Abstract
Many archaea (including all the methanogens, nearly all euryarchaeotes, and some crenarchaeotes) use histones as components of the chromatin that compacts their genomes. The archaeal histones are homo- and heterodimers that pair on DNA to form tetrasomes (as the eukaryotic histones H3 and H4 do). The resulting DNA packaging is known to interfere with assembly of the archaeal transcription apparatus at promoters; the ability of transcriptional activation to function in repressive archaeal chromatin has not yet been explored in vitro. Using four of the Methanocaldococcus jannaschii (Mja) histones, we have examined activation of the model Mja rb2 transcription unit by the Mja transcriptional activator Ptr2 in this simplified-chromatin context. Using hydroxyl radical footprinting, we find that the Ptr2-specific rb2 upstream activating site is a preferred histone-localizing site that nucleates histone: DNA-binding radiating from the rb2 promoter. Nevertheless, Ptr2 competes effectively with histones for access to the rb2 promoter and most potently activates transcription in vitro at histone concentrations that extensively coat DNA and essentially silence basal transcription.
Keywords: chromatin, promoter complex, Ptr2, RNA polymerase, upstream activating site
All cells possess abundant small proteins that contribute to genome compaction by wrapping DNA. Within the archaeal kingdom, nearly all Euryarchaeotes and some Crenarchaeotes encode histones (1–4), small proteins (< 10 kDa) that fold, or are predicted to fold, into three α-helices, joined by two loops. This minimal histone fold (α1-L1-α2-L2-α3), first established for the archetype archaeal histone HMfB from Methanothermus fervidus (5, 6), has since been confirmed for other members of this family (4). The native histone fold is stabilized through dimerization (7), and histones generally are dimers or higher-order oligomers in solution. Most archaea encode > 1 histone variant and are expected to be able to assemble multiple homo- and heterodimers comparable with the eukaryotic histone H3–H4 heterodimer (4). A number of archaea also encode histone variants that represent a departure from the archetype, either adding an approximately 30-residue C-terminal extension (8), or fusing two histone folds into one polypeptide chain (9). Archaeal histone dimers pair on DNA to form tetrasomes (10). The eukaryal (H3–H4)2 tetramer contacts approximately 80 bp of DNA at the center of the nucleosome, and the archaeal tetrasome is thought to make similar contacts (6, 11). Additional archaeal dimers accrete to this tetrasome to form n-some complexes, whose structures are not well defined biochemically and do not yield tetrasome “ladders” upon digestion with micrococcal nuclease. However, tetrasome-localizing DNA sequences (TLS) consisting of DNA segments that wrap more readily around archaeal histone tetramers can be selected (11), and regularly spaced strings of such sequences generate tetrasome arrays that resemble beads-on-a-string nucleosome arrays (12).
The existence of TLS implies the possible presence in archaea of sequence-specific chromatin assemblies that dictate gene-specific or regioselective transcriptional regulation. However, archaeal histones lack the N-terminal and C-terminal “tails” that are the targets chromatin-modifying phosphorylation, acetylation, and methylation in eukaryotes (13, 14). Moreover, archaeal histone tetramers bind much less tightly to DNA than do eukaryotic histone octamers. This distinction and the absence of archaeal homologues of histone H1 imply that the tetrasome-based chromatin superstructures of archaea are not intrinsically static and do not require eukaryal-type ATPase-driven chromatin-remodeling or -mobilizing machineries; indeed, archaeal genomes do not encode such activities.
The packaging of DNA into tetrasomes inhibits archaeal transcription by preventing the assembly of the transcription machinery at promoters. In the Pyrococcus furiosus (15) and Methanothermobacter thermoautotrophicus (16) in vitro systems, a cognate histone (HPyA1 and HMtA2, respectively) has been shown to inhibit preinitiation complex assembly and transcriptional initiation at sufficiently high histone:DNA ratios. The placement in its path of an archaeal tetrasome has also been shown to slow, but not prevent, transcript elongation by M. thermoautotrophicus RNA polymerase (RNAP) (16). Thus, despite its apparent simplicity, the eukarya-like transcription initiation machinery of archaea must be able to overcome constraints imposed by its chromatin. The ability of DNA-binding transcriptional activators to gain access to promoters and genes wrapped within these structures remains unexplored.
In this work, we examine transcriptional activation by the Mja Lrp-family DNA-binding protein Ptr2 in the context of Mja histone-repressed DNA. We find that, although the histones HMjA1-A4 preferentially bind to the Mja rb2 promoter, Ptr2 competes effectively for access to the rb2 upstream activating site (UAS) and exerts its maximal activating capacity in vitro at histone concentrations that extensively coat DNA and essentially shut down basal transcription.
Results
Ptr2 Activates Transcription at the Histone-Repressed rb2 Promoter.
Methanocaldococcus jannaschii (Mja), encodes six histones, five of standard type and one with a long C-terminal tail (Fig. S1). Previous experiments with the Mja transcriptional activator Ptr2 and the Mja in vitro system have utilized relatively small DNA fragments generating short runoff transcripts (17–20). We constructed the plasmid pTerm (Fig. S2) for these experiments to let transcriptional initiation compete with the assembly of extensive arrays of histones on DNA (and avoid consideration of the possibility that proximal DNA ends might affect chromatin assembly and interaction). The plasmid pTerm incorporates a transcription unit defined by the Mja rb2 promoter, the previously designed UAS of Ptr2, a segment of the downstream rb2 gene and two extensively analyzed downstream terminators: tmcrA/M, from Methanothermobacter thermoautotrophicum (Mta), and t21 from the lambdoid coliphage 21 (21, 22). A unique site for cutting the transcribed (template) strand downstream of t21 generates relaxed DNA, allowing the histones to bind to, and wrap, DNA without introducing (or changing) topological strain. The rb2con UAS of pTerm is composed of two SELEX-optimized Ptr2 binding sites (23) spaced three helical DNA turns apart and optimally placed upstream of the TATA box and BRE [the binding site of the Mja TATA box-binding protein (TBP) and the transcription factor B (TFB) recognition clement, respectively] (19). All proteins of the in vitro transcription analysis, including the four histones (Fig. S1) are products of the Mja genome.
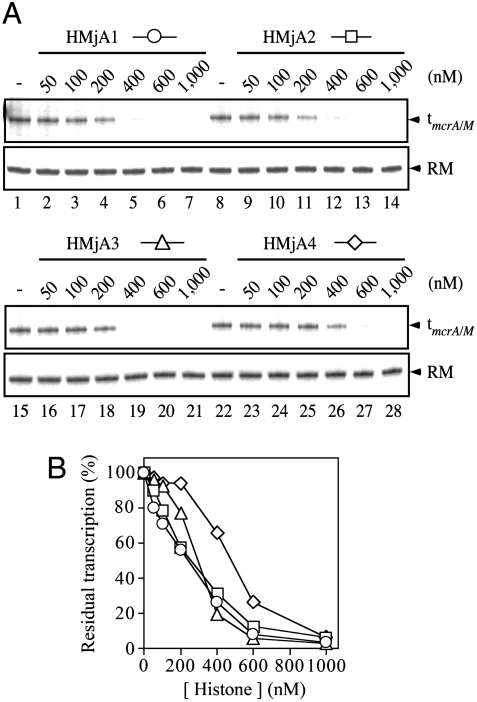
Because this in vitro analysis system involves a competition between histones, initiation factors, and RNAP, we explored different orders of assembly of these components: preformation of chromatin; preequilibration of histones, the initiation factors, and DNA before polymerase recruitment; and simultaneous competition of all components for DNA occupancy (experimental methods presented in SI Text). Transcription was also analyzed in single-cycle as well as multicycle formats. These variations of assembly generated quantitative differences of outcome, principally indicating that the histones repress transcription quantitatively somewhat more effectively when preassembled or competitively prebound together with the initiation factors than when all component proteins are presented together to the transcription template. We also noted that HMjA4 was a consistently less effective transcriptional inhibitor than HMjA1-A3, regardless of the details of the chromatin assembly and transcription protocol. However, the outcome of all experiments was qualitatively the same: All four histones repress transcription, and each one does so essentially completely at sufficiently high concentration (Fig. 1).
Fig. 1.
Repression of transcription by the Mja histones. (A) The relaxed pTerm plasmid template was incubated at 65 °C for 20 min with TBP, TFB, and RNAP, in the absence (Lanes 1, 8, 15, and 22), or with increasing concentrations of individual histones (specified above the panel and measured in terms of the monomer as specified in SI Text), and transcribed for 15 min at the same temperature. The transcript terminating at tmcrA/M and the DNA recovery marker (RM) are shown. (B) residual transcription, data from A, quantified.
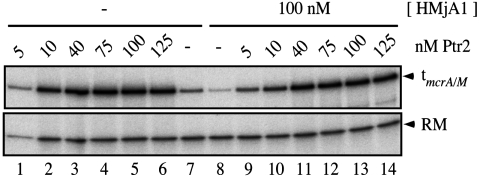
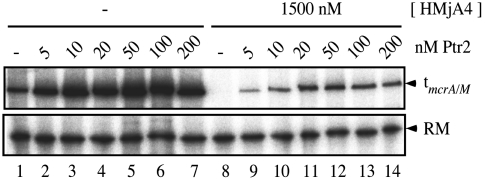
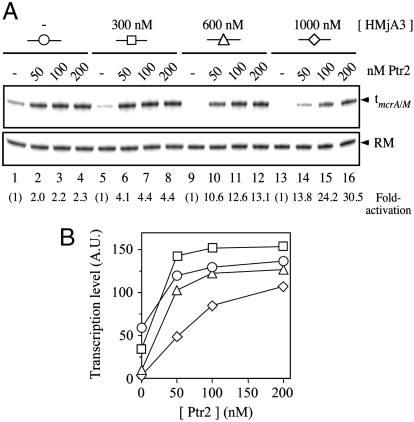
At histone concentrations that substantially repress basal transcription, Ptr2 was able to generate essentially undiminished transcriptional activation: Examples are shown in Fig. 2 (for HMjA1), in Fig. S3 (for HMjA3), and in Table S1 (which summarizes results of several experiments with all four histones). At sufficiently high histone concentrations, Ptr2 was overmatched. An example is shown in Fig. 3: HMjA4 preassembled on pTerm DNA at a concentration of 1.5 μM essentially completely repressed basal transcription, which Ptr2 only restored to approximately 20% of its fully activated level in the absence of histones. An example of a similar experiment with HMjA2 is presented in Fig. S4. The transition from full to only partial activation appears to be a continuous function of the concentration of histones, as indicated by the experiment with HMjA3 shown in Fig. 4: in competition with 300 and 600 nM HMjA3 (which repressed basal transcription respectively 41% and 83%; Fig. 4B), 200 nM Ptr2 secured maximal transcription, but at 1 μM histone, activation was incomplete; in the absence of HMjA3, 50 nM Ptr2 secured nearly maximal activation, but competition by 1 μM HMjA3 diminished this transcription approximately 3-fold (Fig. 4B). A similar experiment with HMjA1 showed a similar effect (Fig. S5).
Fig. 2.
Full activation of transcription in the context of repressive HMjA1. Transcription factors TBP and TFB were incubated with relaxed pTerm DNA at 65 °C for 20 min in the absence (Lanes 1–7) or presence (Lanes 8–14) of 100 nM (monomer) HMjA1 and in the absence (Lanes 7–8) or presence of the indicated (monomer) concentrations of Ptr2 before addition of RNAP for a single round of transcription at the same temperature. The transcript terminating at tmcrA/M and the RM are shown.
Fig. 3.
Partial activation at high HMjA4 concentration. pTerm DNA was incubated at 65 °C for 15 min in the absence (Lanes 1–7) or presence (Lanes 8–14) of 1.5 μM (monomer) HMjA4. The transcription machinery was then added without (Lanes 1 and 8) or with the indicated concentrations of Ptr2 for multiple rounds of transcription at the same temperature.
Fig. 4.
Activation of transcription in the presence of HMjA3. (A) pTerm DNA was incubated at 65 °C for 20 min in the absence (Lanes 1–4), or with increasing concentrations of HMjA3 (specified in terms of the monomer, above the panel), in the absence of His6-Ptr2 (Lanes 1, 5, 9, and 13), or with increasing concentrations of His6-Ptr2 (specified in terms of the monomer, above each lane) and transcribed for 15 min at the same temperature. The transcript terminating at tmcrA/M and the DNA RM are indicated by arrowheads. Fold-activations are shown below each lane. (B) Levels of transcription; data from A, quantified.
The just-presented analysis emphasizes that the ability of Ptr2 to countermand histone-mediated repression makes it an especially effective activator of chromatin transcription. In previous experiments, we emphasized the capacity of Ptr2 for transcriptional activation by focusing on the relatively weak rb2 promoter and reaction conditions (of high ionic strength and low concentrations of the initiation factors) that severely limited basal transcription (17). For the experiments that are presented here, basal transcription was elevated by lowering the ionic strength of the reaction medium and elevating concentrations of the initiation factors, so that even histone-repressed basal transcription could be quantified. Under these conditions, transcriptional activation by Ptr2 in the absence of histones was only a modest 2- to 3-fold for the experiments shown in Fig. 4 and Fig. S5 using the simultaneous assembly protocol, or 4- to 7-fold using the ordered addition protocol of the Table S1 experiments, but allowed the ability to overcome histone-mediated repression to be emphasized. As repression increased, Ptr2-mediated activation increased correspondingly and continuously; when repressed transcription became undetectable, fold-activation by Ptr2 was correspondingly at its indeterminate maximum (Fig. 4 and Fig. S5).
The Competition Between Ptr2 and Histones for Promoter Occupancy.
The ability of Ptr2 to overcome repression by Mja histones prompted a closer examination of histone binding to the rb2 UAS. We turned to hydroxyl radical (•OH) footprinting for this analysis to probe the equilibrium state of histone binding directly, at the temperature and ionic strength of the transcription experiments. The use of long probes minimized boundary effects that DNA ends might exert on binding by proteins that cover extended DNA sites.
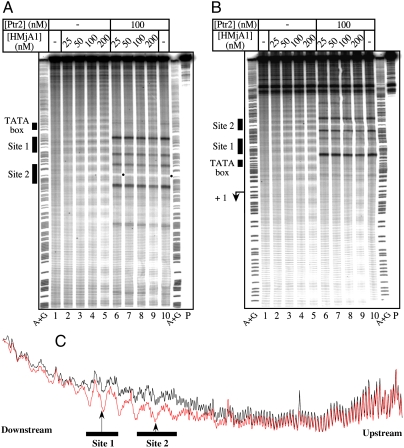
A 460 bp DNA probe (bp -302 to +158 relative to the rb2 start site of transcription), 5′-end labeled on the top (nontranscribed) (Fig. 5A) or bottom (transcribed/template) strand (Fig. 5B), was used to assemble HMjA1 tetrasomes at 65 °C. As shown in Fig. 5 (Lanes 2–5 of each panel), increasing levels of DNA occupancy by HMjA1 are manifested in a highly regular pattern of increasingly full protection against •OH cleavage. This occupancy appears to nucleate within the rb2con UAS at concentrations as low as 25 nM HMjA1 (Fig. 5C), and radiates in both directions at higher HMjA1 concentrations (Lanes 3–5 of Fig. 5 A and B and Fig. S6). HMjA3 generated a similar pattern of protection against •OH cleavage (Fig. S7), as did HMjA2; HMjA4 exhibited a lower affinity for DNA than its paralogues, consistent with its less potent repression of transcription.
Fig. 5.
Promoter occupancy by Ptr2 and histones. A 460 bp DNA probe encompassing positions -302 to +158 relative to the start site of transcription of the rb2con promoter, 5′-end labeled on the top (nontranscribed) strand (A) or the bottom (transcribed/template) strand (B), was generated by PCR using plasmid pTerm as template. This probe was incubated at 65 °C for 20 min in the absence of HMjA1 (Lane 1 of each panel) or with increasing concentrations of HMjA1 (indicated above each panel), and subjected to hydroxyl radical (•OH) cleavage for 30 s at the same temperature. In lanes 6 to 9, HMjA1 binding to DNA is analyzed in reaction mixtures also containing 100 nM His6-Ptr2. In each panel, lane 10 shows His6-Ptr2 binding to DNA in the absence of histone. The TATA box and the two Ptr2 consensus binding sites are identified at the left of each panel. Also shown are the untreated DNA probe (P) as well as the A + G chemical sequencing ladder. The bullets point to the shift of protection at the Site 2 dyad reflecting histone displacement by Ptr2. (C) Aligned density profiles of •OH footprints shown in panel A; profiles in black and red correspond to lanes 1 and 2, respectively. The horizontal bars indicate the two consensus Ptr2 binding sites, with the arrows pointing to their central base pairs, which are the most protected by Ptr2 against •OH cleavage.
His6-Ptr2 binding to the rb2 UAS generates protection of DNA from cleavage at the centers of Sites 1 and 2, flanked by sites of enhanced DNA cleavage (Fig. 5 A and B, Lane 10 in each panel) that stem from tethering of the Fenton reaction-generating metal ion to the immediate vicinity of DNA through chelation by the His6-tag. Removing the tag from Ptr2 abolishes this highly characteristic signal (19, 24). We exploited this feature of the •OH footprint to probe DNA occupancy by His6-Ptr2 in the presence of Mja histones (Fig. 5 A and B, Lanes 6–9). Even at a concentration of 200 nM, HMjA1 was effectively excluded from the promoter region by UAS-bound Ptr2, while remaining free to extensively coat the upstream and downstream regions of the DNA probe (compare Lane 9 to Lane 5 in Fig. 5A and Fig. S7; see also Fig. S8). This vicinal binding of HMjA1 appeared to require a higher histone concentration in the presence than in the absence of Ptr2 (compare Lanes 4 and 5 with Lanes 8 and 9 in Fig. 5A), consistent with the Ptr2 UAS serving as a nucleation site for histone binding. Effective binding by His6-Ptr2 to the rb2con UAS was also observed in the presence of HMjA3 (Fig. S7), HMjA2 and HMjA4. Ptr2 binding, and histone exclusion, are also indicated by the 1–2 bp shift of the protection peak at the Site 2 dyad (marked with a bullet in Fig. 5A and Figs. S7–S10).
These results specify that, in spite of the relatively high and selective affinity of the Mja histones for the rb2con UAS, transcriptional activation by Ptr2 is enabled by its ability to effectively compete for UAS occupancy.
The high affinity of HMjA1 for the rb2con UAS (Kd ∼ 3–5 nM for a histone tetramer, estimated from Fig. S6), and the highly regular nature of the resulting protection pattern, with its approximately 10 bp periodicity, indicate preferred rotational histone positioning within this region. The rb2con UAS incorporates two SELEX-derived high affinity consensus Ptr2 binding sites (Sites 1 and 2) conforming to the palindromic sequence 5′-GGACGATTTTCGTCC-3′, separated by approximately 3 helical turns (31 bp center-to-center). High affinity Ptr2 binding to this UAS stems, at least in part, from its compliance with tight DNA wrapping around the Ptr2 tetramer/octamer, with concomitant bending and narrowing of the minor groove at the dyad centers of the binding sites (Fig. 5C, Vertical Arrows); these are known features of the binding sites of both the bacterial and archaeal Lrp/AsnC family proteins (25–30). It is therefore likely that these same sequence features also accommodate efficient positioning of histones within the synthetically optimized UAS.
This prompted an examination of the rb2 UAS in its wild-type form (which deviates from the Ptr2-binding consensus at the upstream Site 2 and even more strongly at Site 1) for ability to efficiently assemble/position histones. A 457 bp DNA probe (bp -290 to +167, relative to the start site of transcription) was used to assemble HMjA1 tetrasomes. Increasing concentrations of HMjA1 generated a highly regular pattern of protection closely resembling the rb2con promoter footprint (Fig. S9). Histone binding also appeared to nucleate within (and radiate out of) the UAS, albeit at comparatively higher HMjA1 concentrations (compared with Fig. 5A). HMjA3 generated a similar pattern of occupancy (Fig. S10).
We also examined Ptr2’s ability to effectively outcompete the histones for occupancy of the wild-type rb2 UAS. His6-Ptr2 binding was readily detected in the presence of HMjA1 concentrations as high as 200 nM (Fig. S9, Lanes 6–9). Effective binding by His6-Ptr2 to the wild-type rb2 UAS was also observed in the presence of HMjA3 (Fig. S10). These results suggest that Mja histones also exhibit a relatively high affinity for the wild-type rb2 UAS. In vivo, this preferred histone positioning would effectually silence the already weak rb2 promoter, making transcriptional initiation at this promoter even more dependent on Ptr2, as discussed below.
Discussion
Nearly all euryarchaea and some crenarchaea encode histone H4 homologues (31) that bind and wrap DNA. The transcription systems of archaea also fit the eukaryotic mold in that their RNAPs (with accessory subunits whose homologues in the other kingdoms are exclusively eukaryotic) require extrinsic initiation factors. However, archaea lack critical elements of all eukaryotic nuclear transcription and transcriptional regulation [reviewed in (32)] and archaeal histones lack the N-terminal extensions (tails) that are the primary targets of chromatin-modification by acetylation, phosphorylation, and methylation. Moreover, the association of archaeal histones with DNA is not so tight as to require the intervention of chromatin-remodeling machines. Thus, the relationship of archaeal histones to gene regulation may be less like that of their eukaryotic counterparts and more closely follow the functional logic of bacterial chromatin. Nevertheless, it seems reasonable to expect that the presence of histones may confer distinctive mechanisms of gene regulation on the archaea that are not shared by the bacteria. Thus, it is surprising that the issue of how tetrasome-forming and DNA-wrapping archaeal histones affect transcriptional regulation has remained unexplored; the present study constitutes the first direct analysis of the mechanism of activation of transcription by an archaeal RNAP in the context of cognate histones that constitute a major component of its chromatin (33).
Histones and Transcription.
In the Mja in vitro transcription system, histone concentrations that fully occupy the rb2 promoter are not sufficient for complete repression of rb2 transcription (compare Figs. 1 and 5), suggesting that the transcription apparatus itself (TBP, TFB, and RNAP) competes to some extent with histones for access to promoter elements. Ptr2 competes effectively with histones for occupancy of the rb2 UAS and retains nearly full capacity for activation of rb2 transcription at concentrations of histones that almost completely repress basal transcription. In previous work, the capacity of Ptr2 for transcriptional activation was emphasized by focusing on the relatively weak rb2 promoter and on reaction conditions that severely limit basal transcription. In contrast, basal transcription has been elevated here, so that repression by histones could be quantitatively assessed (Fig. 1). Under these conditions, Ptr2 activates the now stronger rb2 transcription by only a modest two-to-three-fold, but its ability to overcome nearly complete repression by histones is further emphasized (Fig. 4 and Fig. S5). Accordingly, we suggest that Ptr2 is generally able to activate transcription from histone-repressed promoters regardless of their intrinsic strength (as determined in vitro by transcription of bare DNA).
The rb2 UAS Is a TLS.
The characteristics of the •OH protection pattern by all four Mja histones are the same. The DNA cleavage pattern in the absence of histones suggests a tendency toward regularly spaced minor groove narrowing in the absence of bound protein. At the rb2 consensus Ptr2 UAS and its immediate vicinity, this pattern coincides with the spacing of DNA protection elicited by each histone, implying an identical rotational setting for all four bound histones (Fig. 5C and Fig. S6). Protection by histones is strongest over the Ptr2 binding sites, with an effective Kd of approximately 3–5 nM estimated for HMjA1 tetrasomes (at 65 °C, in 200 mM NaCl) (Fig. S6). Protection is weaker at the flanks of this preferred central region (in effect acting as a TLS) and spreads out at higher protein concentrations, apparently continuously and in approximately 10 bp increments, rather than quantally, in approximately 80 bp increments (e.g., Fig. 5A). The protection pattern at the lowest histone concentration at which occupancy at the center of the TLS is no more than approximately 50% (Fig. S6) is consistent with an approximately 80 bp binding site centered on the UAS. It also reflects two properties that are known to govern nucleosome binding: greater DNA mobility at the flanks of the nucleosome/tetrasome (34), and less favored but still preferred binding to sites that are displaced by units of the DNA repeat (35).
Global and Gene-Specific Regulation by Archaeal Chromatin.
Because it interferes with the assembly of the transcription machinery at promoters, the packaging of archaeal DNA by histones is inherently inhibitory to transcription (15, 16). In eukaryotes, the dynamic interplay between chromatin and transcription is influenced by enzymes that covalently modify the histones and reposition, reconfigure or eject nucleosomes from important regulatory sequences within promoters (reviewed in refs. 36 and 37). Recent genome-wide studies of nucleosome occupancy (reviewed in refs. 38 and 39) suggest that the inherent sequence-determined properties of their DNA merely help to shape the landscape of nucleosome positioning and density at promoters (both active and repressed), but do not fully dictate it; nucleosome remodelling complexes can override sequence preferences for nucleosome placement, causing specific nucleosomes to encroach into promoter regions that are inherently designed to exclude them.
In contrast, the positioning and density of tetrasomes at archaeal promoters is likely to be directly sequence-determined, and function as an important determinant of the interplay between the transcription apparatus and its regulators. The level of modulation by gene-specific transcriptional regulators (especially activators) would, therefore, depend directly on the dynamic competition with tetrasomes for access to their promoter-proximal binding sites. At the rb2 promoter, a strong UAS that doubles as an efficient TLS imposes a level of repression/silencing by histones in the absence of Ptr2 that enables a wider dynamic range for activation. Thus, the ability of Ptr2 to counter histone-mediated repression is a significant component of achieving the full range of its potential for transcriptional activation. The experiments that are presented here suggest that the context of repressive chromatin may be a generally important component of archaeal gene regulation, at least in those organisms that use histones as components of the chromatin that wraps their genomes and that histone:DNA or chromatin templates (rather than bare DNA) may be the optimal context for analyzing mechanisms of activation in vitro.
Materials and Methods
The purification of proteins for this work; the design and preparation of DNA templates for transcription and of DNA probes for footprinting; single- and multiple-round transcription assays; as well as footprinting with hydroxyl radicals are all described and referenced in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We are grateful to W. Hausner (University of Regensburg) for most generously providing Mja RNA polymerase; to K. Sandman (Ohio State University) for a detailed histone purification protocol; to G.A. Kassavetis for helpful advice and discussion, as always, as well as a critical reading of this manuscript; and to the National Institute of General Medical Sciences for the research grant that supported this work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002360107/DCSupplemental.
References
- 1.Sandman K, Krzycki JA, Dobrinski B, Lurz R, Reeve JN. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc Natl Acad Sci USA. 1990;87:5788–5791. doi: 10.1073/pnas.87.15.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandman K, Reeve JN. Chromosome packaging by archaeal histones. Adv Appl Microbiol. 2001;50:75–99. doi: 10.1016/s0065-2164(01)50004-0. [DOI] [PubMed] [Google Scholar]
- 3.White MF, Bell SD. Holding it together: chromatin in the Archaea. Trends Genet. 2002;18:621–626. doi: 10.1016/s0168-9525(02)02808-1. [DOI] [PubMed] [Google Scholar]
- 4.Sandman K, Reeve JN. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Starich MR, Sandman K, Reeve JN, Summers MF. NMR structure of HMfB from the hyperthermophile, Methanothermus fervidus, confirms that this archaeal protein is a histone. J Mol Biol. 1996;255:187–203. doi: 10.1006/jmbi.1996.0016. [DOI] [PubMed] [Google Scholar]
- 6.Decannière K, Babu AM, Sandman K, Reeve JN, Heinemann U. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J Mol Biol. 2000;303:35–47. doi: 10.1006/jmbi.2000.4104. [DOI] [PubMed] [Google Scholar]
- 7.Arents G, Moudrianakis EN. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li WT, Sandman K, Pereira SL, Reeve JN. MJ1647, an open reading frame in the genome of the hyperthermophile Methanococcus jannaschii, encodes a very thermostable archaeal histone with a C-terminal extension. Extremophiles. 2000;4:43–51. doi: 10.1007/s007920050006. [DOI] [PubMed] [Google Scholar]
- 9.Fahrner RL, Cascio D, Lake JA, Slesarev A. An ancestral nuclear protein assembly: Crystal structure of the Methanopyrus kandleri histone. Protein Sci. 2001;10:2002–2007. doi: 10.1110/ps.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira SL, Grayling RA, Lurz R, Reeve JN. Archaeal nucleosomes. Proc Natl Acad Sci USA. 1997;94:12633–12637. doi: 10.1073/pnas.94.23.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey KA, Marc F, Sandman K, Reeve JN. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J Biol Chem. 2002;277:9293–9301. doi: 10.1074/jbc.M110029200. [DOI] [PubMed] [Google Scholar]
- 12.Tomschik M, Karymov MA, Zlatanova J, Leuba SH. The archaeal histone-fold protein HMf organizes DNA into bona fide chromatin fibers. Structure. 2001;9:1201–1211. doi: 10.1016/s0969-2126(01)00682-7. [DOI] [PubMed] [Google Scholar]
- 13.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 14.Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009;36:75–88. doi: 10.1016/S1673-8527(08)60094-6. [DOI] [PubMed] [Google Scholar]
- 15.Soares D, et al. Archaeal histone stability, DNA binding, and transcription inhibition above 90 degrees C. Extremophiles. 1998;2:75–81. doi: 10.1007/s007920050045. [DOI] [PubMed] [Google Scholar]
- 16.Xie Y, Reeve JN. Transcription by an archaeal RNA polymerase is slowed but not blocked by an archaeal nucleosome. J Bacteriol. 2004;186:3492–3498. doi: 10.1128/JB.186.11.3492-3498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc Natl Acad Sci USA. 2003;100:5097–5102. doi: 10.1073/pnas.0837150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouhammouch M, Hausner W, Geiduschek EP. TBP domain symmetry in basal and activated archaeal transcription. Mol Microbiol. 2009;71:123–131. doi: 10.1111/j.1365-2958.2008.06512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouhammouch M, et al. Promoter architecture and response to a positive regulator of archaeal transcription. Mol Microbiol. 2005;56:625–637. doi: 10.1111/j.1365-2958.2005.04563.x. [DOI] [PubMed] [Google Scholar]
- 20.Ouhammouch M, Werner F, Weinzierl RO, Geiduschek EP. A fully recombinant system for activator-dependent archaeal transcription. J Biol Chem. 2004;279:51719–51721. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- 21.Santangelo TJ, Reeve JN. Archaeal RNA polymerase is sensitive to intrinsic termination directed by transcribed and remote sequences. J Mol Biol. 2006;355:196–210. doi: 10.1016/j.jmb.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 22.Santangelo TJ, Cubonova L, Skinner KM, Reeve JN. Archaeal intrinsic transcription termination in vivo. J Bacteriol. 2009;191:7102–7108. doi: 10.1128/JB.00982-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouhammouch M, Geiduschek EP. A thermostable platform for transcriptional regulation: The DNA-binding properties of two Lrp homologs from the hyperthermophilic archaeon Methanococcus jannaschii. EMBO J. 2001;20:146–156. doi: 10.1093/emboj/20.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchett MA, Wilkinson SP, Geiduschek EP, Ouhammouch M. Hybrid Ptr2-like activators of archaeal transcription. Mol Microbiol. 2009;74:582–593. doi: 10.1111/j.1365-2958.2009.06884.x. [DOI] [PubMed] [Google Scholar]
- 25.de los Rios S, Perona JJ. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J Mol Biol. 2007;366:1589–1602. doi: 10.1016/j.jmb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike H, Ishijima SA, Clowney L, Suzuki M. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc Natl Acad Sci USA. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumarevel T, et al. Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: Implications of effector binding in molecular association and DNA binding. Nucleic Acids Res. 2008;36:4808–4820. doi: 10.1093/nar/gkn456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard PM, et al. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thaw P, et al. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama K, et al. Feast/famine regulation by transcription factor FL11 for the survival of the hyperthermophilic archaeon Pyrococcus OT3. Structure. 2007;15:1542–1554. doi: 10.1016/j.str.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Friedrich-Jahn U, Aigner J, Längst G, Reeve JN, Huber H. Nanoarchaeal origin of histone H3? J Bacteriol. 2009;191:1092–1096. doi: 10.1128/JB.01431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Reich CI, Olsen GJ, Giometti CS, Yates JR., 3rd Shotgun proteomics of Methanococcus jannaschii and insights into methanogenesis. J Proteome Res. 2004;3:538–548. doi: 10.1021/pr034109s. [DOI] [PubMed] [Google Scholar]
- 34.Anderson JD, Thastrom A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol Cell Biol. 2002;22:7147–7157. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennings S, Meersseman G, Bradbury EM. Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 36.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 37.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 38.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 39.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.