Abstract
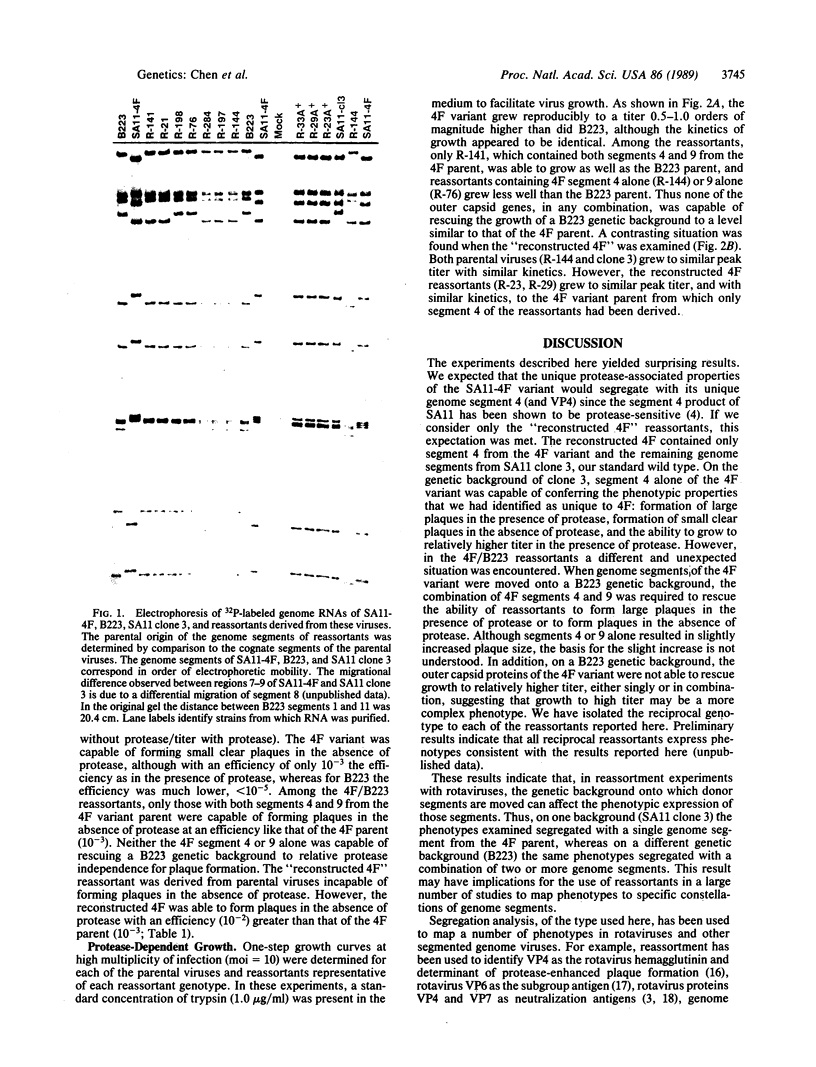
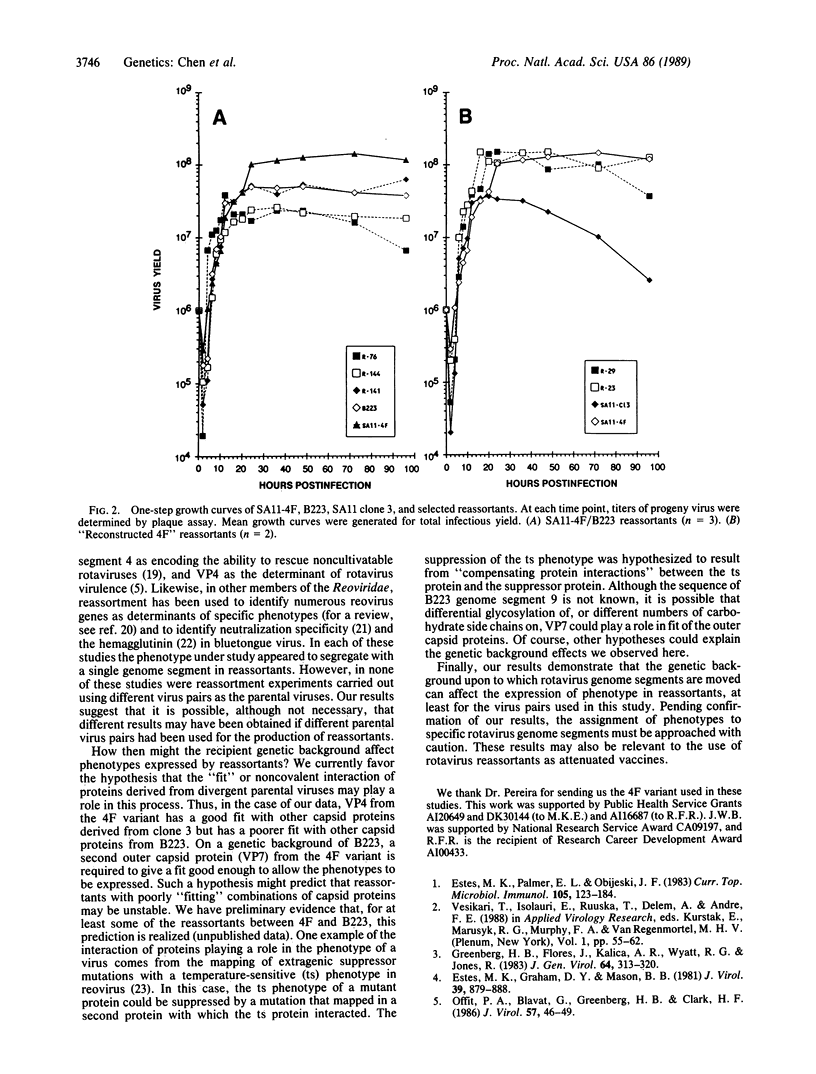
We have previously characterized the biological and immunological properties of a simian rotavirus SA11 variant (4F) with an altered genome segment 4. The SA11-4F variant formed large plaques in the presence of protease, formed small clear plaques in the absence of protease, and grew to high titer in the presence of protease when compared to our standard wild type (SA11 clone 3). To determine the genome segment of the rotavirus SA11 variant 4F that encoded the unique protease-associated phenotypes of the variant, reassortants were generated that segregated the outer capsid genes of 4F onto a genetic background derived from either the bovine rotavirus B223 or our standard SA11 wild type (clone 3), both of which have contrasting protease-associated phenotypes. The parental and reassortant viruses were examined to determine which genes from the 4F variant encoded the ability (i) to form large plaques in the presence of protease, (ii) to form small clear plaques in the absence of exogenous protease, and (iii) to grow to significantly higher titer in the presence of protease. These phenotypes could be transferred to a clone 3 genetic background by a single genome segment from the 4F variant segment 4. However, in the 4F/B223 reassortants a different and unexpected situation was found. On a B223 genetic background the same phenotypes segregated with a combination of a minimum of two 4F genome segments, segments 4 and 9. These results indicate that the recipient genetic background onto which the genes of a donor rotavirus are reassorted can affect the phenotypes conferred by the presence of the donor segments. Thus, the results of segregation mapping experiments using reassortant viruses should be interpreted with caution.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burns J. W., Greenberg H. B., Shaw R. D., Estes M. K. Functional and topographical analyses of epitopes on the hemagglutinin (VP4) of the simian rotavirus SA11. J Virol. 1988 Jun;62(6):2164–2172. doi: 10.1128/jvi.62.6.2164-2172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. K., Penaranda M. E., Crawford S. E., Estes M. K. Two glycoproteins are produced from the rotavirus neutralization gene. Virology. 1986 Jun;151(2):243–252. doi: 10.1016/0042-6822(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Cowley J. A., Gorman B. M. Genetic reassortants for identification of the genome segment coding for the bluetongue virus hemagglutinin. J Virol. 1987 Jul;61(7):2304–2306. doi: 10.1128/jvi.61.7.2304-2306.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M. K., Graham D. Y., Mason B. B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981 Sep;39(3):879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M. K., Palmer E. L., Obijeski J. F. Rotaviruses: a review. Curr Top Microbiol Immunol. 1983;105:123–184. doi: 10.1007/978-3-642-69159-1_3. [DOI] [PubMed] [Google Scholar]
- Gombold J. L., Ramig R. F. Analysis of reassortment of genome segments in mice mixedly infected with rotaviruses SA11 and RRV. J Virol. 1986 Jan;57(1):110–116. doi: 10.1128/jvi.57.1.110-116.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg H. B., Flores J., Kalica A. R., Wyatt R. G., Jones R. Gene coding assignments for growth restriction, neutralization and subgroup specificities of the W and DS-1 strains of human rotavirus. J Gen Virol. 1983 Feb;64(Pt 2):313–320. doi: 10.1099/0022-1317-64-2-313. [DOI] [PubMed] [Google Scholar]
- Greenberg H. B., Kalica A. R., Wyatt R. G., Jones R. W., Kapikian A. Z., Chanock R. M. Rescue of noncultivatable human rotavirus by gene reassortment during mixed infection with ts mutants of a cultivatable bovine rotavirus. Proc Natl Acad Sci U S A. 1981 Jan;78(1):420–424. doi: 10.1073/pnas.78.1.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg H. B., Valdesuso J., van Wyke K., Midthun K., Walsh M., McAuliffe V., Wyatt R. G., Kalica A. R., Flores J., Hoshino Y. Production and preliminary characterization of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J Virol. 1983 Aug;47(2):267–275. doi: 10.1128/jvi.47.2.267-275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlon J., Sugiyama K., Roy P. Molecular basis of bluetongue virus neutralization. J Virol. 1983 Dec;48(3):627–632. doi: 10.1128/jvi.48.3.627-632.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalica A. R., Flores J., Greenberg H. B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983 Feb;125(1):194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- Kapikian A. Z., Cline W. L., Greenberg H. B., Wyatt R. G., Kalica A. R., Banks C. E., James H. D., Jr, Flores J., Chanock R. M. Antigenic characterization of human and animal rotaviruses by immune adherence hemagglutination assay (IAHA): evidence for distinctness of IAHA and neutralization antigens. Infect Immun. 1981 Aug;33(2):415–425. doi: 10.1128/iai.33.2.415-425.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips T. H., Ramig R. F. Extragenic suppression of temperature-sensitive phenotype in reovirus: mapping suppressor mutations. Virology. 1984 Jun;135(2):428–439. doi: 10.1016/0042-6822(84)90198-3. [DOI] [PubMed] [Google Scholar]
- Offit P. A., Blavat G., Greenberg H. B., Clark H. F. Molecular basis of rotavirus virulence: role of gene segment 4. J Virol. 1986 Jan;57(1):46–49. doi: 10.1128/jvi.57.1.46-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit P. A., Blavat G. Identification of the two rotavirus genes determining neutralization specificities. J Virol. 1986 Jan;57(1):376–378. doi: 10.1128/jvi.57.1.376-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira H. G., Azeredo R. S., Fialho A. M., Vidal M. N. Genomic heterogeneity of simian rotavirus SA11. J Gen Virol. 1984 Apr;65(Pt 4):815–818. doi: 10.1099/0022-1317-65-4-815. [DOI] [PubMed] [Google Scholar]
- Ramig R. F. Isolation and genetic characterization of temperature-sensitive mutants of simian rotavirus SA11. Virology. 1982 Jul 15;120(1):93–105. doi: 10.1016/0042-6822(82)90009-5. [DOI] [PubMed] [Google Scholar]
- Sabara M., Gilchrist J. E., Hudson G. R., Babiuk L. A. Preliminary characterization of an epitope involved in neutralization and cell attachment that is located on the major bovine rotavirus glycoprotein. J Virol. 1985 Jan;53(1):58–66. doi: 10.1128/jvi.53.1.58-66.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode G. N., Kelso N. E., Simpson T. F., Gaul S. K., Evans L. E., Babiuk L. Antigenic relationships among some bovine rotaviruses: serum neutralization and cross-protection in gnotobiotic calves. J Clin Microbiol. 1983 Aug;18(2):358–364. doi: 10.1128/jcm.18.2.358-364.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]