Abstract
Ubiquitin carboxyl-terminal hydrolase L1 (UCH-L1) has been implicated in Parkinson’s disease (PD) and is present in neurofibrillary tangles or Lewy bodies. However, the molecular basis for UCH-L1s involvement in proteinacious fibril formation is still elusive, especially in regard to the pathogenicity of the I93M mutation. Here we show that modification of UCH-L1 by cyclopentenone prostaglandins causes unfolding and aggregation. A single thiol group on Cys152 reacts with the α,β-unsaturated carbonyl center in the cyclopentenone ring of prostaglandins, resulting in a covalent adduct. We also show that the PD-associated I93M mutant of UCH-L1 is well-folded, structurally similar to the wild-type protein, and aggregates upon conjugation by cyclopentenone prostaglandins. Our findings suggest a possible mechanistic link between UCH-L1 modification by cyclopentenone prostaglandins and the etiology of neurodegeneration.
Keywords: ubiquitin, NMR, neurodegeneration, protein modification, oxidative damage
The cellular activity of proteins is often regulated through posttranslational modifications that include phosphorylation, acetylation, methylation, glycoslylation, sumoylation, and ubiquitination (1). During the inflammatory response and in response to oxidative stress, proteins can also be modified by various lipid-derived mediators, predominantly arachidonic acid metabolites (2). Covalent Michael adduct formation between the α,β-unsaturated carbonyl center of the cyclopentenone ring with nucleophilic amino acid side-chains of transcription factors and other key proteins can cause potent biological effects, including abrogation of cell proliferation and antiinflammatory and antiviral activities (3–6).
UCH-L1 (also called PARK5 or PGP9.5) is exclusively localized in the brain, where it constitutes ∼2% of the total soluble protein (7, 8). Its 3D structure comprises two lobes, a mainly α-helical domain (α1, α3, α4, α5, and α6), hereafter referred to as the left lobe, and a β-sheet domain that is flanked by two α-helices (α2, β1–β6, α7), hereafter referred to as the right lobe (see orientation in Fig. 1A) (9). It functions predominantly during monoubiquitin recycling in the ubiquitin-proteosome system (UPS), safeguarding balanced protein levels in the cell. The role of UCH-L1 in Parkinson’s (PD) and Alzheimer’s (AD) diseases is still not well understood, although a mutant (I93M) is apparently linked to PD (10, 11), and UCH-L1 is found as a component of Lewy bodies and neurofibrillary tangles of patient brains (12). Biochemical studies suggested the involvement of chemical modification of UCH-L1 caused by neuronal inflammation and oxidative stress, because 4-hydroxy-2-nonenal (HNE) (13, 14), an endogenous neurotoxin and product of lipid hyperoxidation, or Δ12-prostaglandin J2 (Δ12-PGJ2) (15), a dehydrated form of PGJ2 (16, 17) and metabolite of Cyclooxygenase 2 (COX2) (18), reduces UCH-L1’s enzymatic activity. Assessment of the structural differences between free and the HNE-modified UCH-L1 by circular dichroism (CD) spectroscopy suggested loss of α-helical content and increased β-sheet structure (14). In addition, it was suggested that the PD-associated I93M mutant of UCH-L1 mimics HNE-modified UCH-L1 (14). However, no detailed structural or biophysical characterization was carried out, and the structural basis for the modification-mediated loss in hydrolase activity is still unknown.
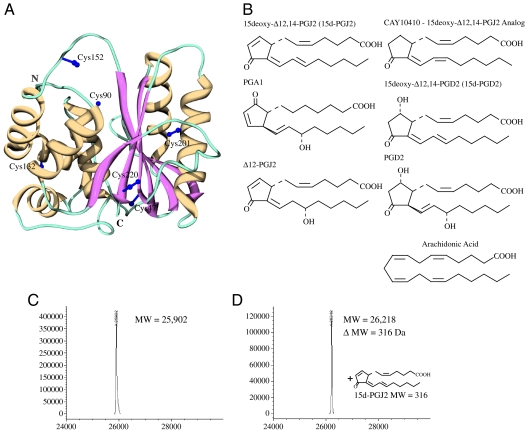
Fig. 1.
3D structure of UCH-L1, chemical structures of prostaglandins and ESI-MS analysis of UCH-L1 modification by 15d-PGJ2. (A) Crystal structure of human UCH-L1 (PDB:2ETL) (9) in ribbon representation (strands: Magenta; helices: Yellow; irregular loops: Teal). The a-helical and b-sheet domains are located on the left and right hand side of the active site cleft, respectively. Side chains for the six Cys residues are depicted in purple ball and stick representation. (B) Prostaglandins used in the present study; molecules containing the α,β-unsaturated carbonyl moiety in the cyclopentenone ring are depicted on the left, those with saturated rings on the right, and arachidonic acid at the bottom. (C–D) ESI-MS analysis of free (C) and 15d-PGJ2 conjugated UCH-L1 (D).
Prompted by previous studies that linked oxidative damage to the neuronal ubiquitination/deubiquitination pathway and reports of increased levels of cyclopentenone prostaglandins in inflammatory fluids (19), we investigated the effects of cyclopentenone prostaglandins, in particular 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), on the highly abundant neuronal protein UCH-L1. We suspected that the structure of UCH-L1 may be influenced in a specific manner by the interaction with 15d-PGJ2. We therefore reacted purified UCH-L1 protein with 15d-PGJ2, identified the presence of singly 15d-PGJ2-modified UCH-L1, determined the modified amino acid as well as the linked carbon atom on 15d-PGJ2, and characterized the structural changes of UCH-L1 upon 15d-PGJ2 conjugation. Our study revealed that 15d-PGJ2 modification of UCH-L1 completely destroyed the folded, 3D structure of the protein and resulted in extensive aggregation of the unfolded, modified polypeptide. Therefore, our results on the destruction and aggregation of UCH-L1, a major neuronal protein in the brain, caused by interaction with the inflammatory response mediator 15d-PGJ2 may provide a molecular explanation for the pathology seen in a number of age-related neurodegenerative diseases, including AD and PD.
Results
UCH-L1 is Modified by 15d-PGJ2.
In order to determine whether UCH-L1 is indeed conjugated by 15d-PGJ2, we reacted purified recombinant UCH-L1 with excess of 15d-PGJ2. (Chemical structures of 15d-PGJ2 and related prostaglandins are provided in Fig. 1B.) The reaction of purified recombinant UCH-L1 with 15d-PGJ2 (2x molar excess) was assessed by liquid chromatography coupled with electrospray ionization mass spectrometry (LC-ESI-MS), revealing the presence of unreacted UCH-L1 protein (experimental mass of 25,902; theoretical mass 25,903 Da—Fig. 1C), as well as a second species with a deconvoluted mass of 26,218 Da, corresponding to the mass of a covalent adduct between one molecule of UCH-L1 and one molecule of 15d-PGJ2, (theoretical mass: 26,217 Da—Fig. 1D). Incubation of UCH-L1 with a 10-fold molar excess of 15d-PGJ2 confirmed the single site modification (only a single product corresponding to a single 15d-PGJ2 conjugation was observed) (Fig. 1D). These data conclusively demonstrate that 15d-PGJ2 indeed covalently reacts with UCH-L1 in a 1∶1 stoichiometry.
The Electrophilic Carbonyl Moiety in the Cyclopentenone Ring of 15d-PGJ2 Is Responsible for UCH-L1 Modification.
15d-PGJ2 contains two electrophilic centers (20), the α,β-unsaturated carbonyl moiety at carbon atom 9 (C9) in the cyclopentenone ring and at carbon atom 13 (C13) in one of the acyl chains (numbering with respect to the first carbon atom of the carboxylic group at one of the acyl chains of 15d-PGJ2—see Fig. 1B). In principle, both carbonyl moieties can readily react with nucleophiles, such as cysteinyl thiol groups of proteins (21). To elucidate which one of the two possible electrophilic centers in 15d-PGJ2 is involved in the reaction with UCH-L1, incubations with a variety of 15d-PGJ2 analogs were performed; CAY10410, 15d-PGD2, and PGD2 (all analogs of 15d-PGJ2 comprising a fully saturated cyclopentenone ring) as well as PGA1 and Δ12-PGJ2 (both analogs of 15d-PGJ2 comprising unsaturated cyclopentenone ring) and arachidonic acid (the precursor in prostaglandin biosynthesis). Reactions with CAY10410, PGD2, 15d-PGD2, and arachidonic acid did not cause modification (only free protein was observed by MS; Fig. S1). Reactions with Δ12-PGJ2 and PGA1 yielded species with masses of 26,235 and 26,239 Da, corresponding addition of a single molecule of Δ12-PGJ2 (mass = 332 Da) or PGA1 (mass = 336 Da), respectively. This demonstrates that only prostaglandin analogs with an α,β-unsaturated carbonyl moiety in the cyclopentenone ring are capable of modifying UCH-L1, confirming that the reactive carbon in 15d-PGJ2 is indeed C9.
15d-PGJ2 Modification Destroys the Native Structure of UCH-L1.
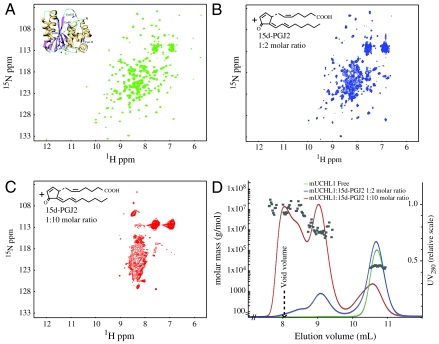
The structural effects of 15d-PGJ2 modification on UCH-L1 were investigated by NMR spectroscopy. The 2D 1H-15N HSQC spectrum of free UCH-L1 displays dispersed resonances, indicative of a well-folded protein (Fig. 2A). Addition of 15d-PGJ2 (2x molar excess) caused a dramatic change in the spectrum (Fig. 2B), suggesting partial unfolding and/or formation of aggregates, based on the presence of severe line broadening for several resonances and their shift into the central, random coil region of the spectrum. It should be noted that identical treatments with CAY10410 (Fig. S2A) and 15d-PGD2 (Fig. S2B) caused no spectral changes whereas 15d-PGA2, a 15d-PGJ2 isomer, also caused similar effects as 15d-PGJ2 (Fig. S2C). This observation is in agreement with the MS results, confirming that UCH-L1 can be modified by any cyclopentenone prostaglandin that contains an electrophilic α,β-unsaturated carbonyl moiety in the ring (prostaglandin J or A series). Addition of excess 15d-PGJ2 (10x molar ratio) resulted in complete unfolding/aggregation as judged by NMR (Fig. 2C), suggesting that the UCH-L1 structure had been completely destroyed by 15d-PGJ2 modification.
Fig. 2.
UCH-L1 aggregation initiated by 15d-PGJ2 modification. (A) 2D 1H-15N HSQC spectrum of UCH-L1 (0.2 mM protein, PBS buffer, pH 7.6, 298 K) exhibiting the expected number of resonances with excellent chemical shift dispersion, indicative of a well-folded protein. (B–C) 2D 1H-15N HSQC spectra of UCH-L1 in the presence of 2 molar (B) and 10 molar (C) excess of 15d-PGJ2, recorded under identical conditions as in A. Substantial resonance broadening is observed and the location of resonances in a narrow spectral region around 8–9 ppm (for 1H) is indicative of unfolded/aggregated protein. (D) Size exclusion chromatographic profiles and light scattering data obtained on the NMR samples whose spectra are shown in A, B, and C. The square symbols are the extracted molar masses based on the light scattering for each fraction during the elution.
Aggregation of the modified UCH-L1 protein was confirmed by subjecting the NMR samples to size exclusion chromatography with in-line multiangle light scattering (SEC-MALS, Fig. 2D). Unmodified UCH-L1 protein behaves as a single monomeric species with a molecular mass of ∼26 kDa (Green). The 15d-PGJ2 modified protein sample exhibits a new peak in the elution profile (Blue), close to the void volume of the column, confirming the presence of high molecular mass aggregates. In the sample that was treated with a 10x molar excess of 15d-PGJ2 only a very small fraction of the protein remained monomeric while the predominant species were high molecular weight aggregates (Red).
In order to evaluate whether our in vitro observations are compatible with in vivo effects caused by 15d-PGJ2, cell-based experiments using midbrain neuronal cells were carried out. Cells were treated with 12 μM biotinylated-15d-PGJ2 (and with PBS as a control), and the endogenous UCH-L1 distribution was examined after 12 h and 24 h (Fig. S3). In untreated cells, no biotin staining (Green) was detected and the UCH-L1 protein (Red) was evenly distributed inside the cells. In the biotinylated-15d-PGJ2 treated cells, colocalization of biotinylated 15d-PGJ2 and UCH-L1 was observed, associated with a dramatic change in the normal distribution of UCH-L1. These effects are even more pronounced at 24 h. Thus, it is very likely that modification and aggregation may also occur in the cellular context during an upsurge of cyclopentenone prostaglandin biosynthesis, such as inflammation or oxidative stress of neuronal cells.
UCH-L1 Modification Involves a Single Cysteine—Cys152.
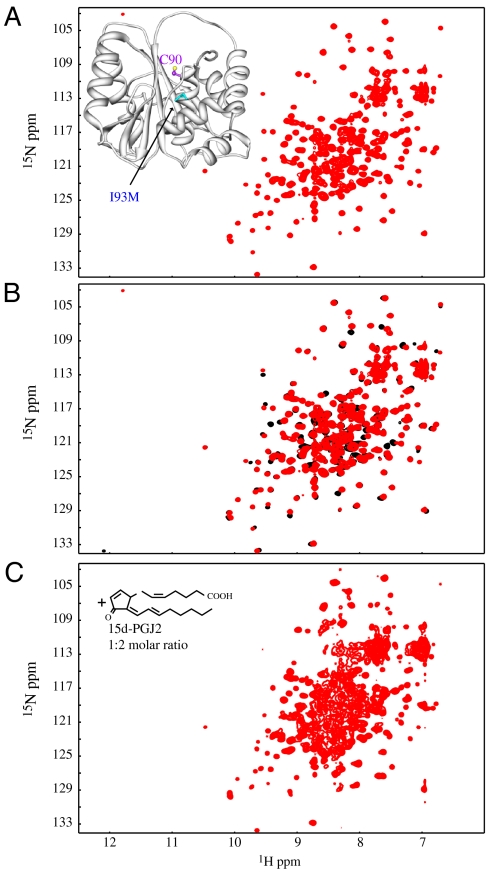
UCH-L1 contains six cysteines in its amino acid sequence, only one of which appears to be involved in modification based on our MS analysis. We therefore created individual Cys-to-Ala mutants and tested their interaction with 15d-PGJ2. Each of the C47A, C90A, C132A, C152A, C201A, and C220A variants was subjected to titration with 15d-PGJ2 and the reaction was followed by 2D 1H-15N HSQC spectroscopy. Spectra for two representative mutants, C90A and C152A, in the absence and presence of 15d-PGJ2 (10x molar excess) are provided in Fig. 3 A and B. (Spectra for the other four mutants are provided in Fig. S4.) For all but the C152A mutant, very similar behavior to wild-type UCH-L1 was observed. For the C152A variant, however, the spectrum appeared essentially unperturbed, and only very minor differences were seen. This suggests that removal of the C152 thiol group by mutation renders the protein refractory to attack by 15d-PGJ2. The spectral results for all other mutants also confirm that 15d-PGJ2 indeed induces unfolding and aggregation of UCH-L1. Note that the Cys-to-Ala mutations do not alter the structures of the mutants, as evidenced by very similar 1H-15N HSQC spectra compared to the wild-type protein (Fig. S5). In a complementary mutant, all Cys residues, except Cys152, were replaced by Ala (denoted as UCH-L1 A47A90A132C152A201A220). The spectral behavior of this variant in response to titration with 15d-PGJ2 is very similar to wild-type UCH-L1 (Fig. 3C). Again, LC-ESI-MS (Fig. S6) confirmed the single prostaglandin modification. Altogether, our MS and NMR data validate that a single thiol modification at position Cys152 by 15d-PGJ2 of UCH-L1 results in unfolding and aggregation of the modified protein.
Fig. 3.
A single Cys in UCH-L1 is conjugated to 15d-PGJ2. 2D 1H-15N HSQC spectra of UCH-L1 mutants in the absence (Left) and presence (Right) of 15d-PGJ2; free C90A (A), free C152A (C), and free A47A90A132C152A201A220 (E) proteins; C90A and C152A in the presence of 10x molar excess of 15d-PGJ2 (B) and (D), respectively, and A47A90A132C152A201A220 in the presence of 5 fold molar excess of 15d-PGJ2 (F). Aggregation is observed for both the C90A (B) and A47A90A132C152A201A220 (F) mutants, while the spectrum of C152A is essentially unaffected. All spectra were recorded as described in the Fig. 2 legend.
The UCH-L1 I93M Mutant Is Well-Folded, Structurally Similar to the Wild-Type Protein, and Aggregates Upon Conjugation to Cyclopentenone Prostaglandins.
The presence of the I93M mutation has been linked to familial PD (10). Molecular properties of the mutant protein were investigated, and it has been suggested that the mutant exhibits similar properties to HNE-modified wild-type protein (14). We prepared I93M mutant protein and analyzed its biophysical properties. As shown in Fig. 4A, the 2D 1H-15N HSQC spectrum of this mutant is well dispersed, indicative of a well-folded protein. Superposition of the wild-type and mutant 2D 1H-15N HSQC spectra reveals no significant changes, demonstrating that the two proteins are structurally very similar (Fig. 4B). Only very minor resonance shifts were observed for residues surrounding the mutation site. We also recorded a 2D 1H-15N HSQC spectrum of the I93M mutant in the presence of 2 fold molar excess of 15d-PGJ2. As illustrated in Fig. 4C, addition of 15d-PGJ2 caused a dramatic change in the spectrum, similar to what was observed for the wild-type protein. This shows that the mutant is also modified by 15d-PGJ2, followed by unfolding and aggregation of the protein.
Fig. 4.
The structure of the I93M mutant is similar to wild-type protein and undergoes 15d-PGJ2 modification, unfolding and aggregation. (A) 2D 1H-15N HSQC spectrum of the UCH-L1 I93M mutant exhibits excellent chemical shift dispersion, demonstrating a well-folded protein. In the inset a ribbon of the x-ray structure (9) is displayed, with the location of the mutated residue marked in cyan. (B) Superposition of the 2D 1H-15N HSQC spectra of wild-type UCH-L1 (Black) and the I93M mutant (Red). Only very small changes in resonance positions were observed, indicating that no significant structural changes between I93M mutant and wild-type proteins are caused by the mutation. (C) 2D 1H-15N HSQC spectrum of the I93M mutant in the presence of 2 fold molar excess of 15d-PGJ2. All spectra were recorded for 0.2 mM protein samples in PBS buffer (pH 7.6) at 298 K.
Discussion
We have shown that UCH-L1 can be modified by 15d-PGJ2 (or other cyclopentenone prostaglandins) at Cys152 only, irrespective of the presence of five other Cys residues in the protein. This also holds true for the I93M mutant that has been linked to familial PD (10) and was thought to exhibit altered structural properties (14). It appears that the observed 15d-PGJ2 modification of UCH-L1 is a rather specific event, not commonly observed with every Cys-containing protein. This is indicated by our results with the Cys-to-Ala UCH-L1 mutants and two other completely unrelated cysteine-containing proteins (Fig. S7). The most striking finding of our study is the complete unfolding of the native 3D UCH-L1 structure upon modification, causing aggregation of the polypeptide chain. Such a dramatic effect caused by a single amino acid modification seems unusual and, to the best of our knowledge, has not been reported previously. All earlier studies of 15d-PGJ2 interactions with proteins reported effects on functional properties (22–25) or computationally modeled conformational effects (26). The only experimental structural report pertains to the recently solved x-ray structure of the 15d-PGJ2/PPARγ complex (27). In that case, the observed conformational changes, however, are small, very different from the complete loss of the native structure observed here for UCH-L1. Similar to PPARγ, treatment of Thioredoxin (Trx), another protein found to be modified by 15d-PGJ2 (28), did not result in major structural pertubations of the Trx structure nor protein aggregation (Fig. S8). Therefore, 15d-PGJ2-mediated unfolding/aggregation seems to be unique to UCH-L1.
Can the location of the modification site in the 3D structure provide clues as to the cause of the dramatic destabilization by the prostaglandin addition? The amino acid sequence of human UCH-L1 differs only by 10 residues from mouse UCH-L1 (used here), suggesting that the x-ray structure of human UCH-L1 (9) (Fig. 1A) is a good model for the mouse structure. Numerous hydrophobic residues are surrounding Cys152. They are conserved between mouse and human UCH-L1. The side chains of these residues are located between the two lobes of the protein, stabilizing the domain interaction in the native, unmodified protein. Upon 15d-PGJ2 conjugation, it is easily possible that the attachment of the prostaglandin to Cys152 (Fig. S9B) will position the acyl chains of 15d-PGJ2 into this interface. The insertion of such a lipophilic moiety may act as a wedge and disrupt the hydrophobic network between the two lobes, initiating the unfolding event. The unfolded chains (partial or fully) will readily interact and form the aggregate. Destruction of the tertiary fold and aggregation of UCH-L1 will abrogate UCH-L1 function, such as enzymatic activity or interactions with other proteins (29).
It is tempting to speculate about UCH-L1’s function in neuronal cells, given its overabundance and widespread distribution. Although an enzymatic function for UCH-L3 has clearly been established, the low hydrolase activity reported for UCH-L1 (30) cast doubts onto its function as an enzyme. Additionally, it was proposed that UCH-L1 acts as a stabilizing factor for monoUb, independent of enzymatic activity (31). Therefore, we hypothesize that, in addition to previously proposed activities of this protein (8, 31, 32), UCH-L1 could operate as a scavenger for highly reactive prostaglandin species, direct metabolites of COX2, or other lipid hyperoxidation products in neuronal cells. In this manner UCH-L1 could contribute to reduce any possible damage caused by oxidative stress or inflammation, endowing neurons with some special protection. If, on the other hand, the level of damage is too great, aggregation and accumulation of insoluble proteinacious material will overwhelm the proteosomal degradation pathway, resulting in the well-known protein deposits that are the hallmark of age-related neurodegenerative diseases including AD and PD.
Furthermore, inflammatory processes that are associated with the activation of COX2 in the brain have been implicated in the pathogenesis of numerous neurodegenerative conditions (33, 34). Unlike oxidative stress that leads to the generation of lipid hyperoxidation byproducts, COX2 activation triggers cyclopentenone prostaglandin production. As shown above, this class of prostaglandins reacts readily with UCH-L1, causing unfolding and aggregation of the protein. We have also shown that the UCH-L1 I93M mutant protein, a mutation reported to be causally associated with PD, exhibits the same fold, has comparable stability, and behaves very similar upon conjugation with 15d-PGJ2 as the wild-type protein. Therefore, this mutation may not be responsible for causing PD. Instead, our results suggest that modification of UCH-L1 by cyclopentenone prostaglandins could be one mechanism by which COX2 contributes to the pathogenesis of PD and other neurodegenerative diseases.
Materials and Methods
Materials.
15d-PGJ2, Δ12-PGJ2, PGA1, CAY10410, PGD2, 15d-PGD2, arachidonic acid, and 15d-PGA2 were obtained from Cayman Chemicals (Ann Arbor, MI). The protein concentration was measured using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE). Human recombinant Trx (hrTrx) was previously purified as described in Qin et al. (35) and was available in the AMG laboratory.
Protein Expression and Purification.
The mouse UCH-L1 gene was cloned into the pET-22b(+) expression vector (Novagen), using NdeI and XhoI restriction sites at the 5′ and 3′ ends, respectively, using primers 5′-GGGAATTCCATATGCAGCTGAAACCGATGGAGATTAACC-3′ (forward) and 5′-AAATTTCTCGAGTTAGGCTGCTTTGCAGAGAGCCAC-3′ (reverse), resulting in a protein with eight additional residues (LEHHHHHH) at its C terminus. Cys-to-Ala and I93M mutants of UCH-L1 were generated by Quickchange site directed mutagenesis (Stratagene). The UCH-L1 A47A90A132C152A201A220 mutant was created using sewing PCR techniques in a similar manner as described for LKAMG (36).
For protein expression, Escherichia coli Rosetta (DE3) cells (Novagen) were transformed with pET-22b-UCH-L1 (or any UCH-L1 mutant) vector. Cells were initially grown at 37 °C, induced with 1 mM IPTG at 16 °C, and grown for further ∼18 h. Isotopic labeling was carried out by growth in modified M9 minimal medium, containing  and/or 13C-glucose as sole nitrogen and/or carbon sources, respectively. For unlabeled samples, cells were cultured in LB medium. Proteins were purified on a Ni2+ affinity column (GE Healthcare), followed by gel filtration on Superdex75 or Superdex200 (GE Healthcare), in PBS buffer, pH ∼ 7.6.
and/or 13C-glucose as sole nitrogen and/or carbon sources, respectively. For unlabeled samples, cells were cultured in LB medium. Proteins were purified on a Ni2+ affinity column (GE Healthcare), followed by gel filtration on Superdex75 or Superdex200 (GE Healthcare), in PBS buffer, pH ∼ 7.6.
Preparation of Prostaglandins for NMR and Mass Spectroscopy.
Prostaglandins from Cayman Chemicals (in methyl acetate or ethanol solution) were gently dried under argon and dissolved in PBS buffer, pH 7.6. Given their chemical instability, freshly prepared prostaglandin solutions were used for all studies.
Mass Spectrometry.
All LC-ESI MS analyses were performed on an Agilent 1100 series (Agilent Technologies) single quadruple electrospray ionization mass spectrometer. Proteins were separated on a Zorbax SB-C3 column (3.0 × 150 mm) using a water acetonitrile gradient with 0.1% formic acid and data were analyzed using Agilent Chemstation.
NMR Spectroscopy.
All NMR spectra were recorded at 298 K on Bruker AVANCE700 and AVANCE600 spectrometers, processed with NMRPipe (37), and analyzed using NMRView (38). All structural figures were generated with Chimera (39).
Cell Cultures and Immunocytochemistry.
Neuronal and glial primary cell cultures were prepared from the midbrain tissue of an embryonic Sprague–Dawley rat, immediately plated into 24-well plates, and grown in 97% Neurobasal Medium (Invitrogen), 2% B-27 (Invitrogen), 0.1% Glutamine (Invitrogen), 0.1% bovine serum albumin (Invitrogen) and 106 U/l penicillin-streptomycin. After 11 days, cells were subjected to either PBS or 15d-PGJ2 treatment. 12 and 24 h after treatment, cells were fixed and washed, and immunocytochemistry (ICC) was performed according to standard procedures. Images were acquired using a laser scanning confocal microscope (Fluoview1000, Olympus), equipped with a spectral detector technology that provides precise wavelength separation of the emitted light.
Supplementary Material
Acknowledgments.
We thank Guillermo Calero, Rieko Ishima, and In-Ja Byeon (Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, PA 15260) for useful discussions and Mike Delk for NMR technical support. This work was a collaboration between the laboratories of S.G. and A.M.G. and was supported by startup funds from the University of Pittsburgh School of Medicine to A.M.G. and by the National Institutes of Health Grant (NS037459-10) to S.G. R.D.M. is supported by a Fondazione Ri.MED (Sicily, Italy) fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002295107/DCSupplemental.
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Boutaud O, Andreasson KI, Zagol-Ikapitte I, Oates JA. Cyclooxygenase-dependent lipid-modification of brain proteins. Brain Pathol. 2005;15:139–142. doi: 10.1111/j.1750-3639.2005.tb00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo M, et al. Cyclopentenone prostaglandins as potential inducers of intracellular oxidative stress. J Biol Chem. 2001;276:12076–12083. doi: 10.1074/jbc.M009630200. [DOI] [PubMed] [Google Scholar]
- 4.Gayarre J, et al. Modification of proteins by cyclopentenone prostaglandins is differentially modulated by GSH in vitro. Ann NY Acad Sci. 2007;1096:78–85. doi: 10.1196/annals.1397.072. [DOI] [PubMed] [Google Scholar]
- 5.Ishii T, Uchida K. Induction of reversible cysteine-targeted protein oxidation by an endogenous electrophile 15-deoxy-delta12,14-prostaglandin J2. Chem Res Toxicol. 2004;17:1313–1322. doi: 10.1021/tx049860+. [DOI] [PubMed] [Google Scholar]
- 6.Sultana R, Perluigi M, Butterfield DA. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer’s disease: Insights into mechanism of neurodegeneration from redox proteomics. Antioxid Redox Sign. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 7.Wilson PO, et al. The immunolocalization of protein gene product 95 using rabbit polyclonal and mouse monoclonal antibodies. Br J Exp Pathol. 1988;69:91–104. [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson KD, et al. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246:670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 9.Das C, et al. Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci USA. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leroy E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 11.Maraganore DM, et al. Case-control study of the ubiquitin carboxy-terminal hydrolase L1 gene in Parkinson’s disease. Neurology. 1999;53:1858–1860. doi: 10.1212/wnl.53.8.1858. [DOI] [PubMed] [Google Scholar]
- 12.Choi J, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa K, et al. Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem Biophys Res Commun. 2003;304:176–183. doi: 10.1016/s0006-291x(03)00555-2. [DOI] [PubMed] [Google Scholar]
- 14.Kabuta T, et al. Aberrant molecular properties shared by familial Parkinson’s disease-associated mutant UCH-L1 and carbonyl-modified UCH-L1. Hum Mol Genet. 2008;17:1482–1496. doi: 10.1093/hmg/ddn037. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, et al. Delta12-Prostaglandin J2 inhibits the ubiquitin hydrolase UCH-L1 and elicits ubiquitin-protein aggregation without proteasome inhibition. Biochem Biophys Res Commun. 2004;319:1171–1180. doi: 10.1016/j.bbrc.2004.05.098. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick FA, Wynalda MA. Albumin-catalyzed metabolism of prostaglandin D2 identification of products formed in vitro. J Biol Chem. 258:11713–11718. [PubMed] [Google Scholar]
- 17.Shibata T, et al. 15-deoxy-delta 12,14-prostaglandin J2. A prostaglandin D2 metabolite generated during inflammatory processes. J Biol Chem. 2002;277:10459–10466. doi: 10.1074/jbc.M110314200. [DOI] [PubMed] [Google Scholar]
- 18.Milne GL, Musiek ES, Morrow JD. The cyclopentenone (A2/J2) isoprostanes—Unique, highly reactive products of arachidonate peroxidation. Antioxid Redox Sign. 2005;7:210–220. doi: 10.1089/ars.2005.7.210. [DOI] [PubMed] [Google Scholar]
- 19.Gilroy DW, et al. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- 20.Straus DS, Glass CK. Cyclopentenone prostaglandins: New insights on biological activities and cellular targets. Med Res Rev. 2001;21:185–210. doi: 10.1002/med.1006. [DOI] [PubMed] [Google Scholar]
- 21.Uchida K, Shibata T. 15-Deoxy-Delta(12,14)-prostaglandin J2: An electrophilic trigger of cellular responses. Chem Res Toxicol. 2008;21:138–144. doi: 10.1021/tx700177j. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, et al. 15-deoxy-Delta12,14-prostaglandin J2 inhibits transcriptional activity of estrogen receptor-alpha via covalent modification of DNA-binding domain. Cancer Res. 2007;67:2595–2602. doi: 10.1158/0008-5472.CAN-06-3043. [DOI] [PubMed] [Google Scholar]
- 23.Oliva JL, et al. The cyclopentenone 15-deoxy-delta 12,14-prostaglandin J2 binds to and activates H-Ras. Proc Natl Acad Sci USA. 2003;100:4772–4777. doi: 10.1073/pnas.0735842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straus DS, et al. 15-deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc Natl Acad Sci USA. 2000;97:4844–4849. doi: 10.1073/pnas.97.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Sala D, Cernuda-Morollon E, Canada FJ. Molecular basis for the direct inhibition of AP-1 DNA binding by 15-deoxy-Delta 12,14-prostaglandin J2. J Biol Chem. 2003;278:51251–51260. doi: 10.1074/jbc.M309409200. [DOI] [PubMed] [Google Scholar]
- 26.Aldini G, et al. Identification of actin as a 15-deoxy-Delta12,14-prostaglandin J2 target in neuroblastoma cells: Mass spectrometric, computational, and functional approaches to investigate the effect on cytoskeletal derangement. Biochemistry. 2007;46:2707–2718. doi: 10.1021/bi0618565. [DOI] [PubMed] [Google Scholar]
- 27.Waku T, et al. Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids. J Mol Biol. 2009;385:188–199. doi: 10.1016/j.jmb.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 28.Shibata T, et al. Thioredoxin as a molecular target of cyclopentenone prostaglandins. J Biol Chem. 2003;278:26046–26054. doi: 10.1074/jbc.M303690200. [DOI] [PubMed] [Google Scholar]
- 29.Kabuta T, et al. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: Ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 31.Osaka H, et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, et al. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 33.Teismann P, et al. Cyclooxygenase-2 is instrumental in Parkinson’s disease neurodegeneration. Proc Natl Acad Sci USA. 2003;100:5473–5478. doi: 10.1073/pnas.0837397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierre SR, Lemmens MA, Figueiredo-Pereira ME. Subchronic infusion of the product of inflammation prostaglandin J2 models sporadic Parkinson’s disease in mice. J Neuroinflamm. 2009;6:18. doi: 10.1186/1742-2094-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin J, Clore GM, Gronenborn AM. The high-resolution three-dimensional solution structures of the oxidized and reduced states of human thioredoxin. Structure. 1994;2:503–522. doi: 10.1016/s0969-2126(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 36.Koharudin LMI, Furey W, Gronenborn AM. A designed chimeric cyanovirin-N homolog lectin: Structure and molecular basis of sucrose binding. Proteins. 2009;77:904–915. doi: 10.1002/prot.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaglio F, et al. NMRPipe—A multidimensional spectral processing system based on Unix pipes. J Biomol Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BA, Blevins RA. NMRView—A computer-program for the visualization and analysis of Nmr data. J Biomol Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.