Abstract
Several prokaryotic ClC proteins have been demonstrated to function as exchangers that transport both chloride ions and protons simultaneously in opposite directions. However, the path of the proton through the ClC exchanger, and how the protein brings about the coupled movement of both ions are still unknown. In this work, we use an atomistic molecular dynamics (MD) simulation to demonstrate that a previously unknown secondary water pore is formed inside an Escherichia coli ClC exchanger. The secondary water pore is bifurcated from the chloride ion pathway at E148. From the systematic simulations, we determined that the glutamate residue exposed to the intracellular solution, E203, plays an important role as a trigger for the formation of the secondary water pore, and that the highly conserved tyrosine residue Y445 functions as a barrier that separates the proton from the chloride ion pathways. Based on our simulation results, we conclude that protons in the ClC exchanger are conducted via a water network through the secondary water pore, and we propose a new mechanism for the coupled transport of chloride ions and protons. It has been reported that several members of ClC proteins are not just channels that simply transport chloride ions across lipid bilayers; rather, they are exchangers that transport both the chloride ion and proton in opposite directions. However, the ion transit pathways and the mechanism of the coupled movement of these two ions have not yet been unveiled. In this article, we report a new finding (to our knowledge) of a water pore inside a prokaryotic ClC protein as revealed by computer simulation. This water pore is bifurcated from the putative chloride ion, and water molecules inside the new pore connect two glutamate residues that are known to be key residues for proton transport. On the basis of our simulation results, we conclude that the water wire that is formed inside the newly found pore acts as a proton pathway, which enables us to resolve many problems that could not be addressed by previous experimental studies.
Introduction
In the early 1980s, a protein from the electric organ of a Torpedo ray was found to be involved in the transport of chloride ions across the cell membrane (1,2). Since then, it has been widely accepted that this protein, identified as a member of the chloride channel (ClC) family, is simply a channel through which a chloride ion is conducted via electrostatic diffusion regulated by an ion gradient or voltage across the cell membrane.
Twenty years later, Dutzler et al. (3,4) successfully identified the three-dimensional structure of a prokaryotic ClC protein and proposed that the glutamate residue serves as a gate to regulate the transport of chloride ions. Inspired by these fascinating results, investigators developed many theoretical and computational approaches, and obtained valuable molecular details about the ion transit pathway and the mechanism of gating (5–13). However, their studies were limited to the transport of chloride ion, and thus the concept of the ClC channel remained unchanged until recently.
Recent reports have shown that the several members of the ClC family are not channels, but instead are chloride ion/proton exchangers in which the flux of chloride ions is coupled to the flux of protons in the opposite direction with a stoichiometric ratio of 2:1 (14–18). However, the pathway for proton conduction and the mechanism of coupled transport for chloride ions and protons have not been elucidated, although some studies have suggested clues for proton transport in the exchangers. Most notably, Accardi and Miller (14) and Accardi et al. (15) proposed that the two glutamate residues (E148, located adjacent to the extracellular side, and E203, exposed to the intracellular solution) of the Escherichia coli ClC exchanger are involved in proton transport in ClC exchangers. They based their findings on experimental results demonstrating that the coupled movement of protons and chloride ions vanishes when these residues are mutated. They also proposed that the proton and chloride ion pathways are separate because the glutamate residue on the intracellular side (E203), which has been suggested to be involved in proton transport, is distant from the putative chloride ion pathway. To prove the validity of their proposal, they attempted to find a residue that could deliver a proton from E203 to E148 (18). Among many candidates, they focused on the highly conserved tyrosine residue Y445 because it has a hydroxyl side chain that lies halfway between the two glutamate residues, E148 and E203. However, they realized that removing the hydroxyl group from Y445 by mutating it to phenylalanine or tryptophan had no significant effect on the protein's electrophysiological behavior.
Our primary objective in this study was to elucidate the proton pathway inside a ClC exchanger and identify the role of E203 in the proton transport. For this purpose, we performed an atomistic molecular-dynamics (MD) simulation, which has proved to be a useful tool for solving problems that cannot be addressed directly by experimental studies (19–22). We used the crystallographic structure of the prokaryotic ClC protein from E. coli (3,4) as a model structure of a ClC exchanger for our simulations. Based on the results from the atomistic MD simulation, we conclude that we have discovered a secondary pathway for proton transport, and we propose a mechanism for the coupled movement of chloride ions and protons.
Materials and Methods
Structural simulations were performed using the wild-type (WT) ClC crystal structure (Protein Data Bank code: 1OTS), which was recently resolved in E. coli by x-ray crystallography at 2.5 Å resolution (4). The two chloride ions at positions Sint and Scen in the crystal were preserved. The positions Sint and Scen were designated according to the notation of Dutzler et al. (4). The initial structure was embedded in a 16:0-palmytoyl,18:1(Δ9)-oleoylphosphatidyl-ethanolamine lipid bilayer, which is generally accepted as a good model for the E. coli membrane (23). The lipid bilayer with the embedded ClC protein was hydrated with ∼30 Å thick water layers.
The final structure contained ∼87,600 atoms and was energy-minimized by the conjugated gradient method. First, only water molecules were minimized, and the other atoms were fixed with constraints. The constraints imposed on the lipid molecules and the protein were then sequentially released so that the ClC protein became free to move. The generated membrane structure was equilibrated by means of the MD simulation for 4 ns with an NPT ensemble. For this equilibration, the integration step of 1 fs was used.
To elucidate the role of E148 and E203 in proton transport, we generated different initial structures by modifying the protonation state of E148 and E203, and the position of the chloride ions. In addition, we investigated the role of the hydroxyl group of Y445 by mutation to a phenylalanine. We also generated the structure with E203 mutated to a glutamine (E203Q) to confirm previous experimental results (15). Each modified structure, summarized in Table 1, was equilibrated for 14 ns with the integration step of 2 fs, and the structural changes caused by the modification were observed.
Table 1.
Conditions for the atomistic MD simulation of the ClC exchanger from E. coli
| Structure | Protonation state of E203 | Protonation state of E148 | Number of Cl− in primary pore | Formation of secondary pore |
|---|---|---|---|---|
| WT | X | X | 2 | X |
| 0 | X | |||
| O | 2 | X | ||
| 0 | X | |||
| O | X | 2 | X | |
| 0 | X | |||
| O | 2 | X | ||
| 0 | O | |||
| Y445F | O | X | 0 | X |
| O | 0 | O | ||
| E203Q | E → Q | X | 0 | X |
| O | 0 | X |
The protonation states of two key glutamates (E203 and E148) are denoted by O (protonated) and X (deprotonated), respectively. The formation of the secondary pore during the 14 ns simulation is also denoted in the last column by O and X. The reference structure (E203 and E148 are deprotonated, and two chloride ions are in their binding sites) is shaded gray. Each configuration was equilibrated for 4 ns and an additional simulation was performed for 14 ns for each configuration to confirm whether the secondary pore had formed.
The Langevin piston algorithm was used for constant pressure and temperature. Pressure and temperature were set to 1 atm and 310 K, respectively. The particle mesh Ewald (PME) summation was used to calculate the electrostatic force without cutoff (24). The electrostatic neutralization of the system, which is a prerequisite for PME, was satisfied by adding an appropriate number of sodium and chloride ions to each modified structure. The periodic boundary condition was applied to the side boundaries. All calculations were performed using the NAMD program for parallel computation (25). The CHARMM force field with all-atom parameters was used (CHARMM22 for protein (26), CHARMM27 for phospholipids (27), and the TIP3P model for water molecules (28)). For sodium and chloride ions (29), parameters in the CHARMM force field were used without modification. The HOLE program (30), which has been widely used to predict the pore in channel proteins, was used for the pore analysis. All simulations were performed on our local Linux cluster.
Results and Discussion
Secondary water pore revealed by an atomistic simulation
According to the crystal structure of the ClC exchanger, water molecules are found in both extra- and intracellular vestibules, but not in the putative ion translocation pathway (3,4). Under these circumstances, it is hypothesized that a proton cannot pass through the pathway due to the absence of a continuous water network, which is a prerequisite for proton conduction. Although the mechanism of proton transport in a ClC exchanger has not been elucidated, a recent study showed that proton transport in a prokaryotic ClC exchanger is impaired when E203 is mutated to a glutamine that functions to mimic the protonated state of a glutamate residue (15).
Inspired by these results, we performed MD simulations by varying the protonation state of E203 to confirm the structural change caused by the protonation state of E203. In this case, the glutamate residue adjacent to the extracellular side, E148, which is known to be a key residue for chloride ion conduction, was kept protonated. Fig. 1 A shows the equilibrated structure of the ClC exchanger with the pore, as predicted by the HOLE program (30). E203 is deprotonated, whereas the two chloride ions are bound to the positions resolved by x-ray crystallography (Sint and Scen). Here, the predicted pore is consistent with the chloride ion pathway, and a water molecule is not observed inside the pore during the equilibration, as previously reported (3,4,31).
Figure 1.
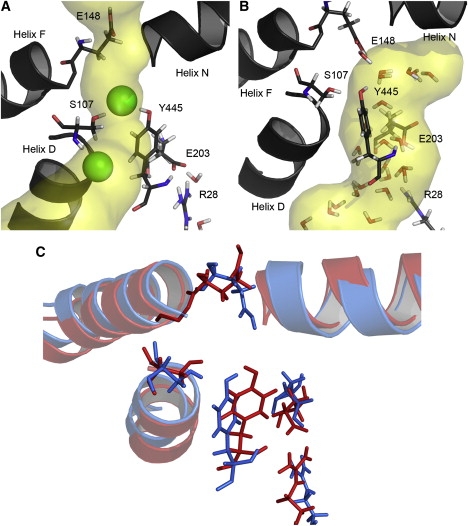
Structural representations of the E. coli ClC exchanger. Equilibrated structures of the ClC exchanger with the primary pore (A) and the secondary pore (B) are shown, and two structures are shown overlapped in C to compare the structural differences between A and B. Two chloride ions are represented as green spheres in A, and the pores predicted by the HOLE program are shown with a yellow transparent color in A and B. The structures in A and B are colored red and blue, respectively, in C. The D-, F-, and N-helices are shown by cartoon representation, and key residues are shown in stick representation. All of the snapshot figures in this work were created with PyMOL (http://www.pymol.org).
However, as E203 is protonated and the two chloride ions inside the chloride ion pathway are manually transported to the outside of the pore, a significant structural change occurs. After a 6 ns equilibration, the region between the Y445 and E203 residues is filled with water molecules originating from the intracellular side, as shown in Fig. 1 B. Consequently, the water molecules form a continuous network that links E203 with E148, a previously unknown region that is predicted to be a pore by the HOLE program, as shown in Fig. 1 B. At first, we assumed this network of water molecules was an intermediate and expected that the water pore would disappear as the system reached equilibrium. However, this water pore was present for an additional 8 ns (see Movie S1 in the Supporting Material).
To identify whether the newly found water pore was an artifact caused by an unstable initial structure and/or an incorrect choice of force field, we generated a new initial structure. In the new structure, two chloride ions were transferred to the extracellular side, and E203 was deprotonated. Under this condition, the water pore was not formed during the 14 ns equilibration. However, when we modified the equilibrated structure by changing the protonation state of E203 from deprotonated to protonated while keeping other conditions unchanged, a water pore was formed after a 6 ns simulation, as it was in the previous simulation. This result proves that the formation of the water pore is reproducible and not the result of an unstable initial structure and/or wrong choice of force field. Hence, we refer to this newly identified water pore as the secondary pore, and designate the chloride ion pathway previously determined by x-ray crystallography as the primary pore.
Here it should be noted that the protonation state of E113, located near E203, is not related to the formation of the secondary water pore, because the secondary water pore was formed regardless of the protonation state of E113 in the WT ClC exchanger. It has been reported, however, that the protonation state of E113 is relevant to the stabilization of chloride ions at their binding sites in the primary pore (10). Hence, the protonation state of E113 was fixed in the deprotonated state to minimize the number of factors to be considered, and therefore the protonation state of E113 was not taken into account for the rest of the simulation.
Role of E203 as a gate
To determine the reason for the formation of the secondary pore, we investigated the structural change caused by protonation of E203 in detail by overlapping the two structures shown in Fig. 1, A and B. As shown in Fig. 1 C, the most notable change in the protein structure was found near E203. Before protonation, the negatively charged side chain of E203 interacted with the positively charged arginine residue (R28) of another subunit of the protein via a salt bridge interaction. However, when E203 lost its negative charge by protonation, the salt bridge interaction between E203 and the R28 was broken. In this case, R28 was separated from E203, resulting in the formation of a cleft between E203 and R28. Water molecules penetrated into the secondary pore through this cleft, and a continuous water network was formed inside the secondary pore, as shown in Fig. 1 B. It seems that E203 acts as a gate that triggers the formation of the secondary water pore by breaking the salt bridge with R28, in a manner reminiscent of E148 acting as a gate for chloride ion conduction by a charge-dipole interaction with α-helix N (12).
The role of E203 as a gate became more evident when we compared the distance between deprotonated E203 and R28 with that between protonated E203 and R28. As shown in Fig. 2, the distance between E203 and R28 remained nearly unchanged at ∼2 Å before protonation. However, these residues moved apart from each other as E203 lost its charge by protonation. Thus, it is clear that the protonation of E203 is a prerequisite for the formation of the secondary pore.
Figure 2.
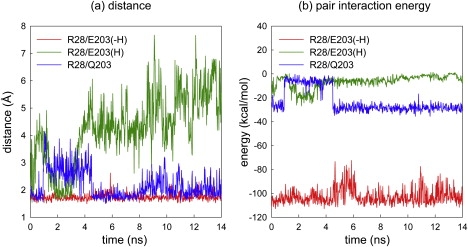
Distance and energy between E203(Q203) and R28 during the 14 ns simulation. Red, green, and blue lines represent the distance and energy between R28 and its pair (deprotonated E203, protonated E203, and Q203), respectively.
It is also possible that several other factors are involved in the formation of the secondary pore. To investigate whether other factors are necessary for the secondary pore formation, we systematically generated structures with an embedded ClC exchanger by varying the position of the chloride ion and the protonation state of the E148 and E203 residues. As summarized in Table 1, the secondary pore was formed only when both E148 and E203 were protonated and the chloride ions at the primary pore were moved to outside of the pore, which implies that the formation of the secondary pore is affected not only by the protonation state of E148 and E203, but also by the position of the chloride ion inside the primary pore. It is interesting to note that the secondary pore was always filled with water molecules originating from the intracellular side, whereas no water molecules were observed to migrate from the extracellular side.
Proton transport via the secondary water pore
Based on the result that the formation of the continuous water network inside the secondary pore is dependent on the protonation state of E203, which is known to be involved with proton transport (15), it is reasonable to assume that this water pore is a pathway for proton transport because a proton is easily transported along the continuous water network by the hop-and-turn, or Grotthuss mechanism (32). The assumption that the secondary water pore is a pathway for proton transport is also supported by experimental results for mutant ClC exchangers. According to a recent experimental result (15), the ClC exchanger loses its ability to transport protons when E203 is mutated to a glutamine (E203Q). Thus, if our assumption is correct, a secondary water pore that transports protons should not be formed in the mutant E203Q ClC exchanger. To determine whether the secondary water pore is formed in the mutant ClC exchanger (E203Q), we equilibrated a ClC exchanger in which E203 was mutated to glutamine for 14 ns using MD simulation. We observed that in the mutated structure, E203Q interacted with R28 via hydrogen bonding (Figs. 2 B and 3 A). We also noted that the hydrogen bonding between E203Q and R28 prevented water molecules on the intracellular side from entering into the secondary pore, as did the salt bridge interaction between E203 and R28 in the WT (see Figs. 1 A, 2, and 3 A). Nonetheless, these two structures (WT and mutated) differ significantly in terms of the mechanism that controls the interaction between E203 (Q203 in the mutant exchanger) and R28. In contrast to the WT ClC exchanger, which is able to control the interaction between E203 and R28 by gaining or losing a negative charge based on the protonation state of E203, the mutant (E203Q) exchanger kept interacting with R28 by hydrogen bonding. As a result, the formation of the secondary pore was not observed in the mutant exchanger, as summarized in Table 1. This result explains why the mutated ClC exchanger lost its function of proton transport when E203 was mutated to glutamine. Based on these results, it can be now concluded that the secondary pore is a pathway for proton transport in ClC exchangers.
Figure 3.
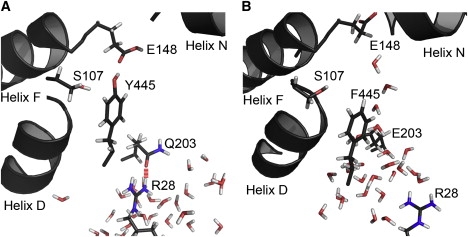
Structural representations of E. coli mutant ClC exchangers. Simulations were performed with mutants E203Q (A) and Y445F (B). The D-, F-, and N-helices are shown by cartoon representation, and key residues are shown in stick representation. Hydrogen bonding between Q203 and R28 is represented by red lines.
This conclusion is consistent with another hypothesis that the proton pathway is separate from the chloride ion pathway, and the two pathways are bifurcated at E148 toward the intracellular side (15). However, this hypothesis assumes that protons would be transported through this imaginary pathway via a protein residue (18) rather than through a continuous water network. This assumption highlights the following unresolved issues: First, proton movement along the protein's residues must be coupled to the occupancy of the binding sites of chloride ions. Second, the distance between E148 and E203 is >15 Å, which is too great a distance for a proton to cross in a single hop. Assuming that the secondary water pore discovered in this work is a proton pathway, these issues are easily explained. First, proton conduction in a ClC exchanger is coupled to chloride ion transport because the secondary pore is formed only when the chloride ions inside the chloride ion pathway are transported to the outside of the pore. Second, a proton can be easily transported between E203 and E148 along the continuous water network by the hop-and-turn, or Grotthuss mechanism (32). In other protein channels and enzymes, such as gramicidin (30), bacteriorhodopsin (33,34), and cytochrome c oxidase (35,36), water networks are utilized as proton pathways.
A plausible proton pathway proceeding from E203 along the aromatic ring of Y445 toward Scen was recently proposed by Kuang et al. (37) based on the results of a searching algorithm. However, they did not show any direct evidence for the presence of a water network. Consequently, it is assumed that protons should be transported via direct movement rather than via the hop-and-turn mechanism. Moreover, the predicted proton pathway proposed by Kuang et al. becomes electronegative only when two chloride ions reside at their binding sites (Sint and Scen), and therefore the coupled transport of chloride ion and proton cannot be clearly explained. This problem with the coupled transport can be resolved by assuming that the secondary water pore revealed in our work is formed as a proton pathway.
In a related study, Wang and Voth (38) performed an MD simulation to prove that protons are transported through the water wire in a ClC exchanger via a mechanism similar to the one proposed here. Using the multistate empirical valence bond method, they successfully showed that protons move directly through the water wire; however, they did not demonstrate the spontaneous formation of the secondary water pore, as shown in this work. They also proposed that an excess proton is transferred during the conformational change of E203 by the rotation and reorientation of its side chain. This movement transfers an excess proton into the interior of the protein because the water-lined proton transport pore they found is not continuous and is disrupted in the vicinity of Y445.
Role of Y445 as a barrier between the proton and chloride ion pathways
The transport of a proton from E203 to E148 requires a series of protein residues or a continuous water network that can mediate the proton movement. Accardi et al. (18) focused on the highly conserved tyrosine residue Y445 as a candidate for proton transport because it is the only residue that has a hydroxyl side chain that lies halfway between E203 and E148. However, they found that mutation of Y445 to phenylalanine (F) or tryptophan (W) had no significant effect on the electrophysiological behavior of the exchanger. It should be noted here that the aromatic side chain of Y445 is similar to that of phenylalanine and tryptophan except for the hydroxyl group.
To determine the role of the Y445 hydroxyl group in proton transport, we generated a ClC exchanger in which the tyrosine residue was mutated to a phenylalanine (Y445F) for MD simulations. As shown in Table 1 and Fig. 3 B, the secondary pore was formed in the structure containing Y445F, as in the WT structure. In addition, we observed no difference in the manner in which the aromatic side chain of both Y445 and Y445F separated the secondary pore from the chloride ion pathway. When the chloride ions inside the primary pore were transported to the outside of the pore, the side chain of the tyrosine residue was rotated slightly toward the primary pore, which resulted in the expansion of the secondary water pore, as shown in Fig. 1, B and C. It is known that the primary role of the tyrosine residue is to stabilize a chloride ion through the hydroxyl group when the ion is bound at the Scen position (3,4). In addition to its primary role, it seems that the tyrosine residue may have a secondary role as a barrier separating the secondary pore from the primary pore via the aromatic side chain. On the basis of these assumptions, we can hypothesize that the removal of the hydroxyl group from the tyrosine residue should not affect the formation of the secondary water pore. This hypothesis is supported by experimental results demonstrating that proton transport is not impaired by the removal of the hydroxyl group from the tyrosine residue by the mutations Y445F and Y445W (7).
Conclusions
In this work, a secondary water pore in a ClC exchanger was revealed with the use of an atomistic MD simulation. The secondary water pore is formed between the intracellular residue E203 and the extracellular residue E148. The systematic research revealed that the secondary water pore is formed only when both E148 and E203 are protonated and the chloride ions inside the chloride ion pathway are transported to the outside. It was also determined that E203 interacts with R28 and functions as a gate that controls the formation of the secondary pore by losing or gaining a negative charge according to its protonation state. In addition, Y445 was demonstrated to function as a barrier that separates the secondary water pore from the chloride ion pathway.
We conclude that the secondary pore is a pathway for proton conduction, on the basis of the following results: First, the secondary pore is formed only when E203, which is known to be a key residue for proton transport (14,15), loses its charge by protonation. Second, a continuous water network inside the secondary pore links E203 and E148, two residues known to be involved in the proton transport in ClC exchangers. Third, assuming the secondary water pore is a proton pathway, two unresolved issues related to the previous proposal (15) are now resolved. Fourth, the highly conserved Y445 functions as a barrier that separates the secondary pore from the chloride ion pathway, as supported by the experimental result (18) that removal of the hydroxyl group from Y445 does not affect the electrophysiological behavior of the exchanger. Finally, considering that the secondary water pore for proton conduction is formed when two chloride ions inside the primary pore are transported to the outside and the secondary water pore is closed again when chloride ions fill the primary pore, it is reasonable to assume that the flux of chloride ions is coupled to the flux of protons in the opposite direction with a stoichiometric ratio of 2:1. It is also probable that more than two chloride ions are transported during the gate-opening time. Nonetheless, the fact that two chloride ions are present in the closed pore prompts us to tentatively conclude that a stoichiometric ratio of 2:1 is present during the ionic exchange process. However, further investigation is necessary to clarify the specific molecular mechanism of the quantization.
In the light of these results, we hypothesize that the transport of chloride ions in a ClC exchanger is coupled to proton transport, as illustrated in Fig. 4. First, the protonation of E148 causes the primary pore to be opened, which results in chloride ion conduction (Fig. 4 A). After the chloride ions pass through the primary pore, the interaction between E203 and the R28 is broken, resulting in the formation of a water network in the secondary pore (Fig. 4 B). Next, the proton belonging to E148 is transferred to a water molecule inside the secondary pore, and subsequently the primary pore is closed by deprotonation of E148 (Fig. 4 C). Finally, the proton is able to pass through the secondary pore via the water network (Fig. 4 D).
Figure 4.
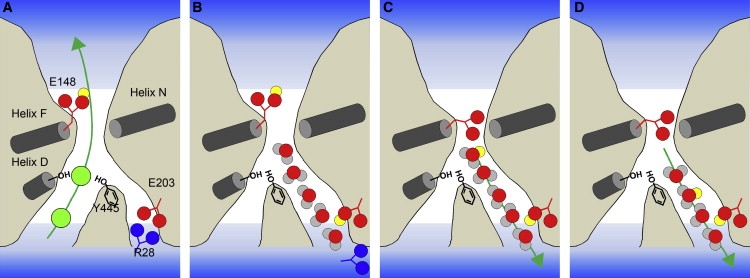
Schematic representation of the coupled movement of chloride ions and protons. The D-, F-, and N-helices are represented by gray cylinders. (A) Chloride ions are represented as green spheres, and protons are represented as yellow spheres. Protonation of E148 causes the primary pore to be opened, which results in chloride ion conduction. (B) After the chloride ions pass through the primary pore, the interaction between E203 and the R28 is broken due to the protonation of E203, resulting in the formation of a water network in the secondary pore. (C) The proton belonging to E148 is transferred to a water molecule inside the secondary pore, and subsequently the primary pore is closed by the deprotonation of E148. (D) The proton passes through the secondary pore via the water network.
Although this work demonstrates that protons are transported in ClC exchangers via the water network formed inside the secondary water pore, rather than by a proton residue, it still remains to be elucidated why the simultaneous protonation of E203 and E148 is required for formation of the secondary pore. Also, additional experimental and theoretical investigations are required to verify that a proton can be transported via this water network. Nonetheless, we believe that the discovery of the secondary water pore in a ClC exchanger is a stepping-stone on the path to understanding the unique characteristics of ClC exchangers in which chloride ion transport is coupled to proton transport, and is a good example of in silico prediction preceding in vivo verification.
Supporting Material
One movie is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00208-0.
Supporting Material
Acknowledgments
This work was supported by funding from the Ministry of Science and Technology of Korea through the Global Research Laboratory program.
References
- 1.Miller C. Open-state substructure of single chloride channels from Torpedo electroplax. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982;299:401–411. doi: 10.1098/rstb.1982.0140. [DOI] [PubMed] [Google Scholar]
- 2.Miller C., White M.M. Dimeric structure of single chloride channels from Torpedo electroplax. Proc. Natl. Acad. Sci. USA. 1984;81:2772–2775. doi: 10.1073/pnas.81.9.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutzler R., Campbell E.B., MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 4.Dutzler R., Campbell E.B., MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 5.Moran O., Traverso S., Pusch M. Molecular modeling of p-chlorophenoxyacetic acid binding to the CLC-0 channel. Biochemistry. 2003;42:5176–5185. doi: 10.1021/bi027368o. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J., Schulten K. Mechanism of anionic conduction across ClC. Biophys. J. 2004;86:836–845. doi: 10.1016/S0006-3495(04)74159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bostick D.L., Berkowitz M.L. Exterior site occupancy infers chloride-induced proton gating in a prokaryotic homolog of the ClC chloride channel. Biophys. J. 2004;87:1686–1696. doi: 10.1529/biophysj.104.042465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corry B., O'Mara M., Chung S.H. Conduction mechanisms of chloride ions in ClC-type channels. Biophys. J. 2004;86:846–860. doi: 10.1016/S0006-3495(04)74160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corry B., O'Mara M., Chung S.H. Permeation dynamics of chloride ions in the ClC-0 and ClC-1 channels. Chem. Phys. Lett. 2004;386:233–238. [Google Scholar]
- 10.Faraldo-Gómez J.D., Roux B. Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli. J. Mol. Biol. 2004;339:981–1000. doi: 10.1016/j.jmb.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 11.Miloshevsky G.V., Jordan P.C. Anion pathway and potential energy profiles along curvilinear bacterial ClC Cl− pores: electrostatic effects of charged residues. Biophys. J. 2004;86:825–835. doi: 10.1016/S0006-3495(04)74158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisset D., Corry B., Chung S.H. The fast gating mechanism in ClC-0 channels. Biophys. J. 2005;89:179–186. doi: 10.1529/biophysj.104.053447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervasio F.L., Parrinello M., Klein M.L. Exploring the gating mechanism in the ClC chloride channel via metadynamics. J. Mol. Biol. 2006;361:390–398. doi: 10.1016/j.jmb.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Accardi A., Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 15.Accardi A., Walden M., Miller C. Separate ion pathways in a Cl−/H+ exchanger. J. Gen. Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picollo A., Pusch M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- 17.Scheel O., Zdebik A.A., Jentsch T.J. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 18.Accardi A., Lobet S., Dutzler R. Synergism between halide binding and proton transport in a CLC-type exchanger. J. Mol. Biol. 2006;362:691–699. doi: 10.1016/j.jmb.2006.07.081. [DOI] [PubMed] [Google Scholar]
- 19.Choi H.S., Huh J., Jo W.H. Electrostatic energy calculation on the pH-induced conformational change of influenza virus hemagglutinin. Biophys. J. 2006;91:55–60. doi: 10.1529/biophysj.105.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi H.S., Huh J., Jo W.H. pH-induced helix-coil transition of amphipathic polypeptide and its association with the lipid bilayer: electrostatic energy calculation. Biomacromolecules. 2006;7:403–406. doi: 10.1021/bm050579z. [DOI] [PubMed] [Google Scholar]
- 21.Ko Y.J., Huh J., Jo W.H. Ion exclusion mechanism in aquaporin at an atomistic level. Proteins. 2008;70:1442–1450. doi: 10.1002/prot.21696. [DOI] [PubMed] [Google Scholar]
- 22.Ko Y.J., Jo W.H. Chloride ion conduction without water coordination in the pore of ClC protein. J. Comput. Chem. 2010;31:603–611. doi: 10.1002/jcc.21432. [DOI] [PubMed] [Google Scholar]
- 23.Tieleman D.P., Berendsen H.J.C. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 1998;74:2786–2801. doi: 10.1016/S0006-3495(98)77986-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 25.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Feller S.E., MacKerell A.D. An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104:7510–7515. [Google Scholar]
- 28.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Beglov D., Roux B. Finite representation of an infinite bulk system—solvent boundary potential for computer-simulations. J. Chem. Phys. 1994;100:9050–9063. [Google Scholar]
- 30.Smart O.S., Goodfellow J.M., Wallace B.A. The pore dimensions of gramicidin A. Biophys. J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutzler R. Structural basis for ion conduction and gating in ClC chloride channels. FEBS Lett. 2004;564:229–233. doi: 10.1016/S0014-5793(04)00210-8. [DOI] [PubMed] [Google Scholar]
- 32.Agmon N. The Grotthuss mechanism. Chem. Phys. Lett. 1995;244:456–462. [Google Scholar]
- 33.Garczarek F., Brown L.S., Gerwert K. Proton binding within a membrane protein by a protonated water cluster. Proc. Natl. Acad. Sci. USA. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garczarek F., Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 35.Xu J., Voth G.A. Computer simulation of explicit proton translocation in cytochrome c oxidase: the D-pathway. Proc. Natl. Acad. Sci. USA. 2005;102:6795–6800. doi: 10.1073/pnas.0408117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J., Sharpe M.A., Voth G.A. Storage of an excess proton in the hydrogen-bonded network of the D-pathway of cytochrome c oxidase: identification of a protonated water cluster. J. Am. Chem. Soc. 2007;129:2910–2913. doi: 10.1021/ja067360s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuang Z., Mahankali U., Beck T.L. Proton pathways and H+/Cl− stoichiometry in bacterial chloride transporters. Proteins. 2007;68:26–33. doi: 10.1002/prot.21441. [DOI] [PubMed] [Google Scholar]
- 38.Wang D., Voth G.A. Proton transport pathway in the ClC Cl−/H+ antiporter. Biophys. J. 2009;97:121–131. doi: 10.1016/j.bpj.2009.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


