weidinger S, et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008;121:1203–1209. doi: 10.1016/j.jaci.2008.02.014. e1. [DOI] [PubMed] [Google Scholar]
- 23.Marenholz I, et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J Allergy Clin Immunol. 2006;118:866–871. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 24.McLean WHI, et al. Filaggrin variants confer susceptibility to asthma. J Allergy Clin Immunol. 2008;121:1294–1295. doi: 10.1016/j.jaci.2008.02.039. author reply 1295–1296. [DOI] [PubMed] [Google Scholar]
- 25.Spergel JM, et al. Epicutaneous sensitization with protein antigen induces localized allergic dermatitis and hyperresponsiveness to methacholine after single exposure to aerosolized antigen in mice. J Clin Invest. 1998;101:1614–1622. doi: 10.1172/JCI1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell MD, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2007;120:150–155. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim BE, Leung DY, Boguniewicz M, Howell MD. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clin Immunol. 2008;126:332–337. doi: 10.1016/j.clim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda H, et al. Development of atopic dermatitis-like skin lesion with IgE hyperproduction in NC/Nga mice. Int Immunol. 1997;9:461–466. doi: 10.1093/intimm/9.3.461. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Overbergh L, Mathieu C, Chan LS. The development of atopic dermatitis is independent of Immunoglobulin E up-regulation in the K14-IL-4 SKH1 transgenic mouse model. Clin Exp Allergy. 2008;38:1367–1380. doi: 10.1111/j.1365-2222.2008.02987.x. [DOI] [PubMed] [Google Scholar]
- 30.Mangan NE, van Rooijen N, McKenzie AN, Fallon PG. Helminth-modified pulmonary immune response protects mice from allergen-induced airway hyperresponsiveness. J Immunol. 2006;176:138–147. doi: 10.4049/jimmunol.176.1.138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.