Abstract
Biologic and genetic differences between HIV-1 clade C in India and clade B in US suggest that the effect of anti-viral therapy in various body compartments may differ between these two clades. We examined the effect of therapy viral loads in semen and blood of HIV-1-clade C infected subjects from India and evaluated whether HIV-1 in the semen is different from that in blood in these subjects. HIV-1 RNA was detected in semen and blood at all stages of the disease. Viral loads in semen and blood were strongly correlated with each other, but not with the CD4+ T cell count. Antiviral treatment reduced viral load drastically in blood and semen within one month of post therapy. Genetic characterization of HIV-1 in the semen and blood demonstrated that they were highly compartmentalized. These data have important implications of sexual transmission of HIV-1 in clade C HIV-1 infected subjects.
Keywords: Semen, Clade C HIV-1, antiretroviral therapy
Introduction
Semen is the major source of infectious virus during sexual transmission of human immunodeficiency virus type 1 (HIV-1). High viral loads in semen have been detected in clade B HIV-1-infected subjects at all stages of the disease (Gupta et al., 1997). However, correlation of seminal viral loads with CD4+ T cell counts is controversial. In some studies, there was a correlation between CD4 cell number and seminal viral loads (Vernazza et al., 1997) and in other studies only a weak association was noticed (Gupta et al., 1997).
There is ample evidence to indicate that HIV-1 populations in semen and blood are biologically and genetically different in clade B-infected US and Western European subjects (Byrn and Kiessling, 1998; Coombs et al., 1998; Delwart et al., 1998; Dyer et al., 1998; Gupta et al., 2000; Zhu et al., 1996). We have recently demonstrated that the HIV-1 population in semen in US subjects is further sub-compartmentalized between seminal leukocytes and seminal plasma (Paranjpe et al., 2002). Such sub-compartmentalization of HIV-1 populations between seminal cells and seminal plasma was detected as early as 3 months after seroconversion and persisted up to 38 months following seroconversion (Paranjpe et al., 2002). Despite these interesting and important findings no studies have been done to date on the characterization of HIV-1 in semen in non-B infected subjects, although there are indications that blood and seminal viral load may be higher in clade C infected subjects compared to clade B infected subjects (Dyer et al., 1998).
India, with predominantly HIV-1 clade C (Cassol et al., 1996; Delwart et al., 1998; Maitra et al., 1999; Mandal et al., 2002; Mandal et al., 2000; Shankarappa et al., 2001; Tripathy et al., 1996) and the rapid spread of HIV-1 infection, provides an excellent opportunity to study the seminal viral load in disease progression in clade C-infected subjects. Furthermore, genetic analysis of HIV-1 circulating in different parts of India indicates that clade C sequences from different parts of India are more closely related to each other than to clade C sequences from Botswana, Burundi, South Africa, Tanzania and Zimbabwe. Hence, the Indian clade C is different than the clade C circulating in other parts of the world (Shankarappa et al., 2001). Interestingly, coreceptor switch from CCR5 to CXCR4 observed with disease progression in clade B infected subjects has never been observed in clade C-infected subjects in India (Cecilia et al., 2000). In addition, more rapid disease progression was observed in HIV-1-infected seroconverters in India than seroconverters in US (Mehendale et al., 2002). These differences in biologic and genetic properties as well as disease progression between clade B and C infected subjects in India raises the possibility that viral dynamics of HIV-1 in various body compartments in clade C-infected subjects in India are different than those in clade B infected subjects in western countries. A number of studies have demonstrated a dramatic reduction of viral load in both semen and blood following antiretroviral therapy in HIV-1 clade B infected subjects (Barroso et al., 2003; Barroso et al., 2000; Craigo et al., 2004; Gupta et al., 1997; Liuzzi et al., 1999; Liuzzi et al., 2003; Vernazza et al., 1997; Zhang et al., 1998). However, there is little information on the effect of antiretroviral therapy on viral load in semen in clade C infected subjects from India.
In this report we describe an in depth analysis of HIV-1 present in semen and its fate following antiretroviral therapy in clade C HIV-1 infected subjects from India. We demonstrate that seminal viral load is positively correlated with blood plasma viral load. Neither Blood plasma viral nor seminal plasma viral load had any significant correlation with CD4 cell number. However, viral load in these two body compartments are reduced drastically following antiretroviral therapy. Additionally, viral variants in semen and blood are highly compartmentalized.
Results
Viral load in semen and blood from therapy naïve patients
HIV-1 viral loads in paired seminal and blood plasma samples collected at individual visits were measured by TaqMan RT Real-Time PCR with Indian clade C specific primers and probe described previously (Ding et al., 2009). This assay has a range of detection of 1 ×103 to 3×1010 copies/ml utilizing 200 µl of plasma or semen. Of the 47 randomly selected therapy-naïve subjects 61.7% (29/47) had detectable HIV-1 RNA in seminal plasma, whereas 84.8% (39/46, 1=NA) subjects had detectable viral RNA in blood plasma. The viral load in this therapy-naïve population ranged from 2.63×103 to 2.69 ×108 copies per ml (median 1.64× 105) in seminal plasma and from 1.85 × 103 to 2.5 × 107 copies per ml (median 2.76× 105) in blood plasma (Table 1). HIV-1 was not detected in 10 seronegative subjects (data not shown). Seminal plasma viral load correlated significantly with blood plasma viral load (r2 =0.59 and p< 0.001) with Spearman rank correlation of 0.81 (data not shown).
Table 1.
Quantitation of HIV-1 RNA in blood and semen (cross-sectional, ART naïve patients, N=47)
| HIV-1 RNA (103copy/ml) | |||
|---|---|---|---|
| Subject No. |
CD4 No.(cells/mm3) |
Blood Plasma |
Seminal Plasma |
| 1 | 487 | 1,157 | 101 |
| 2 | 654 | 5,475 | 220 |
| 3 | 526 | 12, | <ADL |
| 4 | 294 | 1,759 | 64 |
| 5 | 483 | 64 | <ADL |
| 6 | 293 | <ADL | <ADL |
| 7 | 294 | <ADL | <ADL |
| 8 | 265 | 2 | <ADL |
| 9 | 171 | 167 | 22 |
| 10 | 26 | 276 | 12 |
| 11 | 176 | 2,461 | 2,943 |
| 12 | 241 | <ADL | <ADL |
| 13 | 16l | 1,077 | 3 |
| 14 | 263 | 63 | 8 |
| 15 | 203 | 582 | 6 |
| 17 | 263 | <ADL | <ADL |
| 18 | 432 | <ADL | <ADL |
| 21 | 681 | 238 | <ADL |
| 25 | 492 | 1,478 | 282 |
| 27 | 175 | 25,072 | 790 |
| 28 | 212 | 4,563 | 120 |
| 29 | 12 | 4 | <ADL |
| 34 | 156 | 136 | 19 |
| 35 | 57 | 43 | 3 |
| 36 | 92 | 11,619 | 779 |
| 37 | 92 | 2,161 | 30,569 |
| 38 | 468 | 158 | 8 |
| 39 | 76 | 10,174 | 381 |
| 40 | 365 | 13 | <ADL |
| 41 | 294 | 4,016 | 383 |
| 42 | 309 | NA | 3 |
| 43 | 139 | 4,013 | 208 |
| 44 | 640 | <ADL | <ADL |
| 45 | 121 | 1,718 | 3,222 |
| 47 | 200 | 16,841 | 269,481 |
| 48 | 592 | 3,359 | 164, |
| 49 | 312 | 6,146 | 1,710 |
| 50 | 322 | 90 | <ADL |
| 51 | 543 | 15 | <ADL |
| 52 | 522 | 13 | <ADL |
| 53 | 131 | 416 | 29 |
| 54 | 215 | 54 | 11,327 |
| 55 | 458 | 83 | 113 |
| 56 | 471 | 44 | <ADL |
| 58 | 174 | 13 | 181 |
| 59 | 73 | <ADL | <ADL |
| 60 | 132 | 9 | <ADL |
NA sample not available
<ADL below assay detection limit
In this cross-sectional study high seminal viral load is present at all stages of the disease with varying CD4 number (Table 1). Since we do not have information on number of CD4 cell in semen due to low semen sample volume, we determined correlation of seminal and blood viral load with blood CD4. HIV-1 RNA level in seminal plasma and in blood plasma viral load did not correlate significantly with CD4 cell number (r2= 0.05 with p=0.151 and r2= 0.02 with p=0.722, respectively) (data not shown).
Effect of antiretroviral therapy on viral load in semen and blood compartments
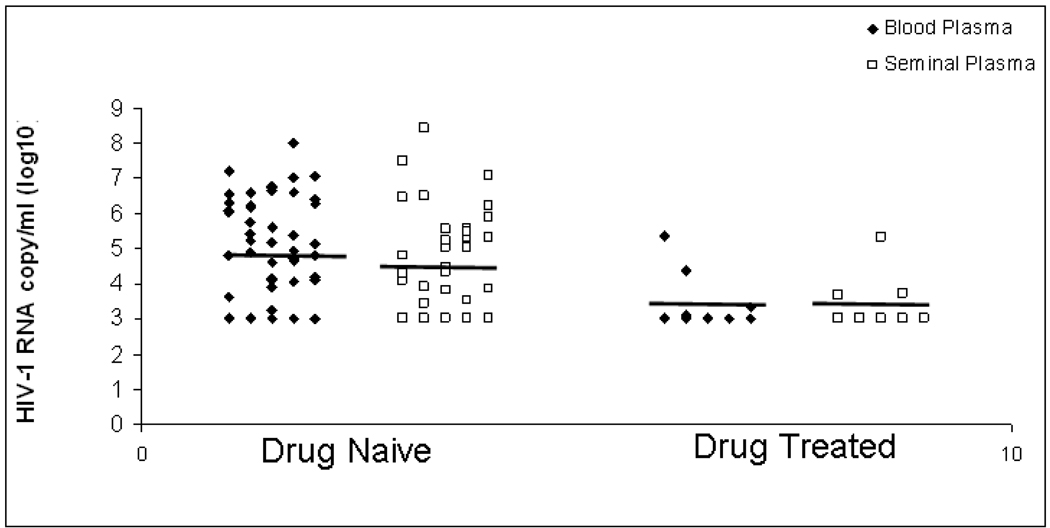
Treatment of HIV-1clade B-infected subjects with potent antiretroviral therapy has been shown to reduce viral load in both semen and blood compartment within 4 weeks of post therapy. To determine whether such therapy reduces viral load in semen in HIV-1 clade C-infected subjects from India, we first examined in a cross-sectional study viral load in 15 subjects following therapy for a varying length of time (0.5 month to 32 months) (Table 2). HIV-1 RNA was detected only 20.0% (3/15) in seminal plasma and 26.7% (4/15) in blood plasma from the drug treated subjects. The viral load in seminal and blood plasma in response to therapy are summarized in Figure 1. In a longitudinal study viral load was measured in paired semen and blood compartments of 4 HIV-1 clade C infected Indian subjects before and after therapy for a period of 1 to 19 months with a combination of Lamivudine, Stavudine and Nevarpine. The level of viral RNA in seminal plasma and blood plasma in these subjects were reduced drastically to undectable level after 4 to 8 weeks of therapy and remained below detectable limit for a period of up to 19 months (Table 3). In one patient (patient 37) viral load in semen did not reduce to undetectable limit within 5 months of treatment, although viral load in blood compartment was reduced to undetectable limit during that period of time. This could be due to difference in drug penetration in male genital organ between patients.
Table 2.
Quantitation of HIV-1 RNA in blood and semen after potent antiretroviral therapy (cross-sectional samples, N=15)
| HIV-1 RNA (103 copy/ml) |
||||
|---|---|---|---|---|
| Subject No. | Month Post-therapy | CD4 No.(cells/mm3) |
Blood Plasma | Seminal Plasma |
| 16 | 28 | 392 | <ADL | <ADL |
| 19 | 2 | 281 | <ADL | <ADL |
| 20 | 9 | 251 | <ADL | <ADL |
| 22 | 11 | 121 | <ADL | <ADL |
| 23 | 32 | 510 | <ADL | <ADL |
| 24 | 17 | 266 | <ADL | 5 |
| 26 | 2 | 89 | 1 | <ADL |
| 30 | 2 | 30 | <ADL | 213 |
| 31 | 17 | 319 | <ADL | <ADL |
| 32 | 17 | 128 | <ADL | <ADL |
| 33 | 21 | 156 | 236 | <ADL |
| 46 | 0.5 | 100 | 24 | <ADL |
| 57 | 7 | 181 | <ADL | <ADL |
| 45-1 | 1 | <ADL | 5 | |
| 53-1 | 1 | 2 | <ADL | |
<ADL below assay detection limit
Figure 1. Viral load of bloods plasma and seminal plasma before and after potent antiretroviral therapy.
Table 3.
Longitudinal paired semen and blood compartments of 4 HIV-1 clade C infected subjects before and after therapy
| HIV-1 RNA (103 copy/ml) |
||||
|---|---|---|---|---|
| Subject No. | Month Post-therapy | CD4 No. (cells/mm3 ) |
Blood Plasma | Seminal Plasma |
| 34-0 | 0 | 156 | 136 | 19 |
| 34-2 | 4 | <ADL | <ADL | |
| 34-3 | 8 | 344 | <ADL | 38 |
| 34-4 | 12 | 331 | <ADL | <ADL |
| 34-5 | 15 | 444 | <ADL | <ADL |
| 34-6 | 17 | 337 | <ADL | <ADL |
| 34-8 | 19 | <ADL | <ADL | |
| 37-0 | 0 | 92 | 2,161 | 30,569 |
| 37-1 | 1 | NA | 15 | |
| 37-2 | 5 | 268 | <ADL | 11 |
| 39-0 | 0 | 76 | 10,174 | 381 |
| 39-1 | 1 | 44 | 37 | |
| 39-2 | 4 | 561 | <ADL | <ADL |
| 39-3 | 11 | 460 | <ADL | <ADL |
| 39-5 | 14 | <ADL | <ADL | |
| 39-6 | 16 | <ADL | <ADL | |
| 58-0 | 0 | 174 | 13 | 181 |
| 58-1 | 2 | <ADL | <ADL | |
| 58-2 | 8 | 237 | <ADL | <ADL |
NA sample not available
<ADL below assay detection limit
Sequence analysis of HIV-1 from cells and plasma from semen and blood in drug naïve patients
Concurrent blood and semen samples obtained from 7 drug naïve subjects were used in the study. Sequences from four compartments: blood plasma and cells, as well as seminal plasma and cells were obtained from four patients (subject number 4, 9, 11, 14), while sequences from three compartments blood plasma, seminal plasma and seminal cells were obtained from three patients (subject number 2, 37, and 40). A total of 490 nucleotide sequences of a 680bp segment encompassing the C2–V5 region of the HIV-1 gp120 from these body compartments were analyzed from the seven patients. All the sequences were screened by the BLAST program from HIV database to rule out potential cross-contamination. Furthermore, sequences derived from each subject of this study were phylogenetically distinct (data not shown), indicating no sample mixing or cross-contamination. Genotyping and recombination analysis showed that all the patients belonged to clade C, and did not have any contamination from other clades.
Compartmentalization of HIV-1 between semen and blood of drug naïve patients
The cladistic method of the Slatkin-Maddison test for compartmentalization among the four body compartments (seminal plasma, seminal cells, blood plasma, blood cells) indicated significant phylogenetic compartmentalization (P< 0.0001) in all seven patients at varying levels of stringency with the majority of patients having ≤ 6 steps resolving the phylogenetic structure of four compartments (Table 4). To confirm the phylogenetic results, we used Gene Flow calculations to estimate genetic migration Fst and Nm values among all the seven patients. Nm values in six of the seven patients was indicative of compartmentalization demonstrating restricted gene flow with Nm values ranging from 1.49 to 8.6 (Table 4). As observed with the phylogenetic analyses this compartmentalization varied among patients at the level of restriction, especially with patient 2 which demonstrated no migration restrictions with a Nm= 109.35. Taken together these data indicate significant compartmentalization in six of the seven patients.
Table 4.
Phylogenetic compartmentalization analysis using MacClade and DnaSP
| DnaSP | MacClade | |||
|---|---|---|---|---|
| Subjects | Nm | Fst | Steps | P |
| 2 | 109.35 | 0.0023 | 2 | <0.0001 |
| 4 | 1.53 | 0.14 | 4 | <0.0001 |
| 9 | 1.61 | 0.13 | 5 | <0.0001 |
| 11 | 8.6 | 0.03 | 8 | <0.0001 |
| 14 | 2.85 | 0.08 | 6 | <0.0001 |
| 37 | 1.85 | 0.12 | 4 | <0.0001 |
| 40 | 1.49 | 0.14 | 3 | <0.0001 |
Genetic diversity of individual compartments in drug naïve patients
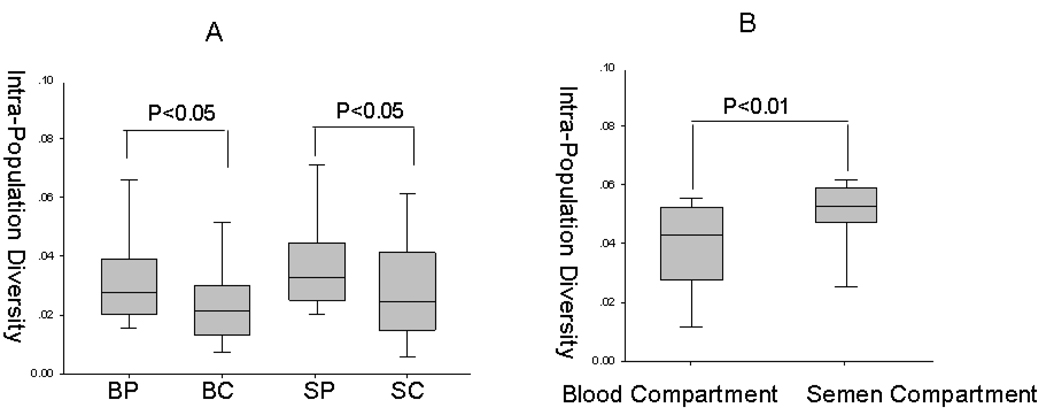
Genetic diversity of individual compartment was calculated based on distance measurments obtained by using the Jukes Cantor matrix from PAUP* software package. The blood plasma and seminal plasma compartments display higher diversity than the blood cell and seminal cell compartments respectively (P<0.05) (figure 2A). Also, the intracompartmental distance within the seminal compartment is higher than the distance within the blood compartment (P<0.01) (figure 2B). Since blood plasma is derived from blood cells, there will be less diversity between them, as compared to seminal cells and seminal plasma which may have different origins (Paranjpe et al., 2002).
Figure 2. Diversity and distance analysis of envelope sequences (C2–V5) of HIV-1 from Indian patients using PAUP* software.
(A) Nucleic acid diversity of corrected intrapopulation diversity was calculated for nucleotide sequences. The quartile (box) differences between the diversity of all sequence clones were plotted as interpopulation distances as a function of different compartments. (B) Corrected population distance between blood derived plasma and cells and semen derived plasma and cells was calculated for nucleotide sequences. Statistical significance of the differences between different compartments for each analysis was performed using nonparametric statistics, the Mann-Whitney test. BP, blood plasma; BC, blood cells; SP, semen plasma; SC, semen cells. Blood compartment, distance between blood plasma and blood cells; Semen compartment, distance between seminal plasma and seminal cells.
Immune selection pressure analysis in drug naïve patients
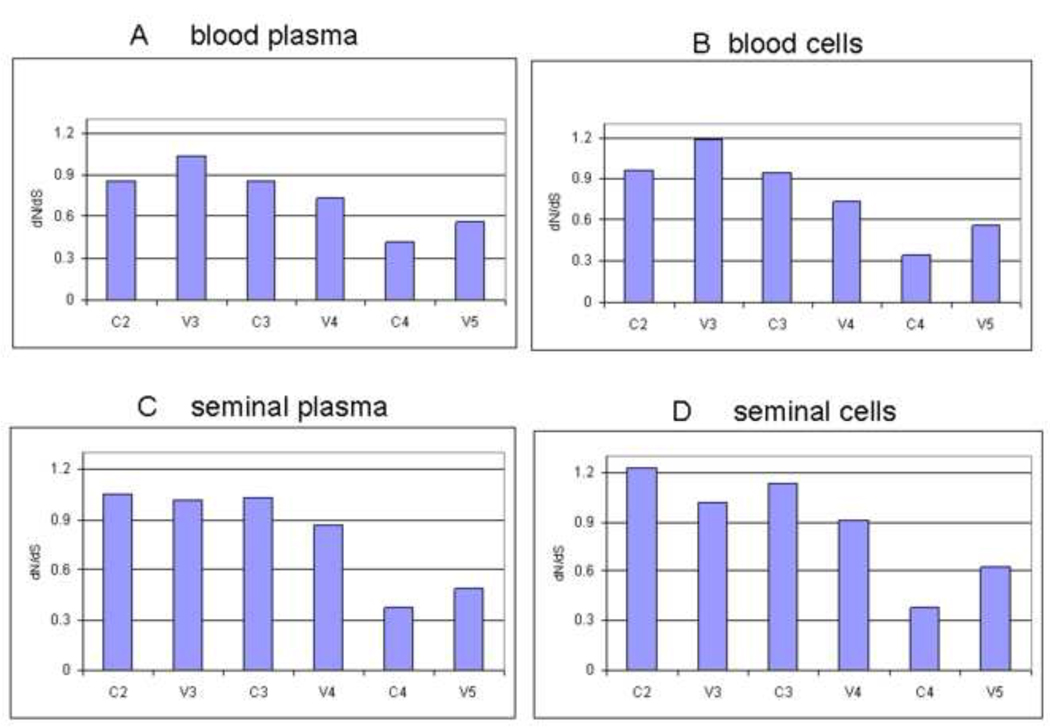
The rates of synonymous (dN) and nonsynonymous (dS) nucleotide substitutions per site of codon aligned nucleotide sequences in the individual compartment populations were calculated by using program DnaSP V4.50.3. The results showed a ratio of dN/dS<1 for the whole C2-V5 sequences in all the four compartments: blood plasma, blood cells, seminal plasma and seminal cell (0.841, 0.828, 0.875, 0.853 respectively), suggesting no immune selection in either semen and blood compartments. However, analysis of various subgenomic regions of the C2-V5 region, such as C2, V3, C3, V4, C4, V5, showed positive immune selection in some regions and such immune selection is different in different compartments (figure 3). As expected V3 region in all 4 compartments showed positive immune selection. In addition, C2 and C3 regions of seminal cells and seminal compartment showed positive selection. In contrast, V4 and V5 regions had no positive immune selection.
Figure 3. Immune selection analysis of subgenome of C2-V5.
Analysis of immune selection at various subgenomic regions (C2, V3, C3, V4, C4, and V5) of the whole C2-V5 sequences from four body compartments (seminal cells and plasma, and blood cells and plasma).
Co-receptor usage prediction and glycoslation in drug naïve patients
Because it is difficult to isolate infectious HIV-1 from semen, co-receptor usage of the HIV-1 in blood and semen compartments was determined based on the deduced basic amino acid substitution in the V3 region using software PSSM and geno2pheno. Consistent with previous study (Cecilia et al., 2000), all of the viruses in all the four compartments are R5 co-receptor usage and NSI.
To determine the variation in immune selection, glycoslation patterns across the C2-V5 region were examined. N-glycoslations have been implicated in the suppression of natural killer (NK) cell–mediated responses, and the suppression of innate immune responses involving NK cells that may be responsible for initiating AIDS pathogenesis (Kottilil et al., 2003; Yoshimura et al., 1996) Cell-specific glycosylation profiles may affect immune susceptibility, and AIDS progression may be related to the glycobiology of the virus (Clark et al., 1997). Surprisingly, we did not find any special pattern of N-linked glycosylation among the four compartments (data not shown).
Discussion
Although there are a number of reports characterizing HIV-1 in semen and the effects of antiretroviral therapy on HIV-1 in semen in HIV-1 clade B infected subjects in the US and in Western Europe, similar information on HIV-1 clade C infected subjects is scarce. Since India harbors mainly HIV-1 clade C characterized by different biologic and genetic properties (Shankarappa et al., 2001), we launched a study investigating the genetics of HIV-1 in semen and the effect of therapy on male genital secretion in 62 HIV-1-infected subjects from India. In therapy naïve subjects we found a high level of HIV-1 RNA in seminal plasma (median 1.64×105) at all stages of the disease with CD4 ranging from 12 cells/mm3 to 681 cells/mm3. The median seminal plasma viral load (1.64×105) is similar to median blood plasma viral load (2.76 ×105), although in approximately 13% of patients (6/47) viral load in seminal plasma is higher than those in blood. Furthermore, neither the seminal nor blood plasma viral load correlated with blood CD4 cell numbers. Similar data has been observed in semen from clade B HIV-1 infected subjects (Gupta et al., 1997).
Three years ago the Indian government launched a limited combination antiretroviral therapy composed of Lamivudine, Stavudine and Nevarpine. Thus far, very little information is available on the effect of such therapy on viral load in blood or semen in HIV-1 infected subjects from India. In this report we have demonstrated that treatment of HIV-infected subjects with highly active combination antiretroviral therapy can promptly reduce the viral load in semen in parallel with that in blood plasma. However, we do not have any information on infectious viral titers or proviral DNA in seminal mononuclear cells from subjects on therapy. This is the first report of a reduction of viral load in semen and blood in clade C HIV-1 infected subjects from India. Since clade C in India is different than the clade Cs circulating in other parts of the world, this information is important to support and extend ongoing therapy intervention program in India. These findings have important implications for the biology of sexual transmission of HIV-1 and its potential reduction by antiretroviral therapy. At present, there is an ongoing HIVNET study to determine whether HIV-1 transmission from the infected Indian subjects treated with antiretroviral therapy is decreased.
Analysis of HIV-1 sequences in semen is critical to the study of the viral variants that are involved in sexual transmission. Analysis of the HIV-1 variants in semen and blood of the majority of the seven drug naïve patients significant compartmentalization of HIV-1 between seminal and blood compartments among six out of seven subjects. However, the degree of compartmentalization varied from subject to subject. Restricted viral migration between semen and blood supports our previous data that HIV-1 in semen is produced locally. Furthermore, subcompartmentalization between seminal cells and seminal plasma support our previous contention with clade B HIV-1 that seminal and blood virus may originate from different genital tissue sites (Paranjpe et al., 2002).
Compartmentalization of HIV-1 between semen and blood suggest that distinct viral variants in these two body compartments may be a product of different rate of evolution due to different selective pressure operating in these two body compartments. We investigated this possibility by analyzing immune selection pressure in semen and blood compartment. Our analysis of pairwise comparisons of synonymous and nonynonymous substitutions by the conservative method of dN/dS indicate differential immune selection between these two compartments. The study by Pillai (Pillai et al., 2005) reported semen specific signature amino acids in clade B HIV-1 infected subjects from US. Using the same methodology we did not find similar semen specific signature amino acids among our 7 clade C infected subjects from India (data not shown). This difference could be due to unique genetic and biologic characteristics of clade C HIV-1 from India.
In one study cellular tropism has been implicated to have a role in compartmentalization between semen and blood derived virus from clade B HIV-1 infected subjects (Delwart et al., 1998). However, our recent study and those reported in the literature indicate that HIV-1 in India are mostly of CCR5 tropic with non-syncytium inducing phenotype even when they are isolated at late stage of infection (Cecilia et al., 2000). In agreement with these reports only CCR5 genotype was detected in both semen and blood compartment and therefore cellular tropism did not have any role in compartmentalization.
In summary, this report for the first time describes high levels of HIV-1 in semen from clade C HIV-1 infected Indian subjects at all stages of the disease and potent antiretroviral therapy rapidly reduces viral load in semen in parallel with that in blood. In addition, this report provides detailed genetic characterization of HIV-1 in semen from clade C HIV-1 infected Indian subjects, showing that viral variants present in semen are different than those in blood and difference in evolution of HIV-1 variants in these two compartments could be due to difference in immune selection pressure.
Methods
Study population, semen and blood samples
This study was conducted according to the IRB approval of the University of Pittsburgh. Cryopreserved paired semen and blood specimens from a total of 62 HIV-1 infected individuals were received from the Virology Department of the School of Tropical Medicine (STM) in Calcutta, India from May 2006 to September 2008. The subjects age distribution were 23 to 54 years old (mean=35). Of these 62 subjects, cross-sectional samples were obtained from 47 antiretroviral therapy -naïve subjects and from 15 subjects treated with combination antiretroviral drugs Lamivudine, Stavudine and Nevarpine for varying period of time (from 0.5 to 32 months). Detailed clinical history and CD4 numbers were available from most of the subjects. Therapy naïve subjects had CD4 counts ranging from 681 cells/mm3 to 12 cells/mm3. Drug treated subjects had CD4 counts ranging from 510 cells/mm3 to 30 cells/mm3. Longitudinal samples were collected from 4 subjects before and after therapy for a period of 1 to 19 months post-therapy.
Samples were processed within 4 hrs of collection. Blood plasma was prepared by centrifuging EDTA treated blood at 1200g at room temperature. Semen was processed within 4 hr of collection. After 30 min incubation in ice, liquefied semen was centrifuged for 10 minutes at 800–1000g at room temperature to collect seminal cells and seminal plasma. Seperated seminal cells and seminal plasma were kept at −70°C and later transported in dry ice from Calcutta to India. Preliminary results indicate that transportation of semen and blood plasma samples in dry ice does not cause any significant change in viral load in semen and blood (data not shown).
Quantitation of HIV-1 RNA in semen and blood
Total RNA was isolated from two hundred microliters of seminal or blood plasma by Nuclisens Isolation Kit according to manufacturer instructions (Biomerieux) and subjected to Real Time RT-PCR specific for Indian HIV-1 clade C as described previously (Ding et al., 2009). To rule out the potential PCR contamination, a no-template control was included in each PCR run in which the template was replaced with same volume of Nuclease-free water. Each sample was run in triplicate. A standard curve generated by known copy number of in vitro transcribed into HIV-1pol HIV-1 was used for copy number estimation.
Cloning and sequencing of C2–V5 region of HIV-1 env RNA
Total RNA was extracted from blood plasma, blood cells, semen plasma and semen cells, and subjected to reverse transcription and polymerase chain reaction (RT-PCR) of the C2–V5 region of the HIV-1 envelope as described previously (Delwart, 1993; Liu, 1997). Negative controls were applied with each PCR run to detect any possible contamination. For each sample, PCR products from five independent PCR reactions were cloned into the TOPO XL vector from the TOPO TA-Cloning system (Invitrogen, Carlsbad, CA). DNA from 15–20 screened clones were purified using the QIAprep® Spin Miniprep Kit (Qiagen) and sequenced using primers for forward and reverse directions in an ABI Prism 3700 DNA Sequencer.
Analysis of HIV envelope sequences in blood and semen
Sequences were assembled and error checked utilizing software Vector-NTI (Invitrogen). Nucleotide and deduced amino acid sequences from each clone were aligned using the Clustal W multiple sequence alignment program from the MEGA4.0 software and edited manually where necessary.
Genotyping and phylogenetic reconstruction
Genotyping was carried out in viral genotyping tools for Recombinant Identification Program: RIP 3.0 and Genotyping from NCBI -http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi. The neighbor-joining method of Jukes Cantor corrected nucleotide distances was employed to construct the phylogenetic trees with the optimality criterion set to distance as measured in PAUP4* with a transition-to-transversion ratio of 2. Statistical significance of branchings and various clustering were assessed by the bootstrap resampling method using 1000 replicate. The trees were viewed and edited for publication using FigTree version 1.1.2.
Evaluation of compartmentalization
Two separate methods to test for compartmentalization of HIV-1 between the four body compartments: blood plasma, blood cells, seminal plasma and seminal cells as described previously (Paranjpe et al., 2002). First, software DnaSP V4.50.3.was utilized to estimate the standard population genetic parameters. Genetic differentiation among populations was evaluated by estimating Fst, the fraction of nucleotide diversity as a result of genetic variation between populations. Nm, the average level of gene flow, among the four compartments was confirmed from Fst. The value of Nm equal or less than 5 indicated significant population subdivision among the four compartments. Second, a cladistic method of Slatkin-Maddison analysis (Slatkin and Maddison, 1989; Slatkin and Maddison, 1990) as implemented in the MacClade 4 program was also used to assess compartmentalization among these four compartments . A MacClade data file listing all of the specimens was constructed for each patient and each compartment was treated as a four-state unordered character. The number of migration events needed to postulate the observed spatial distribution of HIV-1 sequences in the phylogenetic trees was estimated using MacClade. The null model of this analysis is that sequences from one compartment would be as likely to be evolutionarily related to sequences from other compartments as to itself. The frequency of distribution under this null model was obtained by constructing 10,000 random trees made by random joining/splitting of the `true` phylogenetic tree in MacClade. The number of migration events on the true tree was compared to the null distribution and the probability that the true tree came from a population lacking compartmentalization was determined. Tissue specific compartmentalization was considered statistically significant if fewer steps were seen in the true tree than in 95% of the random trees.
Analysis of selection pressure at different regions and different sites
The rates of synonymous and nonsynonymous substitution in the individual compartment populations were calculated by using program DnaSP V4.50.3. First, we analyzed the whole C2-V5 region. Second, we analyzed six regions C2, V3, C3, V4, C4, and V5 of the C2-V5 region each separately using the same program.
Co-receptor usage prediction and glycosylation
Software PSSM and geno2pheno were employed to predict co-receptor usage and syncytium-inducing. They are available at http://ubik.microbiol.washington.edu/computing/pssm/(Jensen et al., 2003) and http://coreceptor.bioinf.mpi-inf.mpg.de/index.php(Sing et al., 2007). Potential of N-linked glycosylation sites within each sequence was predicted by software N-GlycoSite available at the HIV database website (http://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html).
Statistical analysis
Virologic data were measured in copies per milliliter and immunologic data were measured as cell number per millimeter cubed. All variables were summarized using means and standard deviations as well as medians and ranges. For statistical analyses, log base 10 transformation of viral load data was used. In comparison of viral loads in semen and plasma all values less than 1000 were set to equal 1000. Correlations were calculated using a non-parametric method (i.e., Spearman rank) and reported with associated p-values. Further, linear regression was used to calculate an estimate of rate of change (i.e., slope) between each pair of continuous variables (e.g., CD4 and semen viral load). Slopes were reported with associated p-values and r-squared values. All analyses were conducted in STATA version 9
Acknowledgement
We thank Dr. Milka Rodriguez and Ms. Kathy Kulka for handling semen samples from India. This work was supported by a grant R21AI65392 from National Institute of Health. Dr. Chengli Shen was also partially supported by an AIDS International Fogarty grant TW001038-06.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Nucleotide sequence accession numbers
Sequences were deposited in GenBank under the accession no. FJ541508-FJ541996.
References
- Barroso PF, Schechter M, Gupta P, Bressan C, Bomfim A, Harrison LH. Adherence to antiretroviral therapy and persistence of HIV RNA in semen. J Acquir Immune Defic Syndr. 2003;32(4):435–440. doi: 10.1097/00126334-200304010-00014. [DOI] [PubMed] [Google Scholar]
- Barroso PF, Schechter M, Gupta P, Melo MF, Vieira M, Murta FC, Souza Y, Harrison LH. Effect of antiretroviral therapy on HIV shedding in semen. Ann Intern Med. 2000;133(4):280–284. doi: 10.7326/0003-4819-133-4-200008150-00012. [DOI] [PubMed] [Google Scholar]
- Byrn RA, Kiessling AA. Analysis of human immunodeficiency virus in semen: indications of a genetically distinct virus reservoir. Journal of Reproductive Immunology. 1998;41(1–2):161–176. doi: 10.1016/s0165-0378(98)00056-4. [DOI] [PubMed] [Google Scholar]
- Cassol S, Weniger BG, Babu PG, Salminen MO, Zheng X, Htoon MT, Delaney A, O'Shaughnessy M, Ou CY. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Research & Human Retroviruses. 1996;12(15):1435–1441. doi: 10.1089/aid.1996.12.1435. [DOI] [PubMed] [Google Scholar]
- Cecilia D, Kulkarni SS, Tripathy SP, Gangakhedkar RR, Paranjape RS, Gadkari DA. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology. 2000;271(2):253–258. doi: 10.1006/viro.2000.0297. [DOI] [PubMed] [Google Scholar]
- Clark GF, Dell A, Morris HR, Patankar M, Oehninger S, Seppala M. Viewing AIDS from a glycobiological perspective: potential linkages to the human fetoembryonic defence system hypothesis. Mol Hum Reprod. 1997;3(1):5–13. doi: 10.1093/molehr/3.1.5. [DOI] [PubMed] [Google Scholar]
- Coombs RW, Speck CE, Hughes JP, Lee W, Sampoleo R, Ross SO, Dragavon J, Peterson G, Hooton TM, Collier AC, Corey L, Koutsky L, Krieger JN. Association between culturable human immunodeficiency virus type 1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: evidence for compartmentalization of HIV-1 between semen and blood. Journal of Infectious Diseases. 1998;177(2):320–330. doi: 10.1086/514213. [DOI] [PubMed] [Google Scholar]
- Craigo JK, Patterson BK, Paranjpe S, Kulka K, Ding M, Mellors J, Montelaro RC, Gupta P. Persistent HIV type 1 infection in semen and blood compartments in patients after long-term potent antiretroviral therapy. AIDS Res Hum Retroviruses. 2004;20(11):1196–1209. doi: 10.1089/aid.2004.20.1196. [DOI] [PubMed] [Google Scholar]
- Delwart EL, Shpaer EG, McCutchan FE, Louwagie J, Grez M, Rübsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- Delwart EL, Mullins JI, Gupta P, Learn GH, Jr, Holodniy M, Katzenstein D, Walker BD, Singh MK. Human immunodeficiency virus type 1 populations in blood and semen. Journal of Virology. 1998;72(1):617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Tarwater P, Rodriguez M, Chatterjee R, Ratner D, Yamamura Y, Roy P, Mellors J, Neogi D, Chen Y, Gupta P. Estimation of the predictive role of plasma viral load on CD4 decline in HIV-1 subtype C-infected subjects in India. J Acquir Immune Defic Syndr. 2009;50(2):119–125. doi: 10.1097/QAI.0b013e3181911991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JR, Eron JJ, Hoffman IF, Kazembe P, Vernazza PL, Nkata E, Costello Daly C, Fiscus SA, Cohen MS. Association of CD4 cell depletion and elevated blood and seminal plasma human immunodeficiency virus type 1 (HIV-1) RNA concentrations with genital ulcer disease in HIV-1-infected men in Malawi. Journal of Infectious Diseases. 1998;177(1):224–227. doi: 10.1086/517359. [DOI] [PubMed] [Google Scholar]
- Gupta P, Leroux C, Patterson BK, Kingsley L, Rinaldo C, Ding M, Chen Y, Kulka K, Buchanan W, McKeon B, Montelaro R. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. Journal of Infectious Diseases. 2000;182(1):79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- Gupta P, Mellors J, Kingsley L, Riddler S, Singh MK, Schreiber S, Cronin M, Rinaldo CR. High viral load in semen of human immunodeficiency virus type 1-infected men at all stages of disease and its reduction by therapy with protease and nonnucleoside reverse transcriptase inhibitors. Journal of Virology. 1997;71(8):6271–6275. doi: 10.1128/jvi.71.8.6271-6275.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Li FS, van 't Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77(24):13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottilil S, Chun TW, Moir S, Liu S, McLaughlin M, Hallahan CW, Maldarelli F, Corey L, Fauci AS. Innate immunity in human immunodeficiency virus infection: effect of viremia on natural killer cell function. J Infect Dis. 2003;187(7):1038–1045. doi: 10.1086/368222. [DOI] [PubMed] [Google Scholar]
- Liu SL, Schacker T, Musey L, Shriner D, McElrath MJ, Corey L, Mullins JI. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi G, Chirianni A, Bagnarelli P, Clementi M, Piazza M. A combination of nucleoside analogues and a protease inhibitor reduces HIV-1 RNA levels in semen: implications for sexual transmission of HIV infection. Antivir Ther. 1999;4(2):95–99. [PubMed] [Google Scholar]
- Liuzzi G, D'Offizi G, Topino S, Zaccarelli M, Amendola A, Capobianchi MR, Perno CF, Narciso P, Antinori A. Dynamics of viral load rebound in plasma and semen after stopping effective antiretroviral therapy. Aids. 2003;17(7):1089–1092. doi: 10.1097/00002030-200305020-00022. [DOI] [PubMed] [Google Scholar]
- Maitra A, Singh B, Banu S, Deshpande A, Robbins K, Kalish ML, Broor S, Seth P. Subtypes of HIV type 1 circulating in India: partial envelope sequences. AIDS Research & Human Retroviruses. 1999;15(10):941–944. doi: 10.1089/088922299310656. [DOI] [PubMed] [Google Scholar]
- Mandal D, Jana S, Bhattacharya SK, Chakrabarti S. HIV type 1 subtypes circulating in eastern and northeastern regions of India. AIDS Research & Human Retroviruses. 2002;18(16):1219–1227. doi: 10.1089/08892220260387968. [DOI] [PubMed] [Google Scholar]
- Mandal D, Jana S, Panda S, Bhattacharya S, Ghosh TC, Bhattacharya SK, Chakrabarti S. Distribution of HIV-1 subtypes in female sex workers of Calcutta, India. Indian Journal of Medical Research. 2000;112:165–172. [PubMed] [Google Scholar]
- Mehendale SM, Bollinger RC, Kulkarni SS, Stallings RY, Brookmeyer RS, Kulkarni SV, Divekar AD, Gangakhedkar RR, Joshi SN, Risbud AR, Thakar MA, Mahajan BA, Kale VA, Ghate MV, Gadkari DA, Quinn TC, Paranjape RS. Rapid disease progression in human immunodeficiency virus type 1-infected seroconverters in India. AIDS Res Hum Retroviruses. 2002;18(16):1175–1179. doi: 10.1089/08892220260387913. [DOI] [PubMed] [Google Scholar]
- Paranjpe S, Craigo J, Patterson B, Ding M, Barroso P, Harrison L, Montelaro R, Gupta P. Subcompartmentalization of HIV-1 quasispecies between seminal cells and seminal plasma indicates their origin in distinct genital tissues. AIDS Research & Human Retroviruses. 2002;18(17):1271–1280. doi: 10.1089/088922202320886316. [DOI] [PubMed] [Google Scholar]
- Pillai SK, Good B, Pond SK, Wong JK, Strain MC, Richman DD, Smith DM. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J Virol. 2005;79(3):1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Chatterjee R, Learn GH, Neogi D, Ding M, Roy P, Ghosh A, Kingsley L, Harrison L, Mullins JI, Gupta P. Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. Journal of Virology. 2001;75(21):10479–10487. doi: 10.1128/JVI.75.21.10479-10487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Low AJ, Beerenwinkel N, Sander O, Cheung PK, Domingues FS, Buch J, Daumer M, Kaiser R, Lengauer T, Harrigan PR. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir Ther. 2007;12(7):1097–1106. [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123(3):603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. Detecting isolation by distance using phylogenies of genes. Genetics. 1990;126(1):249–260. doi: 10.1093/genetics/126.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy S, Renjifo B, Wang WK, McLane MF, Bollinger R, Rodrigues J, Osterman J, Essex M. Envelope glycoprotein 120 sequences of primary HIV type 1 isolates from Pune and New Delhi, India. AIDS Research & Human Retroviruses. 1996;12(12):1199–1202. doi: 10.1089/aid.1996.12.1199. [DOI] [PubMed] [Google Scholar]
- Vernazza PL, Dyer JR, Fiscus SA, Eron JJ, Cohen MS. HIV-1 viral load in blood, semen and saliva. AIDS. 1997;11(8):1058–1059. [PubMed] [Google Scholar]
- Yoshimura M, Ihara Y, Ohnishi A, Ijuhin N, Nishiura T, Kanakura Y, Matsuzawa Y, Taniguchi N. Bisecting N-acetylglucosamine on K562 cells suppresses natural killer cytotoxicity and promotes spleen colonization. Cancer Res. 1996;56(2):412–418. [PubMed] [Google Scholar]
- Zhang H, Dornadula G, Beumont M, Livornese L, Jr, Van Uitert B, Henning K, Pomerantz RJ. Human immunodeficiency virus type 1 in the semen of men receiving highly active antiretroviral therapy. N Engl J Med. 1998;339(25):1803–1809. doi: 10.1056/NEJM199812173392502. [DOI] [PubMed] [Google Scholar]
- Zhu T, Wang N, Carr A, Nam DS, Moor-Jankowski R, Cooper DA, Ho DD. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. Journal of Virology. 1996;70(5):3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]