Abstract
Human DNA glycosylase NEIL1 exhibits a superior ability to remove oxidized guanine lesions guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) from duplex DNA in comparison to other substrates. In the present work, Gh and Sp lesions in bubble, bulge and single-stranded DNA were found to be good substrates for NEIL1 but were typically excised at much slower rates than from canonical duplex substrates. A notable exception was the activity of NEIL1 on removal of Gh in bubble structures which approaches that of the normal duplex substrate. The cleavage of Gh in the template strand of a replication or transcription bubble may prevent mutations associated with Gh during replication or transcription. However, hydantoin lesion removal in the absence of an opposite base may also result in strand breaks and potentially deletion and frameshift mutations. Consistent with this as a potential mechanism leading to an N-1 frameshift mutation, the nick left after the removal of the Gh lesion in a DNA bulge by NEIL1 was efficiently religated in the presence of polynucleotide kinase (PNK) and human DNA ligase III (Lig III). These results indicate that NEIL1 does not require a base opposite to identify and remove hydantoin lesions. Depending on the context, the glycosylase activity of NEIL1 may stall replication and prevent mutations or lead to inappropriate removal that may contribute to the mutational spectrum of these unusual lesions.
Reactive oxygen species (ROS) are generated in mitochondria as byproducts of oxygen respiration. These radicals can be over-produced in cells under oxidative stress, due to inflammation, or as a result of exposure to toxic agents including ionizing radiation (1, 2). Although ROS are produced as normal products of cellular metabolism, excessive free radicals can react with DNA, RNA, lipids and proteins, resulting in deleterious effects. Oxidative DNA damage, including abasic sites, base lesions, DNA strand breaks and DNA-protein cross-links, are implicated in a number of diseases such as cancer, aging and neurological diseases (3–6).
Due to its low redox potential, guanine is the most susceptible to oxidation leading to 8-oxo-7,8-dihydro-guanine (OG) as one of the major products (7). OG is an abundant damaged base in vivo, being generated at ~2000 lesions per human cell per day (8, 9) (10); however, it is also extremely sensitive to further oxidation because its redox potential is approximately 600 mV below that of guanine (11). Two major secondary oxidation products of OG, guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) (12, 13), have garnered the most attention. Sp was detected by Hailer et al. in Nei-deficient Escherichia coli (E. coli) after chromate treatment (14). In vitro, these hydantoin lesions can be generated from G or OG with a variety of oxidants including one-electron oxidants, singlet oxygen, peroxyl radicals and transition metal complexes (15–17). Their formation in oligonucleotides is highly dependent on pH and temperature, with Sp being the major product in single-stranded DNA while Gh is formed in duplex DNA or in nucleosides under acidic conditions (12, 18). However, it is not clear if Sp and Gh can be formed from hydroxyl-radical mediated oxidation of OG.
Oxidative DNA base damage is repaired primarily by the base excision repair (BER) pathway (5). This repair pathway is initiated by damage-specific DNA glycosylases that recognize and remove the base at the damaged nucleotide (5, 19). Full repair requires the activity of several other enzymes. In many cases the abasic site is further processed by AP endonucleases (APE) and phosphodiesterases that cleave out the remaining sugar fragments to yield the proper ends for DNA polymerases. DNA polymerases then act accordingly to incorporate a correct base opposite the complementary strand, and DNA ligase seals the nick as the last step of BER. Many glycosylases involved in repair of oxidative damage possess associated β- and δ-lyase activities that mediate DNA strand scission at the site of the damaged nucleotide (19). In this case, the action of polynucleotide kinase (PNK) rather than APE can provide the proper ends for the DNA polymerase (20). The repair of 8-oxoguanine has been extensively characterized in both bacterial and mammalian cells (5). In E. coli, the Fpg glycosylase removes OG from OG:C base pairs while MutY removes A from OG:A mispairs formed during replication. The functional homolog to Fpg in mammalian cells is OGG1 that acts mainly on OG and formamidopyrimidine (FapyG, FapyA) lesions (21, 22).
Two glycosylases, endonuclease VIII (Nei) and endonuclease III (Nth), have been identified in prokaryotes for excising oxidized pyrimidines (5, 19, 23). Several “Nei-like” glycosylase enzymes from mimivirus (MvNEIL), murine (mNEIL) and human (NEIL1) have also been identified and characterized (14, 24, 25). These glycosylases are structural homologs to Nei and Fpg and have a wide substrate range. NEIL1 can cleave ring-opened formamidopyrimidines (FapyG, FapyA), thymine glycol (Tg), 5-hydroxycytosine (5-OHC) and 5-hydroxyuracil (5-OHU) from double-stranded DNA (26–28). Recently, we established that the hydantoin lesions Gh and Sp are superior substrates for NEIL1 (29). Gh and the two Sp diastereomers, Sp1 and Sp2, are cleaved 100-fold faster than the initially reported substrates Tg and 5-OHC (29). Previous studies demonstrated that these hydantoin lesions are removed indiscriminate of the opposite base in double-stranded DNA, a process that would promote mutations when the lesions are mispaired (29).
Frameshift mutations have been observed in plasmid DNA treated with methylene blue and light, and these authors demonstrated that the lesion responsible could not be OG (30). Sp was later found to be formed under these singlet oxygen conditions (31), and an in vitro polymerase study showed that hydantoins may participate in sequence-directed primer slippage, forming a bulged lesion structure. These results suggested that Sp and/or Gh may be responsible for the frameshift mutations observed during singlet oxygen damage (32, 33). Recent in vivo studies with hydantoin lesions showed that the lesion bypass efficiency and the mutation type varied in different sequence contexts, although the hydantoins were highly mutagenic in any sequence, leading to G→T and G→C transversion mutations. (34, 35).
Given the role of glycosylases in initiating repair of base lesions, the impact of the base removal activity on mutagenesis is highly influenced by the context in which it occurs. Facile repair of a lesion in an inappropriate context may contribute to the mutagenic potential of a given lesion. Thus, in order to elucidate the influence of NEIL1 on the mutation spectrum of Gh and Sp, we evaluated the kinetics of removal of Gh, Sp1 and Sp2 in single-stranded DNA, bulge and bubble-containing duplexes and several different DNA duplex sequence contexts. Gh and Sp1 are removed from single-stranded DNA, albeit with a significantly reduced rate compared to the corresponding duplex substrates. However, the hydantoin lesions, most notably Gh, are good substrates in bubble and bulge-containing duplex structures. Quantitative measurements revealed that the removal of the hydantoin lesions from bubble and bulge structures is slower than from double-stranded DNA, but faster than from single-stranded DNA. The sequence context of the lesion does not appear to influence the removal of hydantoin lesions from duplex DNA substrates. Lastly, we demonstrate here that reconstitution of a BER system comprising NEIL1, polynucleotide kinase (PNK) and human DNA ligase III (Lig III), leads to an N-1 deletion when the Gh lesion is present in a bulge structure. Based on these observations, the repair of Gh, Sp1 and Sp2 lesions by NEIL1 in these contexts may lead to frameshift and point mutations.
Materials and methods
General materials and instrumentation
All buffers were prepared with distilled, deionized water from a Barnstead ultrapure water system and passed through a 0.45 µm filter before use. Storage phosphor autoradiography was performed on a Typhoon 9400 phosphorimager. Gel electrophoresis was analyzed using ImageQuaNT software (version 5.2) and the rate constants were determined using GraFit 5.0 software. [γ-32P]-ATP was purchased from ICN. T4 Polynucleotide kinase and Mung Bean Nuclease (MBN) was obtained from New England BioLabs. Human ligase III was purchased from Enzymax. Oligonucleotides containing Tg were purchased from Midland Reagents. G-25 spin columns were obtained from Amersham Pharmacia. All other chemicals used for these experiments were purchased from Fisher Scientific, VWR, or Sigma.
NEIL1 purification
NEIL1 was purified from Rosetta (DE3) pLysS cells strains (Novagen) using a pET30a plasmid containing the NEIL1 gene as previously described (29). The active enzyme concentration was measured and reaction conditions were optimized as previously reported (29).
DNA preparation
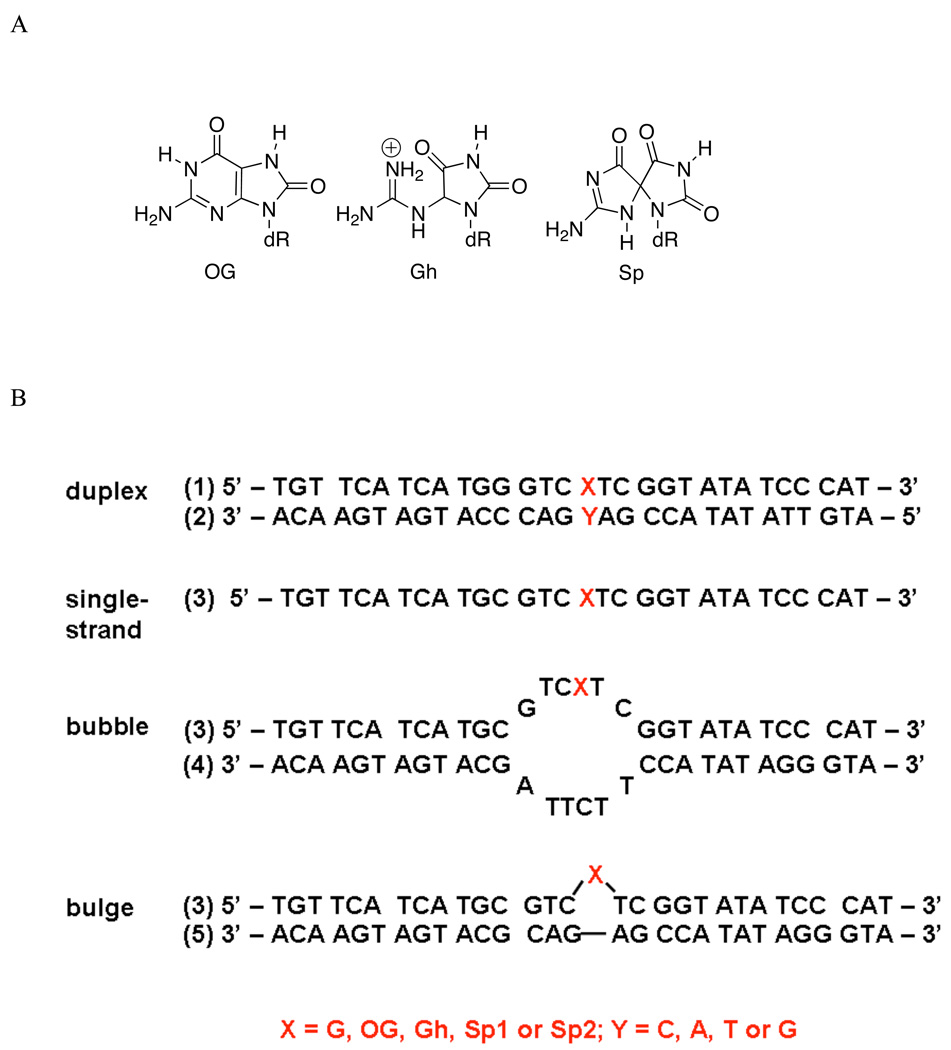
A 30-nt sequence (3) that was used for constructing the bubble, bulge and single-stranded DNA substrates is shown in Table 1. A partial complement (4) was annealed to form a six-nucleotide bubble structure; similarly a 29-nt complement (5) was annealed to the 30-nt strand to form a single-base bulge structure. Figure 1 shows the resulting bubble and bulge structures used in the study. Three 40-nt strands were annealed with their complementary strands to form duplexes with G opposite the lesion Gh, (6•7), (8•9), and (10•11).
Table 1.
Sequences used in the glycosylase study
| (1) 5’ – TGTTCATCATGGGTCXTCGGTATATCCCAT – 3’ |
| (2) 3’ – ACAAGTAGTACCCTGNAGCCATATAGGGTA – 5’ |
| (3) 5’ – TGTTCATCATGCGTCXTCGGTATATCCCAT – 3’ |
| (4) 3’ – ACAAGTAGTACGATTCTTCCATATAGGGTA – 5’ |
| (5) 3’ – ACAAGTAGTACGCAGAGCCATATAGGGTA – 5’ |
| (6) 5’ – TCATGGGTCYTCGGTATATCAGTGCTATCACATTAGTGTA – 3’ |
| (7) 3’ – AGTACCCAGGAGCCATATAGTCACGATAGTGTAATCACAT – 5’ |
| (8) 5’ – TCATGGGTGYACGGTATATCAGTGCTATCACATTAGTGTA – 3’ |
| (9) 3’ – AGTACCCACGTGCCATATAGTCACGATAGTGTAATCACAT – 5’ |
| (10) 5’ – TCATGGGTTYGCGGTATATCAGTGCTATCACATTAGTGTA – 3’ |
| (11) 3’ – AGTACCCAAGCGCCATATAGTCACGATAGTGTAATCACAT – 5’ |
X = OG, Tg, Gh, Sp1 and Sp2; Y = Gh; N = C, A, T or G
Figure 1.
DNA lesion structures and contexts used in glycosylase assays. A) Chemical structures of lesions examined in various contexts B) A six-nucleotide mispair was used to form a bubble in the middle of a 30-nucleotide duplex. For the bulge-containing duplex, a 29-nucleotide complement was annealed with the lesion-containing 30-nucleotide strand to form a bulge in the middle of the duplex as shown.
All oligonucleotides (except those containing Tg) were synthesized by the DNA/Peptide Core Facility (University of Utah). The Tg-containing oligonucleotide was used as provided by Midland Reagents as a mixture of possible diastereomers (36). The OG-containing oligonucleotides were cleaved from the column overnight in fresh 30% NH4OH containing 0.25% β-mercaptoethanol and then deprotected for 17 h in 55°C water bath. Gh and Sp-containing oligonucleotides were prepared by oxidation of OG-containing oligonucleotides using K2IrCl6 and confirmed by ESI-MS as previously described (29). Sp1 and Sp2 were collected separately and named according to their order of elution on anion exchange HPLC. All the oligonucleotides were 99% pure after the purification via HPLC with a Dionex DNAPac PA-100 ion exchange column (4 mm × 250 mm). Separation and purification were achieved with buffer solutions consisting of 35% solvent B and 65% A initially, with a gradient to 100% solvent B. (Solvent A contained 10% acetonitrile and 90% H2O, while solvent B contained 10% acetonitrile with 1.5 M ammonium acetate, pH 7, flow rate = 1 mL min−1). UV spectra were recorded at 260 nm. Lesion-containing oligodeoxynucleotides were radiolabeled using [γ-32P]-ATP with T4 polynucleotide kinase at 37 °C. After purification with a ProbeQuant G-25 spin column, labeled oligodeoxynucleotides were annealed with a 20% excess of the complementary strand in annealing buffer (10 mM EDTA, 150 mM NaCl and 20 mM Tris-HCL, pH 7.6) at 90 °C for 5 min and gradually cooled to room temperature.
Characterization of bubble, bulge and single-stranded oligodeoxynucleotides
Both strands used to form the bubble and bulge structures were labeled with [γ-32P]-ATP and then annealed. Single-stranded DNA, bubble and bulge structures with or without lesions were detected by non-denaturing PAGE. A Mung Bean Nuclease (MBN) assay was performed by incubating 20 units of MBN with 100 nM solutions of different DNA structures at 25 °C for 30 min in 1X Mung Bean Nuclease reaction buffer (50 mM sodium acetate, 30 mM NaCl, 1 mM ZnSO4, pH 5.0 at 25°C). The reaction mixtures were quenched with EDTA and then loaded onto 15% denaturing PAGE.
Glycosylase assay
The glycosylase activity of NEIL1 was evaluated with single-turnover experiments as previously described (29). The rate constants of removal of hydantoin lesions from single-stranded oligonucleotides were measured at 37 °C while the reaction in bubble and bulge structures were measured at 25 °C.
Reconstitution of BER for bulge structure
The reconstitution of base excision repair of the bulge structure was evaluated with NEIL1, PNK and Lig III. The reaction was started with 20 nM Gh-containing bulge DNA and 200 nM active NEIL1 in an assay buffer (20 mM Tris-HCl, pH 7.6, 10 mM EDTA, 0.1 mg/mL BSA, and 150 mM NaCl) at 37 °C for 10 min. The reaction was continued with the addition of 20 units PNK, 10 mM MgCl2 in a kinase buffer (70 mM Tris-HCl, 10 mM MgCl2, 5 mM dithiothreitol, pH 7.6) with extra 10 mM MgCl2 for 10 min. Then 1.25 nmoles active Lig III was added to the reaction with ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM dithiothreitol, pH 7.5), and the reaction was allowed to continue for 20 min. and finally quenched with 5 µL loading buffer (5% bromophenol blue and 5% xylene cyanol FF in 6 M urea) and loaded onto a 20% denaturing PAGE gel.
RESULTS
Characterization of Bubble and Bulge Structures
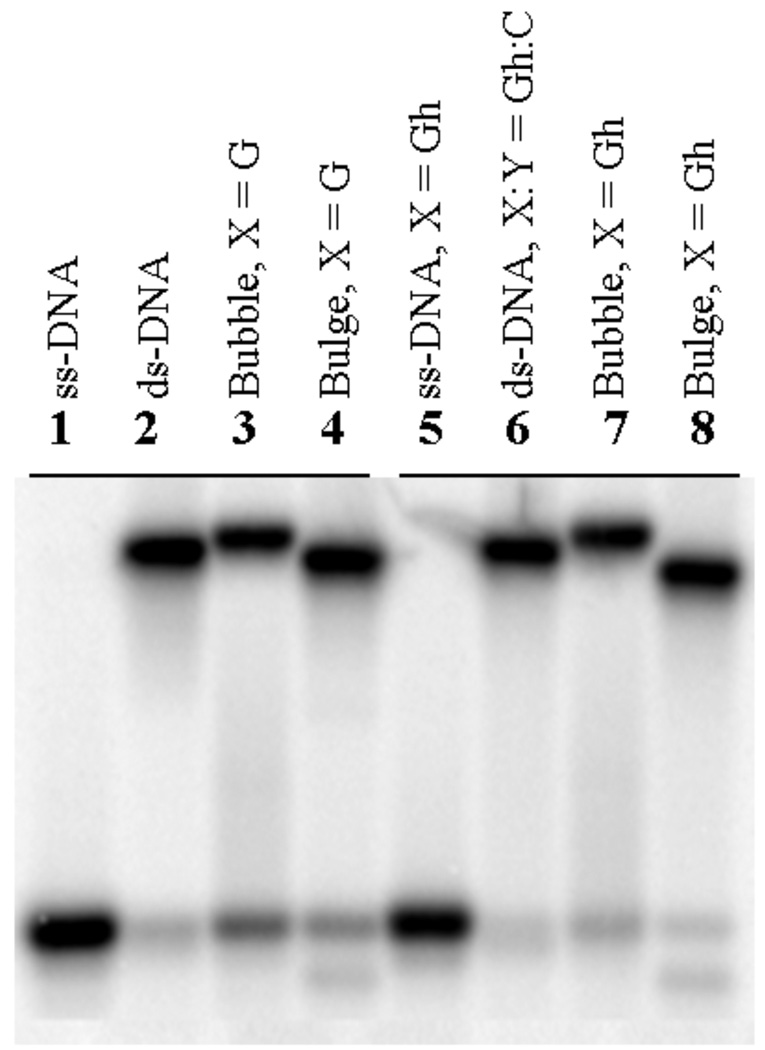
Sequences used in these experiments (Figure 1; Table 1) were designed analogous to the duplex sequence (1•2) used in a previous NEIL1 study for the purposes of comparison (29). In order to characterize the bubble and bulge structures, both the Gh-containing strand and the complementary strand were [γ-32P]-ATP labeled and then annealed. Figure 2 shows the separation of single-stranded DNA, double-stranded DNA, bubble and bulge structures on a non-denaturing PAGE. Lanes 5–8 represent the lesion-containing oligonucleotides while lanes 1–4 are oligonucleotides comprised of normal nucleotides for comparison. Single-stranded oligonucleotides (lanes 1 and 5) migrated faster than other structures. Double-stranded oligonucleotides (lanes 2 and 6) migrated slightly faster than the bubble structure (lanes 3 and 7) though slightly slower than the bulge structure (lanes 4 and 8). The difference in the migration in the native PAGE was more obvious in the Gh-containing oligonucleotides than in the normal oligonucleotides.
Figure 2.
Characterization of bubble, bulge, single-stranded and double-stranded oligonucleotides by non-denaturing gel electrophoresis. Lanes 1 to 4 represent [γ-32P]-ATP labeled oligonucleotides without lesions. Lane 5–8 represent [γ-32P]-ATP labeled oligonucleotides with the Gh lesion at position X (see Fig. 1). Lanes 1 and 5 are single-stranded DNA. Lanes 2 and 6 are double-stranded DNA. Lane 3 and 7 are bubble structures. Lane 4 and 8 are bulge structures.
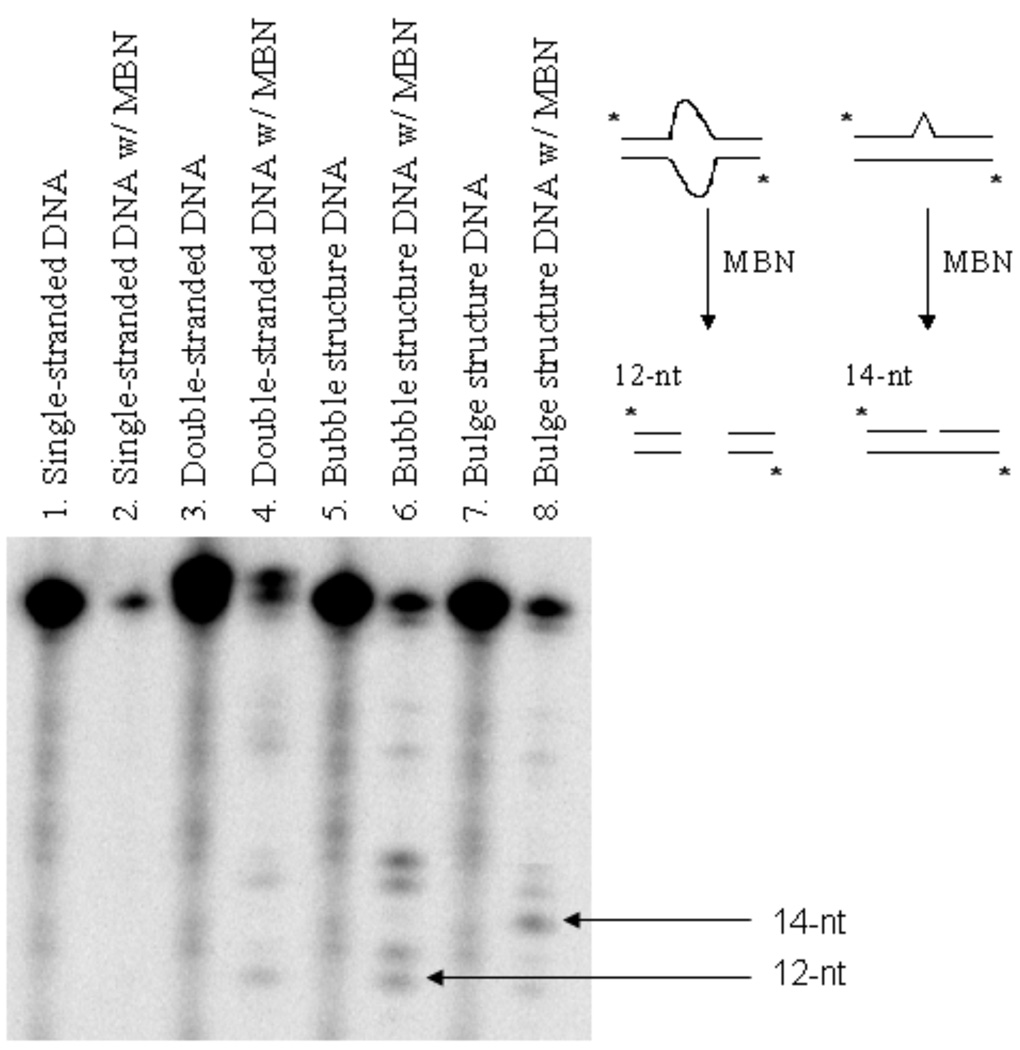
Further characterization was performed using Mung Bean Nuclease (MBN) to interrogate the single-stranded regions of the structures. Figure 3 shows that MBN only minimally cleaved the double-stranded oligonucleotide (lane 4), although this single-strand specific nuclease was able to cleave the centrally located bubble structure leaving two short double-stranded oligonucleotides. A 12-nucleotide fragment is observed (Figure 3, lane 6) as the product of complete single-strand digestion, together with some incomplete digestion products. The bulge structure can also be cleaved at the Gh site resulting in a 14-nucleotide cleavage product (Figure 3, lane 8). Together, the non-denaturing gel study and MBN assay indicate that the bubble and bulge structures proposed in Figure 1 are reasonable.
Figure 3.
Probing structures of bubble and bulge substrates with Mung Bean Nuclease (MBN). Lane 4 shows that double-stranded DNA cannot be cleaved by MBN. Lane 6 shows the mispaired part in the bubble structure can be cleaved by MBN to shorter oligonucleotides (12-nts to 18-nts). Lane 8 shows that in the bulge structure, Gh-containing oligonucleotides can be cleaved by MBN to yield a 14-nt oligonucleotide.
Glycosylase activity of NEIL1 in single-stranded DNA
The lesion-containing sequence (3) used in this study was similar to the one used in a previous investigation of duplex DNA (29). To avoid any self-folding that appeared possible for sequence (1), the new lesion-containing sequence was slightly changed with only one base difference from the sequence corresponding to that in the 30-base pair duplex. This high similarity in sequence context allows us to compare the glycosylase activity of NEIL1 in the different DNA structures. The lesion-containing 30-nucleotide strand (3) was analyzed by M-fold software to test its predicted folding and hybridization (37). No significant self-folding was predicted for any of the sequences for the single-stranded DNA study.
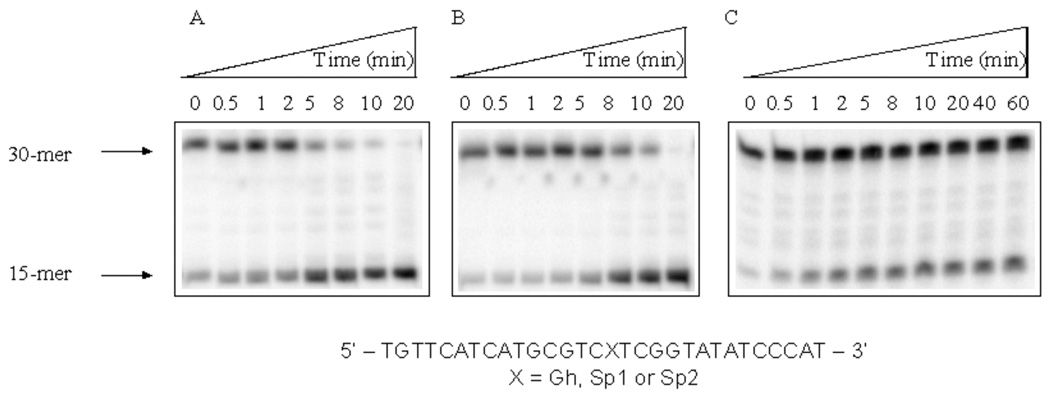
The glycosylase NEIL1 has been shown to remove 5-OHU from single-stranded DNA (14, 38). Single-stranded DNA with OG, Tg, Gh, Sp1 and Sp2 lesions were investigated to obtain the rate constants of glycosylase activity of NEIL1 under single-turnover conditions at 37°C. The PAGE analysis of the NEIL1 reaction with OG-containing oligonucleotides showed only background cleavage (data not shown) indicating that NEIL1 is esentially inactive towards OG in single-stranded DNA. For the corresponding Tg-containing oligonucleotide, approximately 20% product was obtained at the longest time point of 60 min (data not shown) allowing for an upper limit estimate of the cleavage rate (< 0.02 min−1). Gh, Sp1 and Sp2 were expected to be good substrates for NEIL1 in single-stranded DNA due to their rapid removal from duplex DNA. As illustrated in representative storage phosphor autoradiograms in Figures 4A and 4B, NEIL1 mediates cleavage at Gh and Sp1 that is complete within a 20-min time course. The rate constants at 37°C under single-turnover conditions (STO) determined from several experiments were 0.40 ± 0.03 min−1 and 0.30 ± 0.02 min−1 for Gh and Sp1, respectively (Table 2). Surprisingly, Sp2 in the single-stranded oligonucleotide (Figure 4C) cannot be completely removed by NEIL1 and the extent of cleavage levels out at 38% during the one-hour reaction time. Based on the fraction of substrate cleaved, we estimate an upper limit for the rate constant (kest < 0.17 min−1). This indicates a preference for lesion removal by NEIL1 within the context of single-stranded oligonucleotides of the following order: Gh, Sp1 > Sp2 ≫ Tg > OG.
Figure 4.
Storage phosphor autoradiograms of NEIL1-mediated excision of hydantoin lesions in single-stranded 30-mer oligonucleotides. A: Gh-containing ss-DNA over a 20-min time course; B: Sp1-containing ss-DNA over a 20-min time course; C, Sp2-containing ss-DNA over a 60-min time course. All reactions were performed at 37°C.
Table 2.
Rate constants determined at 37°C for cleavage of Gh, Sp1 and Sp2 in bubble, bulge and single-stranded DNA as well as duplex data for comparison.
| Gh (min−1) | Sp1 (min−1) | Sp2 (min−1) | |
|---|---|---|---|
| Bubble (3•4)* | 35 ± 1.6 | 1.9 ± 0.1 | 0.23 ± 0.02 |
| Bulge (3•5)* | 1.8 ± 0.2 | 1.4 ± 0.1 | 0.26 ± 0.01 |
| ssDNA (3) | 0.40 ± 0.03 | 0.30 ± 0.02 | n.d. |
| Duplex (1•2), Y = G§ |
189 ± 18 | 356 ± 13 | 152 ± 11 |
| Duplex (1•2), Y = A§ |
98 ± 10 | 82 ± 7 | 63 ± 8 |
| Duplex (1•2), Y = C§ |
104 ± 14 | 177 ± 11 | 139 ± 8 |
| Duplex (1•2), Y = T§ |
356 ± 36 | > 500 | 224 ± 27 |
| Duplex (6•7) | 153 ± 19 | n.d. | n.d. |
| Duplex (8•9) | 147 ± 16 | n.d. | n.d. |
| Duplex (10•11) | 212 ± 13 | n.d. | n.d. |
The rate constants in bubble and bulge were measured at 25°C.
n.d.: Not determined. Data cannot be fitted to calculate the rate constant because the reaction was incomplete.
Duplex data from a previous study (29).
Glycosylase Activity of NEIL1 in Bubble and Bulge Structures
DNA containing a bubble structure in the middle of a duplex was constructed by the synthesis of a 30-nucleotide duplex (3•4) with 6 mispaired bases in the center. The lesion was placed inside the bubble structure with a G as the opposite base. The bulge structure was constructed by annealing a 30-nucleotide sequence with a 29-nucleotide complementary strand (3•5) with no nucleotide opposite the lesion. The glycosylase activities of NEIL1 towards lesions in the bubble and bulge structures were initially measured under STO conditions at 37°C using manual pipetting methods. Bubble and bulge structures containing Gh and Sp1 lesions undergo complete cleavage at the damaged base site within the first time point of 20 sec indicating rate constants that are too fast to measure accurate manually (> 4 min−1). Sp2 in both bubble and bulge structures was also examined using the same methods, and the lesion was removed with a much lower rate constant (~ 0.8 min−1). Accordingly, Gh and Sp1 were studied further using a rapid quench instrument for a more precise measurement.
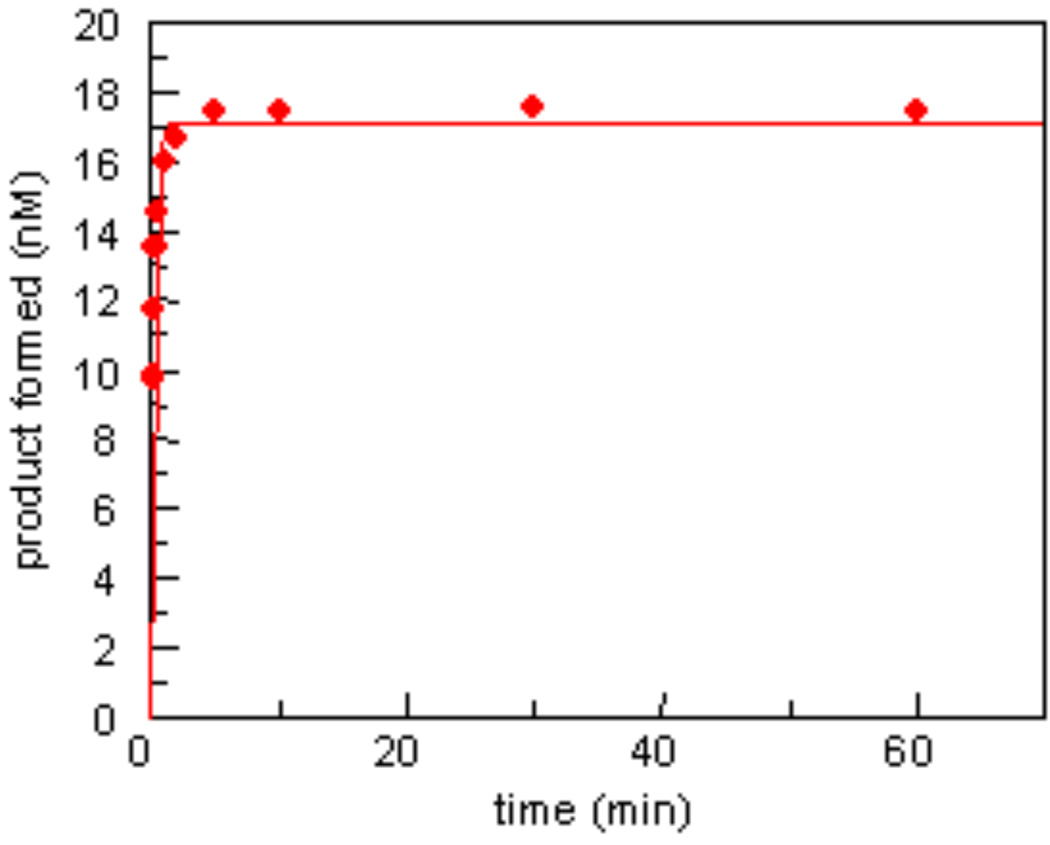
Experiments using the rapid quench instrument at 37 °C were performed under the same reaction conditions as in manual reactions. Surprisingly, only the substrate containing Gh in the bubble structure went to completion. Figure 5 shows data for product formation when Gh in a bulge structure was cleaved in a manual reaction. In the manual experiment, 17 nM product (85%) was formed in a 2-minute time course. However, a total product formation of approximately 8 nM (40%) was formed in 3 minutes using the rapid quench instrument. In analogous experiments with the duplex substrate 90% cleavage of lesions in duplex DNA is observed during the first 15 sec (29). This abnormality of bubble and bulge structures in rapid quench experiments could be due to the instability of the bubble and bulge structures, which may easily dissociate or form a larger bubble structure under the shear pressure of the rapid quench instrument.
Figure 5.
Representative plot of product for by excision of Gh from the bulge structure by NEIL1 over a 60-min period at 37°C. The reaction proceeds to 85% completion in 2 min.
The rate constants for removal of Gh, Sp1 and Sp2 from bubble and bulge structures were then measured at 25 °C to slow down the reaction rate, so that the experiments could be performed manually and allow for comparisons under similar conditions (Table 2). The rate constants, k2, for removal of Gh (1.8 ± 0.2 min−1) and Sp1 (1.4 ± 0.1 min−1) from the bulge structure are similar, while the removal of Sp2 was nearly 6-fold slower (0.26 ± 0.2 min−1). In the bubble structure, the rate of excision of Sp1 (1.9 ± 0.1 min−1) and Sp2 (0.23 ± 0.1 min−1) was similar to that observed in the bulge structures. The removal of Gh was too fast to measure manually but this substrate behaved well on the rapid quench and the measured rate was quite high (35 ± 1.6 min−1). In all cases, NEIL1 can differentiate between Gh and Sp, as well as between the two diasteromers of Sp. Compared to the Sp1 lesion, rate constants of cleavage of Sp2 are 8-fold lower in the bubble and 6-fold lower in the bulge structures. Notably, however, the rate constants for Sp removal in the duplex compared to those in the corresponding bubble and bulge substrates are approximately 2 orders of magnitude faster. In the case of Gh, the rate constant measured in the bubble structures is approaching that for the corresponding duplex substrate. However, since the reaction rate measurements were performed at lower temperatures (25° C) with the bubble and bulge sequences, they may be more similar to the rates of excision in the duplex if they were measured at 37° C.
Rate Constants (k2) for Gh Lesion in Different Sequence Contexts
Three oligonucleotides containing different flanking sequences around the lesion CXT, GXA and TXG (X = Gh) were synthesized and annealed with their complementary strands (sequences 6 – 11) containing a guanine opposite the lesion in order to study sequence effects on the activity of NEIL1. The rate constants of excision of the Gh lesion from different surroundings followed the pattern TXG (duplex 10•11) > CXT (duplex 6•7) ~ GXA (duplex 8•9) (Table 2). The rate of cleavage of Gh from TXG (k2 = 212 ± 13 min−1 at 37°C) is very close to the previous double strand data (k2 = 189 ± 18 min−1) (29). Gh was cleaved 1.5-fold slower in CXT and GXA contexts than in the TXG sequence with the rate constant k2 of 153 ± 19 min−1 and 147 ± 16 min−1, respectively. It is also notable that the rates are similar to the 30 bp duplexes examined previously (29) indicating that the location of the lesion and length of the duplex do not greatly affect the efficiency of the excision activity by NEIL1.
Reconstitution of BER with Gh lesion in bulge structure
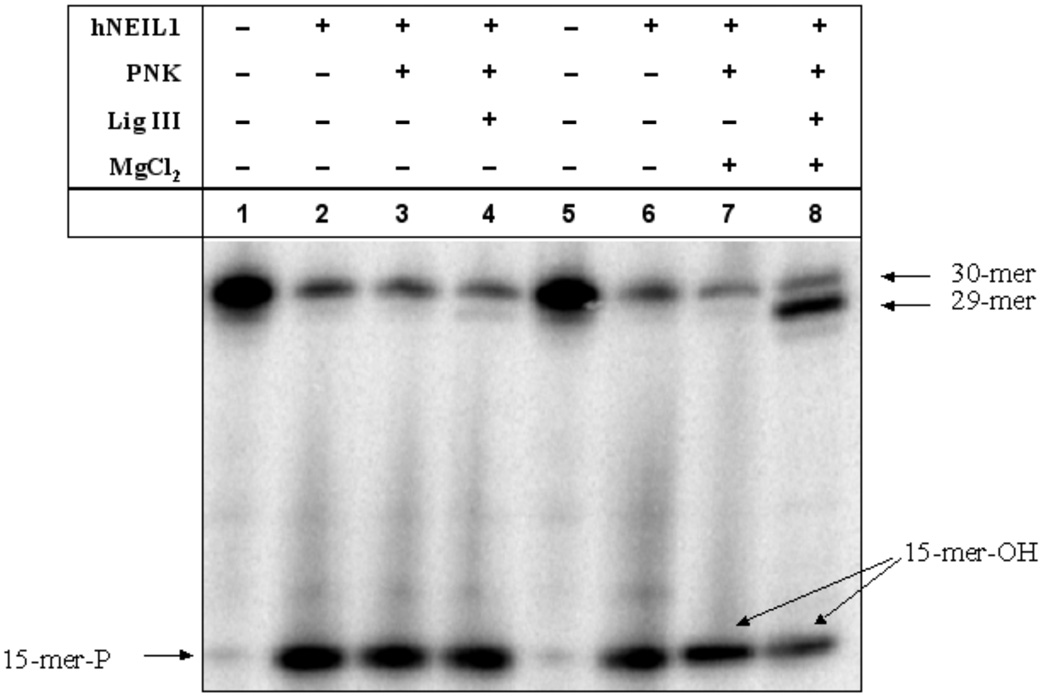
In E. coli, the 3’ phosphate group generated by DNA glycosylase in BER is removed by AP endonuclease (APE1) (39, 40). However, in mammalian cells, removal of the 3’ terminal phosphate group is dependent on PNK instead of APE1 after strand scission by NEIL1 or NEIL2 (41). Here, we attempted to reconstitute an in vitro BER system by the sequential use of NEIL1, PNK and Lig III. Gh in the bulge structure was used to test the repair ability of this BER system to remove the lesion and to reseal the ends surrounding a single-strand nick site. In the first step, NEIL1 cleaved Gh in the DNA bulge structure. Figure 6 shows that upon addition of additional MgCl2, Lig III ligates the nick after PNK removed a phosphate group on the 3’end of the nick. The ligation product is a 29-mer, which migrates a little faster by PAGE than the original 30-mer. The ligation is very efficient with around 80% product formed in a 20-min reaction time. Since the assay buffer for NEIL1 contains 10 mM EDTA, additional MgCl2 was required for the activity of PNK and Lig III. No ligation product was detected without the addition of MgCl2.
Figure 6.
Reconstitution of BER with NEIL1, PNK and Lig III. NEIL1 cleaved the Gh-containing bulge structure and left a single-strand nick with phosphate groups on both 5’and 3’ termini (lanes 2 and 6). With extra MgCl2 added, PNK removed the phosphate group on the 3’end of the nick, thus preparing the single-nucleotide deletion for ligation (lane 7). Lig III then ligated the two strands to form a 29-mer oligonucleotide (lane 8).
Discussion
Oxidative guanine lesions are closely associated with DNA mutations that result from misinsertion during DNA replication past a lesion (35, 42, 43). Our previous kinetic study showed robust activity of NEIL1 towards Gh and Sp hydantoin lesions in defined DNA duplexes (29), which suggests that NEIL1 might be responsible for removal of hydantoin lesions in vivo. However, NEIL1 removal of the hydantoin lesions, indiscriminate of the base opposite the lesion, may lead to mutations that are enhanced by repair, particularly if repair does not occur until after translesion synthesis. For example, NEIL1 cleaves Gh and Sp efficiently when the opposite base is G or A, the nucleotides that are misinserted by polymerases in preference to C (18). The present work demonstrates that NEIL1 can also remove the hydantoin lesions from non-duplex DNA structures including single-stranded DNA, a six-nucleotide bubble and a single-base bulge, in which there is no base pair formation involving the lesion. The consequences of this activity will depend greatly on the circumstances in which it occurs and implicates the likelihood that NEIL1 is actively recruited at appropriate times. However, this also suggests that the activity of NEIL1 on these lesions may contribute to the mutation spectrum that is observed for these unusual lesions.
The common guanine oxidation product OG was found not to be a substrate for NEIL1 in these non-canonical contexts, which is consistent with the fact that OG in a duplex showed only background levels of excision. Tg was a better substrate than OG in single-stranded DNA, but the reaction did not go to completion. The fact that Sp1 is removed more readily and faster than Sp2 in the single-stranded DNA is consistent with the results from double-stranded DNA (29). Rate constants for removing hydantoin lesions in single-stranded DNA, in comparison with those in the duplex, were almost 3 orders of magnitude lower with NEIL1. The preference of NEIL1 for duplex substrates is consistent with recent structural studies of a NEIL1 ortholog from mimivirus, MvNei1, bound to a duplex containing an abasic site analog, tetrahydrofuran (THF) (44). The structural studies reveal significant interactions with the phosphodiester backbone and bases of the strand opposite the THF. These results and the large preference for duplex DNA based on the rate constant measurements suggest that the glycosylase NEIL1 may also be more likely to remove lesions from a DNA duplex in vivo.
The bubble-containing duplex substrates examined herein are particularly interesting since these structures resemble those formed during transcription or replication. Previous work showed that NEIL1 cleaved 5-OHU in bubble structures at levels that are comparable to those in single-stranded and double-stranded DNA contexts suggesting a potential role of NEIL1 in replication or transcription associated repair (20, 38). Notably, 5-OHU (like 5-OHC) is a rather poor substrate for NEIL1 in all contexts compared to Gh and Sp (29). In our previous work with duplex substrates containing Gh and Sp, the two hydantoin lesions are removed at comparable rates, with the action upon Sp1 being higher than both Sp2 and Gh, in general. In the bubble structures, we observe a greater resolution of differential processing of the three hydantoins with respect to each other than with other lesion DNA contexts. In the bubble substrates, Gh is removed almost 20-fold faster than Sp1, which in turn is removed 8-fold faster than Sp2. Moreover, the rate of Gh removal at 25° C in these contexts approaches that in duplex substrates at 37° C (~3-fold slower) providing evidence that Gh removal in these contexts would be comparable to that in a duplex substrate in cellular settings.
The activity of NEIL1 on the hydantoin lesions in various contexts may also be further modulated by the activity of other proteins involved in replication and transcription. Indeed, NEIL1 expression increases in S-phase (27) and NEIL1 has been shown to interact with several key replication proteins suggesting involvement in replication-associated repair (20). For example, NEIL1 has been shown to interact with proliferating cell nuclear antigen (PCNA), the sliding clamp for DNA replication (45). Moreover, PCNA stimulated the 5-OHU excision by NEIL1 in single-stranded and bubble substrates, but not the corresponding duplex substrate. The Flap endonuclease I (FEN1) also has been shown to interact with NEIL1 and stimulate its 5-OHU removal activity with all substrates, which provides additional evidence for a role in replication associated repair as well as long-patch BER (46). In addition, other proteins involved in damage sensing and replication, such as the 9-1-1 complex and Werner helicase (47, 48) have been shown to interact with NEIL1 and stimulate the glycosylase activity with 5-OHU. A very recent study showed that Cockayne syndrome B (CSB) protein, a protein known to be involved in transcription coupled repair, stimulated the FapyG and 5-OHU removal activity mediated by NEIL1 (49). These findings suggest that repair of oxidative damage by NEIL1 is highly coordinated with other cellular processes involved in DNA damage response and genome stability.
Notably, the stimulatory affects afforded by the protein partners identified for NEIL1 depends on the nature of the lesion context, suggesting that some enzymes may participate in stimulating NEIL1 activity in general, while others may act to facilitate the activity of NEIL1 only in conjunction with specific cellular processes. These protein modulators may also exhibit different stimulatory activities that are dependent on the type of lesion, i.e. Gh versus Sp. In addition, the stimulation of the activity may be due to enhancing different aspects of the lesion removal process, for example, lesion recognition, lesion excision or DNA product release. A detailed examination of the effects of proteins on the activity of NEIL1 with the hydantoins may be particularly revealing as to the role played by NEIL at distinct stages in replication, transcription, damage signaling and response. NEIL1 activity for removal of Gh or Sp in single-stranded substrates or partially single stranded substrates may appear to be pro-mutagenic since the absence of the opposite base would not lead to appropriate repair. However, if NEIL1 is recruited to the template strand that is transiently single stranded during replication or transcription, the removal of Gh or Sp may serve as a mechanism to stall replication and facilitate repair of the template strand prior to replication (20). This may be particularly important for Gh which is only partially blocking of DNA replication, and completely miscoding during replication events (43).
The bulged hydantoin substrates for NEIL1 were examined to explore the possibility that NEIL1 may promote mutations in some contexts. Indeed, Wagner et al. previously demonstrated that in vitro treatment of double-stranded plasmid DNA with methylene blue and white light forms lesions that efficiently promote –1 and –2 frameshift mutagenesis in E. coli, and that the frameshift was not due to the formation of OG (30). We and others have shown that Gh and Sp nucleosides can be formed during photooxidation from G via several pathways including singlet oxygen, the product of methylene blue photochemistry (31, 32, 50) Taking these results together, Gh and/or Sp lesions in DNA might be responsible for the frameshift mutations observed in E. coli. Pertinent to this, a previous experiment showed that extension of a primer past template Gh or Sp lesions by Klenow fragment could lead to sequence-dependent deletions (33). For example, insertion of a G opposite Gh or Sp when the 5’base flanking the lesion was a complementary C led to slippage of the primer to form a better G:C base pair (rather than G:X), and further extension continued to yield ultimately an N-1 extension product (33). During this process, the lesion is flipped out of the helix to create a single-base bulge. In that experiment, we found that bulge formation triggered exonuclease activity. However, if the bulged hydantoin persists, it could be a substrate for NEIL1. Indeed, in the present set of experiments, both Gh and Sp1 were completely released by the glycosylase/lyase activity of NEIL1 in the bulge substrate duplex. The rates of the removal activity of the bulged hydantoin lesions is greater than that in single-stranded DNA but reduced compared to duplex DNA. However, instead of helping to restore an undamaged duplex, NEIL1 removal of a bulged base may lead to a single-base deletion by initiating an APE-independent BER pathway that requires subsequent action PNK and Lig III for complete repair(41). With the bulge structure substrate, NEIL1 removed the bulged Gh, PNK action removed the 3’-phosphate, and the gap was sealed using Lig III. The action of these three enzymes fully restores the base paired duplex, leading to a product consistent with a frameshift mutation.
Recently, two in vivo lesion bypass studies with Gh, Sp1 and Sp2 lesions demonstrated that the hydantoin products are highly mutagenic, although the bypass efficiencies and mutation types differed in the two studies. Henderson et al. (43) and Delaney et al. (35) used sequences containing 5’-GXA-3’ and 5’-TXG-3’ sequence contexts, respectively, to assess site-specific lesion bypass efficiency and mutation type. Surprisingly, using the same research method, they obtained different mutation rates and types for the Gh lesion. In the sequence GXA, the Gh lesion showed 75% bypass efficiency (compared to G as a control) while this figure was less than 20% for Gh in the sequence context TXG. In addition, in the sequence GXA, the well-bypassed Gh caused 98% G → C mutations. However, 57% G → C and 40% G → T mutation were found in the sequence TXG. The G → C mutation is presumably caused by a misinsertion of dGMP opposite template Gh to form a Gh:G mispair; this was confirmed by an in vitro insertion study with Klenow fragment (18). To determine if the differences in bypass efficiency and mutation type were related to BER, we determined rate constants for the glycosylase activity of NEIL1 towards the Gh lesion in these two sequence contexts under STO conditions. Gh in both sequences was cleaved extremely efficiently but with no significant discrimination. Thus, glycosylase activity is not a factor that influences the bypass rate and the difference in the in vivo assay might be caused by the polymerase only.
In summary, NEIL1 exhibits efficient activity towards Gh and Sp in duplex as well as non-canonical DNA structures. Although the activity for removal of these lesions in most cases was significantly reduced in single-stranded, bubble and bulge substrates compared to duplex DNA, the superior cleavage of hydantoins compared to other oxidized bases lends support to the hypothesis that Sp and Gh are physiologically relevant substrates in vivo. Curiously, the activity of NEIL1 is not diminished when an incorrect base (A, G, or T) is present opposite the lesion, and some activity remains when no base is opposite the lesion, as in the non-duplex substrates studied here. In the particular case of a single-base bulged lesion, the serial activities of NEIL1, PNK, and Lig III cooperate to insure a single-base deletion mutation. In contrast, the robust activity for removal of Gh in replication bubble-like structures, coupled with the high mutagenic potential of Gh and the known network of interactions of NEIL1 with transcription and replication proteins, suggests that replication/transcription associated repair may be a critical and unique aspect of the activity of this BER glycosylase.
Acknowledgement
We gratefully acknowledge Dr. Viswananth Bandaru and Prof. Susan Wallace (University of Vermont) for providing the NEIL1 expression plasmid and for helpful suggestions. We also thank Dr. Sucharita Kundu for help with the purification of the NEIL1 enzyme.
This work was supported by the National Cancer Institute (CA090689).
Abbreviations
- APE1
AP endonuclease
- BER
base excision repair
- bp
base pair
- EDTA
ethylenediaminetetraacetic acid
- Fpg/MutM
bacterial formamidopyrimidine glycosylase
- FapyG
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- FapyA
4,6-diamino-5-formamidopyrimidine
- Gh
guanidinohydantoin
- 5-OHC
5-hydroxycytosine
- Lig III
human DNA ligase III
- Nei
endonuclease VIII
- NEIL1
human Nei-like glycosylase 1
- nt
nucleotide
- OG
8-oxo-7,8-dihydro-guanine
- PAGE
polyacrylamide gel electrophoresis
- 5-OHU
5-hydroxyuracil
- PNK
polynucleotide kinase
- ROS
reactive oxygen species
- Sp
spiroiminodihydantoin
- STO
single turnover
- Tg
5,6-dihydroxy-5,6-dihydrothymine (thymine glycol)
REFERENCES
- 1.Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1989;86:9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 3.Fuss JO, Cooper PK. DNA repair: Dynamic defenders against cancer and aging. PLoS Biol. 2006;4:899–903. doi: 10.1371/journal.pbio.0040203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 5.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 7.Shigenaga MK, Aboujaoude EN, Chen Q, Ames BN. Assays of oxidative DNA damage biomarkers 8-oxo-2'-deoxyguanosine and 8-oxo-guanine in nuclear DNA and biological fluids by high-performance liquid chromatography with electrochemical detection. Methods Enzymol. 1994;234:16–33. doi: 10.1016/0076-6879(94)34073-0. [DOI] [PubMed] [Google Scholar]
- 8.Beckman KB, Ames BN. Oxidative decay of DNA. J. Biol. Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 9.Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, Klungland A, Olinski R. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radic. Biol. Med. 2004;37:1449–1454. doi: 10.1016/j.freeradbiomed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Collins AR, Cadet J, Moeller L, Poulsen HE, Vina J. Are we sure we know how to measure 8-oxo-7,8-dihydroguanine in DNA from human cells? Arch. Biochem. Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Steenken S, Jovanovic SV, Bietti M, Bernhard K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2'-deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J. Am. Chem. Soc. 2000;122:2372–2374. [Google Scholar]
- 12.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 2000;2:613–617. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 13.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem. Res. Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 14.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Gimisis T, Cismas C. Isolation, characterization, and independent synthesis of guanine oxidation products. Eur. J. Org. Chem. 2006;6:1351–1378. [Google Scholar]
- 16.Pratviel G, Meunier B. Guanine oxidation: one- and two-electron reactions. Chem. Eur. J. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 17.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc. Chem. Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 18.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow Fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 19.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 20.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: A new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst) 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 27.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Nat. Acad. Sci. U.S.A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano T, Katafuchi A, Shimizu R, Terato H, Suzuki T, Tauchi H, Makino K, Skorvaga M, Van Houten B, Ide H. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner J, Fuchs RPP. Frameshift mutagenesis induced in Escherichia coli after in vitro treatment of double-stranded DNA with methylene blue plus white light: Evidence for the involvement of lesion(s) other than 8-oxo-7,8-dihydro-2'-deoxyguanosine. Chem. Res. Toxicol. 1997:568–574. doi: 10.1021/tx960169j. [DOI] [PubMed] [Google Scholar]
- 31.DeFedericis H-C, Patrzyc HB, Rajecki MJ, Budzinski EE, Iijima H, Dawidzik JB, Evans MS, Greene KF, Box HC. Singlet oxygen-induced DNA damage. Radiat. Res. 2006;165:445–451. doi: 10.1667/rr3533.1. [DOI] [PubMed] [Google Scholar]
- 32.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin is the major product of 8-oxo-7,8-dihydroguanosine reaction with peroxynitrite in the presence of thiols, and guanosine photooxidation by methylene blue. Org. Lett. 2001;3:763–766. [PubMed] [Google Scholar]
- 33.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow Fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 34.Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 35.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 36.Iwai S. Chemistry and thermodyanamic studies of oligonucleotides containing the two isomers of thymine glycol. Chemistry. 2001;7:4343–4351. doi: 10.1002/1521-3765(20011015)7:20<4343::aid-chem4343>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 37. mfold. http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi.
- 38.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 39.Pascucci B, Maga G, Hubscher U, Bjoras M, Seeberg E, Hickson ID, Villani G, Giordano C, Cellai L, Dogliotti E. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002;30:2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J. Biol. Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- 41.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: Insights from studies using defined lesions. Mutat. Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 43.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 44.Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J. Biol. Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dou H, Theriot CA, Das A, Hegde ML, Matusmoto Y, Doldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the Human DNA Glycosylase NEIL1 with proliferating cell nuclear antigen. J. Biol. Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 46.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional Interaction between human oxidized base-specific DNA glycosylase NEIL1 and Flap endonuclease I. J. Biol. Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das A, Boldagh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Pinto Souza N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA Glycosylase NEIL1. J. Biol. Chem. 2007;2007:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 49.Muftouglu M, de Souza-Pinto N, Dogan A, Aamana M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockanye syndrome Group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo W. Ph. D. dissertation. University of Utah; 2001. Characterization of purine oxidation products from one-electron oxidants, superoxide, and singlet oxygen. [Google Scholar]