Abstract
Formation of a complex between the XRCC1 N-terminal domain (NTD) and DNA polymerase β (Pol β) is central to base excision repair of damaged DNA. Two crystal forms of XRCC1-NTD complexed with Pol β have been solved, revealing that the XRCC1-NTD is able to adopt a redox-dependent alternate fold, characterized by a disulfide bond, and substantial variations of secondary structure, folding topology, and electrostatic surface. Although most of these structural changes occur distal to the interface, the oxidized XRCC1-NTD forms additional interactions with Pol β, enhancing affinity by an order of magnitude. Transient disulfide bond formation is increasingly recognized as an important molecular regulatory mechanism. The results presented here suggest a paradigm in DNA repair in which the redox state of a scaffolding protein plays an active role in organizing the repair complex.
Keywords: disulfide switch, DNA repair, scaffolding protein
Maintenance of the integrity of DNA is a critically important cellular function that depends on the coordinated activities of enzymes that recognize and interpret DNA damage, prepare the damaged site for corrective biochemistry, and implement the required reactions. The scaffold protein XRCC1 plays a central role in base excision repair (BER) and single strand break repair, coordinating the binding and activities of enzymes involved in the repair process. XRCC1 interacts with DNA polymerase β (Pol β), DNA ligase III, PARP1 and 2, polynucleotide kinase, and apurinic/apyrimidinic endonuclease 1 (APE1) (1, 2), and evidence for additional binding partners continues to be reported (3–5). Although a substantial amount of structural data for the individual components involved in the various repair processes is now available, structural information for the corresponding XRCC1 complexes is currently rather limited (6–8).
The N-terminal domain (NTD) of XRCC1 is known to bind to Pol β with high affinity (9), and its solution structure has been determined with NMR (10). Mutagenesis studies have identified the XRCC1 epitope involved in complex formation with Pol β (11). Chemical shift mapping studies localized the Pol β/XRCC1-binding interface on the thumb and palm subdomains, which led to the suggestion that the complex adopts a sandwiched structure, in which the fragile gapped DNA intermediate is surrounded by both Pol β and XRCC1 (8, 10).
The present study provides detailed structural characterization of the molecular interface connecting the XRCC1-NTD and the catalytic domain of Pol β. In contrast with earlier models, the XRCC1-NTD interacts exclusively with the thumb subdomain of Pol β; the arrangement of this complex excludes direct interactions with the DNA in the immediate vicinity of the gap. Additionally the characterization of the XRCC1-NTD/Pol β complex revealed an unexpected variation in the structure of XRCC1-NTD, which undergoes dramatic changes in topology, secondary structure, and electrostatic surface, concomitant with formation of a disulfide bond. The oxidized form of XRCC1-NTD exhibits enhanced affinity for Pol β, suggesting that formation of the disulfide bond regulates repair pathways involving the XRCC1/Pol β complex. XRCC1 has been thought to play an indirect, organizational role in DNA repair; however, the results presented here identify a direct, redox-dependent pathway for modulating the activity of XRCC1 and, in turn, the DNA repair process.
Results
Analysis by Small-Angle X-Ray Scattering.
We initially focused on the characterization of the complex formed between the XRCC1-NTD and the Pol β catalytic domain (Pol β CD), because it appears that the N-terminal Pol β lyase domain does not participate in this interaction, and because the position of this domain can be variable in the absence of a gapped DNA substrate (12). The interaction of XRCC1-NTD with the Pol β CD was first investigated using small-angle x-ray scattering (SAXS) (Table S1). The XRCC1-NTD model has an overall shape of a prolate ellipsoid, consistent with the NMR structure (Fig. S1A and Table S1). The Pol β CD model has an overall crescent or “C” shape (Fig. S1B and Table S1), indicating that the domain organization seen in crystal structures is maintained in solution. XRCC1-NTD complexed with the Pol β CD is a large extended structure and appears to be less globular and compact than would be predicted from previous NMR chemical shift mapping studies (8). This model can be divided into two regions of shape and size that are similar to the respective SAXS and crystal structure models of the two component proteins (Fig. S1C).
Crystal Structure of the XRCC1-NTD.
The x-ray crystal structure of the XRCC1-NTD was solved by molecular replacement using the solution structure of the XRCC1-NTD as a search model (10). Eight intact molecules were found in the asymmetric unit and the structure was refined to Rcryst/Rfree values of 20.0%/23.7%. Data collection, stereochemistry, and refinement statistics are summarized in Table S2.
The XRCC1-NTD structure closely matches the known NMR structure, and has a 1.3-Å backbone atom rmsd with the average NMR structure [Protein Data Bank (PDB) code 1XNA] (Fig. S2A). Although the secondary structural elements of the XRCC1-NTD are also in close agreement with the results of the solution structure determination, some loops show greater variation. A hydrophobic pocket occupied by a buried Tyr136 residue in the NMR structure contains Phe142 in the crystal structure, and Tyr136 is solvent exposed (Fig. S2B). Additionally, in the crystal structure, the face of the Phe142 phenyl ring is positioned near Ala35 Cβ in the hydrophobic pocket, leading to a predicted upfield shift of the 1HB resonance. The conformation of these residues observed in the crystal structure is consistent with the NMR assignments of this residue (10).
Structure of a Reduced and Oxidized XRCC1-NTD/Pol β CD Complex.
The x-ray crystal structures of two different crystal forms (oxidized and reduced) of the XRCC1-NTD complexed to the thumb subdomain of the Pol β CD were obtained. The two complexes were refined to Rcryst/Rfree values of 24.2%/29.5% and 18.9%/25.0%. Data collection, stereochemistry, and refinement statistics are summarized in Table S2. Although both were crystallized under identical conditions, utilizing the same batch of protein, the two structures revealed a significant variation in the structure of the XRCC1-NTD.
The reduced structure (Fig. 1A), solved to a resolution of 2.95 Å, contains four molecules in the asymmetric unit, corresponding to two molecules of Pol β CD and two molecules of XRCC1-NTD. The Pol β CD overlays closely with previously determined Pol β CD structures (0.5-Å backbone rmsd with PDB code 1RPL) (13).
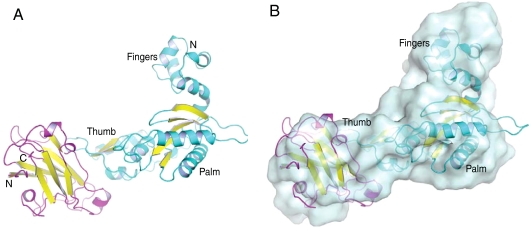
Fig. 1.
Structure of the XRCC1-NTD complex with Pol β catalytic domain. (A) Structure of reduced XRCC1-NTD (magenta ribbon representation with yellow β-strands), bound to Pol β CD (cyan ribbon representation with yellow β-strands). (B) The structure shown in A superimposed with the SAXS-derived model of the complex (surface representation).
Although the structure of the XRCC1-NTD observed in the complex is in generally close agreement with the previously determined solution structure (1.2-Å backbone rmsd with PDB code 1XNA) (10), a more significant difference is found between the crystallographically determined reduced complex interface and the results of chemical shift mapping (CSM) studies (8). The previous CSM studies showed shift perturbations in both the thumb and palm domains; however, the amide shifts in the palm domain were significantly smaller than those in the thumb and had to be analyzed separately. Although these results were interpreted to support a model in which the XRCC1-NTD interacts simultaneously with both the palm and thumb subdomains, the SAXS-derived model does not support this conclusion. Therefore, the crystallographic model chosen for the biological unit of the reduced complex was that which best matched the SAXS data (Fig. 1B). Additionally, the same interface is observed in the oxidized structure. No other interface involving symmetry-related molecules is common to both the reduced and oxidized XRCC1-NTD. The structure of the complex is also in excellent agreement with the results of a mutagenesis mapping study of XRCC1-NTD (11). This structure has a CRYSOL-determined (14) χ2 value of 2.5 referenced to the postulated biological reduced complex (Table S1), supporting the conclusion that the crystal structure matches the SAXS solution data.
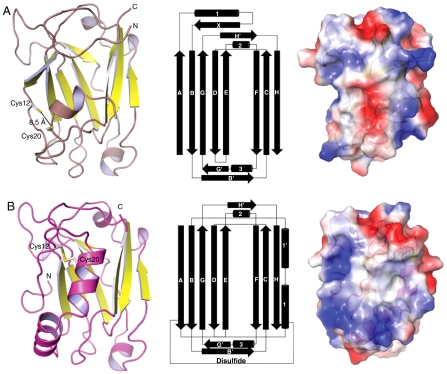
The second structure derived from these studies, the oxidized complex, was solved to a resolution of 2.35 Å (Table S2) and contains one complex in the asymmetric unit. In this structure, the fingers subdomain of the Pol β CD has been proteolyzed, postulated to result from contaminating proteases from the host expression organism, so that only the palm and thumb subdomains of Pol β are observed. The fingers subdomain is not directly involved in the interaction with the XRCC1-NTD; therefore its absence is not expected to significantly influence the results. The Pol β components of the reduced and oxidized complexes are in essentially identical conformations (0.5 Å all atom rmsd). The conformation of a β-hairpin in the thumb subdomain of Pol β (residues 300–309), referred to here as the V303 hairpin, is perturbed slightly, possibly resulting from the altered XRCC1 interface. The XRCC1-NTD structure observed in these crystals was unexpectedly altered in comparison with the first structure obtained. The structure of the oxidized complex is characterized by a disulfide bond between Cys12 and Cys20, along with a significant change in the folding topology of XRCC1-NTD (Fig. 2 A and B). In the reduced form of the protein, the sulfhydryls are separated by 8.5 Å; upon oxidation, they are separated by 2.1 Å with geometry consistent with the most prevalent form of disulfide bond found in proteins (15). This has the effect of positioning the N terminus of the protein in such a manner that contacts with Pol β are formed that are not present in the reduced state (Fig. 3). Additionally, van der Waals interactions formed between the N terminus and Glu81/Gln82 perturb the conformation of the loop containing these residues (Fig. 3). The length of α-helix 1, containing Cys20, is extended by five amino acids. Additionally, in the reduced form, residues 30–36 have β-strand character, β-strand X, whereas in the oxidized form they form α-helix 1’. The cumulative effect of the structural rearrangement is a drastically altered electrostatic surface, with the rearranged structural elements forming an apolar area surrounded by both electropositive and electronegative regions, reminiscent of a canonical protein interaction site (16) (Fig. 2 A and B).
Fig. 2.
Reduced and oxidized XRCC1-NTD. (A) (Left) XRCC1-NTD from the reduced complex shown as a ribbon diagram with yellow β-strands; (Center) topology diagram (10); (Right) surface representation color-coded by electrostatic potential (blue positive, red negative), relative to the left panel, the view has been rotated +90° about a vertical axis. (B) (Left) XRCC1-NTD from oxidized complex shown as a magenta ribbon with yellow β-strands; (Center) topology diagram; (Right) surface representation colored by electrostatic potential (blue positive, red negative), relative to the left panel, the view has been rotated +90° about a vertical axis.
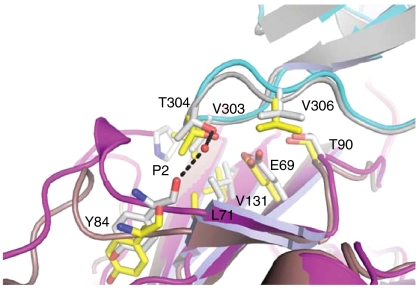
Fig. 3.
Protein interface region showing structural differences between the reduced and oxidized complex of XRCC1-NTD with Pol β. In the reduced/oxidized complex, XRCC1-NTD is shown in brown/magenta and Pol β CD in gray/cyan. Water molecules are shown as red spheres and hydrogen bonds are represented by black dashed lines. Several of the important residue interactions are illustrated for the reduced complex in yellow ball and stick representations, and for the oxidized complex in gray ball and stick representations.
Reduced and Oxidized XRCC1-NTD/Pol β Interface.
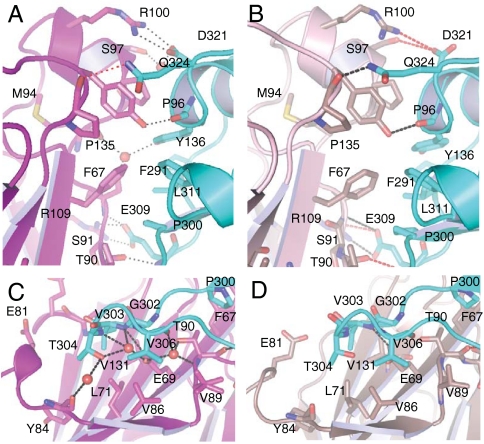
The reduced complex is characterized by a large number of specific interprotein interactions (Fig. 4 B and D). A total of 1181/1231 Å2 (molecules BD/CE) of solvent accessible surface area is buried in the protein∶protien interface. A total of nine hydrogen bonds are formed, four of which are side-chain/side-chain interactions, three being contributed by two salt bridges between Arg109/Glu309 and Arg100/Asp321 in XRCC1-NTD/Pol β CD (Table S3). Nevertheless, the H-bond lengths indicate that four of these bonds are weak, particularly the interactions between Asp321 (Pol β) with Ser97 and with Arg100 (XRCC1-NTD).
Fig. 4.
Close-up views of the XRCC1-NTD/Pol β PT interface. (A and C) oxidized complex, (B and D) reduced complex; hydrogen bonds less than 3.4 Å are indicated with black dashed lines; hydrogen bonds greater than 3.4 Å are indicated with red dashed lines, and water molecules are shown as red spheres. XRCC1-NTD shown in magenta tones with Pol β CD shown as a cyan ribbon.
No ordered water molecules are observed at the interface, presumably as a consequence of the lower resolution of this crystal form. The interface of XRCC1-NTD with Pol β CD involves multiple interactions with the 40 C-terminal residues in Pol β CD, including the V303 hairpin (Fig. 4). In addition to significant chemical shift perturbations, the V303 hairpin was found to experience a large dynamic perturbation as a result of XRCC1-NTD binding (8). A significant constraint on the flexibility of the loop is predicted from the interactions observed in the complex.
Although the overall features of the reduced and oxidized interface are very similar, the oxidized complex has a more developed interface. A total of 1294 Å2 of solvent accessible surface area is buried upon complexation. Nine hydrogen bonds are formed, four of which correspond to the two salt bridges between Arg109/Glu309 and Arg100/Asp321 (Fig. 4 and Table S3). In contrast with the many weak H bonds characterizing the reduced complex, eight of the nine H-bonding distances are < 3.1 Å, suggesting the oxidized form of XRCC1 binds to Pol β with higher affinity than the reduced form (Fig. 4). There are also additional hydrophobic interactions involving Tyr84 and Pro2 of XRCC1, and Thr304 and Val303 on Pol β, respectively.
Five ordered water molecules are observed at the interface mediating indirect interprotein hydrogen bonds in the oxidized complex (Fig. 4 A and C). The position of the side-chain and main-chain atoms that coordinate these water molecules suggests that the same water network is present in the interface of the reduced complex, with the exception of one water molecule. This water molecule is coordinated by amino acids that are located in the loop between β-strands D and E in the XRCC1-NTD, and the V303 hairpin in the Pol β (Fig. 3). This loop in XRCC1, although distal to the disulfide bond, contacts the N terminus of XRCC1 in the oxidized state. The loop also adopts a different conformation in the two forms; the conformation of the backbone atoms of the reduced form precludes the interprotein water-mediated hydrogen bonds found in the oxidized form (Fig. 3).
Structural Switching of XRCC1-NTD.
Treatment of the XRCC1-NTD with 5 mM H2O2 or other oxidizing agents resulted in the formation of multiple molecular species, as indicated by the NMR heteronuclear single quantum coherence (HSQC) spectrum of U-[15N] XRCC1-NTD (Fig. S3). In order to study the behavior of the oxidized form of the protein more easily, we designed stabilized forms of oxidized XRCC1-NTD based on changes in side-chain solvent accessibility. Residue Ile4, which moves from a buried position in the reduced form to a solvent-exposed position in the oxidized structure, was one of the residues identified as a target for mutagenesis. Ile4 was subsequently mutated to an aspartic acid residue to favor the solvent-exposed conformation found in the oxidized protein. The presence of the disulfide bond was confirmed spectrophotometrically in the I4D mutant protein using Ellman’s reagent. A comparison of the effects of oxidation and mutation on the 1H-15N HSQC spectra of U-[15N] XRCC1-NTD indicates that a species which coresonates with I4D XRCC1-NTD is produced by treatment with 5 mM H2O2; however additional resonances are observed that correspond to unreacted protein and to protein showing evidence of additional oxidative changes (Fig. S3). Almost all of the resonances in the 1H-15N HSQC spectrum of the I4D XRCC1-NTD mutant overlay with the spectrum obtained for XRCC1-NTD after treatment with oxidizing agents, supporting the conclusion that I4D XRCC1-NTD is a close structural homolog of one of the XRCC1-NTD oxidation products. The high amount of H2O2 needed to achieve only partial conversion suggests that activation of the XRCC1 disulfide switch is more complex than a simple sensing of the cellular oxidation potential.
Determination of XRCC1-NTD/Pol β Dissociation Constants.
The interaction of XRCC1-NTD with Pol β was evaluated using a Pol β CD P300C mutant that allows covalent attachment of an environmentally sensitive 4-fluoro-7-aminosulfonylbenzofurazan (ABD) fluorescent probe. Addition of saturating amounts of XRCC1-NTD alters the emission maximum of an ABD-labeled Pol β CD by 300% (Fig. S4). This change in fluorescence was used to determine a dissociation constant (Kd) of 110 ± 10 nM by direct titration of Pol β CD with XRCC1-NTD (Fig. S4). This result is in reasonable agreement with the previously determined binding constant of 300 nM (11), with the difference possibly resulting from the presence of the fluorophore. Addition of saturating amounts of I4D XRCC1-NTD, produced a similar change in fluorescence in ABD-labeled Pol β CD; a Kd of 12 ± 2 nM was determined for this complex (Fig. S4). This ∼10-fold increase in affinity is consistent with the enhanced Pol β binding site observed structurally in the oxidized XRCC1-NTD complex.
Discussion
The structural studies described in this report significantly alter the current view of how XRCC1 interacts with one of its critical protein partners, Pol β. In contrast with earlier results that supported a sandwich type of complex in which gapped DNA is held between Pol β and XRCC1 (8, 10, 17), the present studies indicate that both the oxidized and reduced forms of the XRCC1-NTD bind specifically to the thumb subdomain of Pol β; no direct interactions with the palm subdomain are formed. Additionally, superposing the catalytic domain in the structure of the XRCC1 complex with the structure of the Pol β complex with gapped DNA indicates that there are no direct contacts between the XRCC1-NTD and the gapped DNA (Fig. 5). This result has several interesting implications. First, conformational activation of Pol β by an incoming nucleotide in the presence of gapped DNA results in a significant repositioning of the thumb subdomain. Interaction of XRCC1-NTD with both the palm and thumb subdomains would likely alter or interfere with this repositioning, and hence it could alter the kinetic behavior of the enzyme. Second, the present structure does not support a role of the XRCC1-NTD in protecting the fragile gapped DNA intermediate by formation of a sandwich type complex, although it remains possible that a segment of the DNA beyond the gap can make contact with XRCC1. In this context, it was recently concluded that the XRCC1-Pol β interaction is important for repair of damage caused by alkylating agents, whereas the interaction of XRCC1 with DNA is biologically less relevant (18).
Fig. 5.
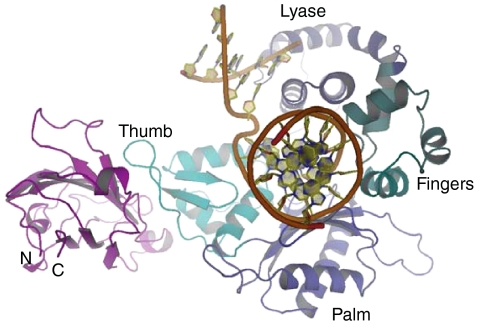
Overlay of the reduced XRCC1-NTD-Pol β complex with the gapped DNA-Pol β complex. The structures of the two complexes were aligned based on the catalytic domain of Pol β, and the overlapping Pol β catalytic domain from the complex with XRCC1-NTD has been omitted for clarity. The XRCC1-NTD is shown in magenta; in the gapped DNA-Pol β complex (PDB code 1BPX), the DNA is shown as an orange ribbon, and the domains of Pol β, shown in various shades of blue, are indicated. As is apparent from the figure, there are no direct contacts between the XRCC1-NTD and the gapped DNA.
Of greater significance is the discovery of an oxidized form of the XRCC1-NTD, characterized by altered folding topology, a significantly altered electrostatic epitope, an enhanced Pol β binding interface, and a Cys12–Cys20 disulfide bond. The importance of reversible disulfide bond formation as a central feature of redox regulation is increasingly recognized (15, 19–24). We have verified the predicted enhancement in Pol β binding affinity using a fluorescence binding assay and a stabilized analog of oxidized XRCC1-NTD. The mutated XRCC1-NTD residue, Ile4, is located far from the Pol β binding site in the oxidized XRCC1-NTD structure, and so should not have any direct effect on the interaction. Thus, one consequence of disulfide switch activation in XRCC1 is an enhanced affinity for Pol β, which would be consistent with the known importance of BER in the response to oxidative stress (25, 26).
Because, as noted above, most of the structural changes are remote from the Pol β binding site, it is unlikely that this is the sole or even the major biological consequence of the oxidative activation. A key feature of the structural change is an altered electrostatic epitope, forming new surfaces reminiscent of a canonical protein∶protein interaction site (16). This result suggests that the oxidation of the XRCC1-NTD plays a role in organizing additional factors (protein or DNA), that cannot bind to the reduced form of the protein. Because APE1 is a known binding partner for XRCC1 (27) and a redox modulator of the activities of many transcription factors (28), it is intriguing to speculate that it may play a role in the activation or deactivation of the proposed XRCC1 disulfide switch. The results presented here suggest a paradigm in base excision repair and single strand break repair of DNA, in which the redox state of scaffolding proteins play active roles in modulating the behavior of the repair complexes.
Materials and Methods
Cloning Overexpression and Purification.
The human xrcc1 and the rat DNA polymerase β gene were kind gifts from the laboratory of Sam Wilson National Institute of Environmental Health Sciences (NIEHS). Three XRCC1-NTD constructs were made: XRCC1-NTDf corresponding to amino acids 1–181, XRCC1-NTDtr corresponding to amino acids 1–151, and XRCC1-NTD I4D corresponding to XRCC1-NTDf with an I4D substitution. The Pol β CD construct corresponds to amino acids 91–335. Each construct was amplified and subcloned into a pET21a plasmid. The coding sequence was cloned in-frame with an ATG start codon and a hexa-histidine affinity tag was fused in-frame at the carboxy terminus (29). The rat and human Pol β amino acid sequence and structure are highly conserved; two differences occur in the 85 residues at the C terminus of Pol β, Q317K, and R326K, neither of which is positioned at the XRCC1-NTD binding interface. Thus, the interface with human Pol β should be identical to that with rat Pol β.
The XRCC1-NTD and Pol β CD plasmids were transformed into BL21-DE3-Rosetta cells (Novagen). Bacteria were grown in terrific broth (TB) overnight at 37 °C. The overnight culture was diluted into TB, grown to an A600 of 0.8 at 37 °C, and induced by the addition of 1 mM IPTG. The cells were grown at 37 °C for 3–4 h, harvested by centrifugation (5000 g for 10 min), resuspended in 20 mM imidazole, 20 mM MOPS, 500 mM sodium chloride (pH 7.5), and lysed by sonication. A lysate was prepared by centrifugation (30,000 g for 30 min). Proteins were purified by immobilized metal affinity chromatography: 20 mL of lysate was loaded onto nitrilotriacetate resin charged with loaded Ni+2 (GE Amersham). Following a wash with 20 mL of 20 mM imidazole, 20 mM MOPS, 500 mM sodium chloride (pH 7.5), the protein was eluted with a step gradient of 75 and 400 mM imidazole [20 mM Mops, 500 mM sodium chloride (pH7.5)]. Protein-containing fractions were concentrated to 10 mL and loaded on a Superdex 26/60 S75 preparative grade gel filtration column (GE Amersham) preequilibrated with 20 mM Tris·HCl (pH 7.8), 300 mM NaCl.
Fluorescence Titrations.
Binding affinities of XRCC1-NTDf and its I4D analog with the Pol β CD were determined using a Pol β CD P300C mutant, which had been derivatized with a fluorescent reporter group, ABD (Anaspec). XRCC1-NTD was added to a sample containing 200 nM Pol β in 20 mM Tris, pH 7.5, 40 mM NaCl. Steady-state fluorescence measurements were performed at room temperature in an Aminco Bowman 1 fluorimeter. Fluorescence was measured by recording the emission spectra from 420 to 540 nm with a fixed excitation wavelength of 395 nm (8 nm slit width). Emission fluorescence intensities from three separate titrations were corrected for dilution by aliquot addition, normalized, averaged, and the values fit to a two-state hyperbolic binding isotherm. The data were fit using two parameters corresponding to the dissociation constant and to a correction for the incomplete reaction of the enzyme with the fluorophore.
SAXS Data Acquisition and Processing.
SAXS data were collected at room temperature on the X9 beamline at the National Synchrotron Light Source (Brookhaven National Laboratory). The wavelength of the beam was 0.855 Å and the sample to detector distance was 2 m. XRCC1-NTDtr (4.5–13.7 mg/mL), Pol β CD (2.3–7.2 mg/mL), and the complex of XRCC1-NTDtr and Pol β CD (4.1–12.3 mg/mL) collected from a gel filtration column were concentrated and dialyzed into a 10 mM Tris, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) , and 150 mM NaCl pH 7.5 buffer for SAXS analysis; SAXS data were collected on three to five different concentrations of protein. Scattering data were circularly averaged and scaled to obtain a relative scattering intensity (I) as a function of momentum transfer vector, q (q = [4π sin θ]/λ), after subtraction of buffer scattering contributions.
SAXS Data Analysis and Model Construction.
All scattering data were analyzed using the Primus software package (30); the GNOM45 software package (31) was used for all P(r) and I0 analyses. The radius of gyration, Rg, and forward scattering, I0, were calculated from the second moment and the start of P(r) (32), respectively, where Rg is the rms of all elemental volumes from the center-of-mass of the particle, weighted by their scattering densities, and I0 is directly proportional to the molar particle concentration multiplied by the square of the scattering particle molecular weight for particles with the same mean scattering density (Table S1) (32). Guinier plots for XRCC1-NTDtr, Pol β CD, and the XRCC1-NTDtr complexed with Pol β CD were linear over a q range of 0.0105–0.052, 0.0105–0.057, and 0.0105–0.046 Å-1 respectively, indicating samples were monodisperse.
Three-dimensional shapes of the XRCC1-NTDtr, Pol β CD, and the XRCC1-NTDtr complexed with Pol β CD were constructed from their respective SAXS data using the GASBOR22IQW program (q-range input for each analysis was from 0.01 to 0.4 Å-1) (33), by calculating the distribution of linearly connected 1.9-Å spheres that best fit the scattering data. Each calculation was repeated at least five times with different random starting points for the Monte Carlo optimization algorithm; no predefined shape or symmetry constraints were used. From these runs, the predicted structure with the lowest deviation of the calculated scattering profile from experimental data was used for interpretation. To compare the SAXS-based models with the atomic structures, the SUPCOMB13 (34) program was used.
Crystallization and Data Collection.
XRCC1-NTDtr and XRCC1-NTDf/Pol β CD complex were concentrated to 12 and 24 mg/mL, respectively, and dialyzed into 10 mM Tris, 40 mM NaCl 0.5 mM TCEP for crystallization. Crystals of XRCC1-NTDtr were grown by hanging drop vapor diffusion in drops containing 2 μL of the protein solution mixed with 2 μL of 0.2–0.3 M MgCl2, 25–30% (wt/vol) PEG 3350 equilibrated against 900 μL of the same solution. Crystals of the reduced complex were grown by hanging drop vapor diffusion in drops containing 2 μL of the protein solution mixed with 2 μL of 0.2 M tripotassium citrate, 20–25% (wt/vol) PEG 3350 equilibrated against 900 μL of the same solution. Crystals of the oxidized complex were grown by streak seeding week-old preequilibrated drops of the reduced XRCC1-NTDf complexed with Pol β CD that failed to produce crystals in the expected 2–3 days, with the reduced complex crystals. Crystals were transferred stepwise to at least 30% (wt/vol) PEG 3350 for cryoprotection, mounted in a nylon loop, and flash frozen in liquid nitrogen. All data were collected at 100 K on a Rigaku 007HFmicromax x-ray generator with a Saturn92 CCD detector. The diffraction data were scaled and indexed using x-ray detector software (35).
Structure Determination Methods, Model Building, and Refinement.
All three crystal structures were solved by molecular replacement using the Phaser program. The crystal structure of Pol β [PDB code 1ZQY (36)] and the solution structure of XRCC1-NTD [PDB code 1XNA (10)] were used as the initial models for fitting the x-ray data. Manual model building was carried out in COOT and refined using REFMAC5 and PHENIX (37, 38). The models exhibit good stereochemistry as determined by PROCHECK and MolProbity; final refinement statistics are listed in Table S2 (39, 40). The final XRCC1-NTD model has Ramachandran plot values of 97.3% favored and 100% allowed. The final reduced complex model has Ramachandran plot values of 93.0% favored and 99.1% allowed. The final oxidized complex model has Ramachandran plot values of 96.8% favored and 100% allowed. Additional electron density near the N-terminal proline is observed in the oxidized complex. PDB coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 3K75, 3LQC, and 3K77 for the reduced complex, oxidized complex, and free XRCC1-NTD, respectively.
NMR Spectroscopy.
Labeled proteins were expressed in minimal media supplemented with 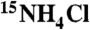 and were purified as described above. Prior to NMR measurements, proteins were concentrated to ∼0.3 mM and exchanged into 20 mM Tris-d11 pH 7.5, 40 mM NaCl, 5% D2O buffer. The 1H-15N HSQC experiments were performed at 25 °C on a Varian UNITY INOVA 600 MHz NMR spectrometer, equipped with a 5 mM 1H triple resonance probe with actively shielded z-axis gradients. The NMR data were processed using NMRPipe (41) and the spectra were analyzed using NMRView (42).
and were purified as described above. Prior to NMR measurements, proteins were concentrated to ∼0.3 mM and exchanged into 20 mM Tris-d11 pH 7.5, 40 mM NaCl, 5% D2O buffer. The 1H-15N HSQC experiments were performed at 25 °C on a Varian UNITY INOVA 600 MHz NMR spectrometer, equipped with a 5 mM 1H triple resonance probe with actively shielded z-axis gradients. The NMR data were processed using NMRPipe (41) and the spectra were analyzed using NMRView (42).
Supplementary Material
Acknowledgments.
The authors would like to thank the laboratory of Sam Wilson (NIEHS) for the human XRCC1 gene and the rat polymerase β gene, Drs. Eugene DeRose and Joseph Krahn, NIEHS for helpful input on the NMR and crystallography studies, Dr. Lin Yang of the X9 beamline, at the National Synchrotron Light Source at Brookhaven National Laboratory, for assistance with data collection. Use of the X9 beamline is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914077107/DCSupplemental.
References
- 1.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 2.Horton JK, et al. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo H, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol Cell Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements PM, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair. 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, et al. JWA regulates XRCC1 and functions as a novel base excision repair protein in oxidative-stress-induced DNA single-strand breaks. Nucleic Acids Res. 2009;37:1936–1950. doi: 10.1093/nar/gkp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali AA, Jukes RM, Pearl LH, Oliver AW. Specific recognition of a multiply phosphorylated motif in the DNA repair scaffold XRCC1 by the FHA domain of human PNK. Nucleic Acids Res. 2009;37:1701–1712. doi: 10.1093/nar/gkn1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beernink PT, et al. Specificity of protein interactions mediated by BRCT domains of the XRCC1 DNA repair protein. J Biol Chem. 2005;280:30206–30213. doi: 10.1074/jbc.M502155200. [DOI] [PubMed] [Google Scholar]
- 8.Gryk MR, et al. Mapping of the interaction interface of DNA polymerase beta with XRCC1. Structure. 2002;10:1709–1720. doi: 10.1016/s0969-2126(02)00908-5. [DOI] [PubMed] [Google Scholar]
- 9.Kubota Y, et al. Reconstitution of DNA base excision-repair with purified human protiens: Interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 10.Marintchev A, et al. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat Struct Biol. 1999;6:884–893. doi: 10.1038/12347. [DOI] [PubMed] [Google Scholar]
- 11.Marintchev A, Gryk MR, Mullen GP. Site-directed mutagenesis analysis of the structural interaction of the single-strand-break repair protein, x-ray cross-complementing group 1, with DNA polymerase beta. Nucleic Acids Res. 2003;31:580–588. doi: 10.1093/nar/gkg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawaya MR, et al. Crystal structure of rat DNA polymerase beta: Evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 13.Davies JF, II, et al. 2.3 A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 14.Svergun DI, Barberato C, Koch MHJ. CRYSOL—A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 15.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 16.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 17.Rice PA. Holding damaged DNA together. Nat Struct Biol. 1999;6:805–806. doi: 10.1038/12257. [DOI] [PubMed] [Google Scholar]
- 18.Wong HK, Wilson DM., III XRCC1 and DNA polymerase beta interaction contributes to cellular alkylating-agent resistance and single-strand break repair. J Cell Biochem. 2005;95:794–804. doi: 10.1002/jcb.20448. [DOI] [PubMed] [Google Scholar]
- 19.Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28:210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 20.Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 21.Linke K, Jakob U. Not every disulfide lasts forever: Disulfide bond formation as a redox switch. Antioxid Redox Sig. 2003;5:425–434. doi: 10.1089/152308603768295168. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez R, Riddle M, Woo J, Momand J. Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Protein Sci. 2008;17:473–481. doi: 10.1110/ps.073252408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan SW, et al. Conformational changes in redox pairs of protein structures. Protein Sci. 2009;18:1745–1765. doi: 10.1002/pro.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voehler MW, et al. Modulation of the structure, catalytic activity, and fidelity of African swine fever virus DNA polymerase X by a reversible disulfide switch. J Biol Chem. 2009;284:18434–18444. doi: 10.1074/jbc.M109.012542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Barzilai A, Yamamoto K. DNA damage responses to oxidative stress. DNA Repair. 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA absic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yip TT, Hutchens TW. Immobilized metal-ion affinity chromatography. Methods Mol Biol. 2004;244:179–190. doi: 10.1385/1-59259-655-x:179. [DOI] [PubMed] [Google Scholar]
- 30.Konarev PV, et al. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 31.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 32.Ashish, et al. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J Biol Chem. 2008;283:2761–2772. doi: 10.1074/jbc.M708325200. [DOI] [PubMed] [Google Scholar]
- 33.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from x-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2000;34:33–41. [Google Scholar]
- 35.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 36.Pelletier H, et al. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Johnson BA, Blevins RA. NMR View—A computer-program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.