Abstract
Different signaling pathways are deployed in specific developmental contexts to generate sexually dimorphic traits. Recently, Sex-lethal (Sxl), the female determinant in Drosophila melanogaster, was shown to down-regulate Notch (N) signaling to accomplish sex-specific patterning. Paradoxically, however, both Sxl and N are ubiquitously expressed in all of the female cells. This raises a key question as to how, during monomorphic female development, N signaling escapes the negative impact of Sxl. Here, we uncover a regulatory loop involving Hrp48, an abundant Drosophila hnRNP, Sxl and N. Phenotypic consequences of the partial loss of hrp48 resemble that of N but are more pronounced in females than in males. Likewise, N levels are drastically diminished only in females. Interestingly, monomorphic female tissues including wing, eye and antennal discs display considerable increase in Sxl amounts. Finally, female-specific attenuation of N signaling is rescued upon simultaneous removal of Sxl. Thus, our data demonstrate that in monomorphic contexts, Hrp48 functions as a moderator of Sxl expression to achieve adequate levels of N receptor production and signaling. We propose that it is critical to modulate the activities of the master determinant underling sexual dimorphism, to ensure that it does not function inappropriately in monomorphic tissues and disrupt their development.
Keywords: sex determination, dosage compensation
During metazoan development, a small number of signaling pathways are iteratively used to orchestrate processes such as cell division, cell fate specification, and survival. In various different developmental contexts, these signaling pathways are deployed to elicit diverse and yet specific responses. The coordinated temporal and spatial regulation of these pathways underlies the final cellular makeup, size, and shape of organs.
N signaling constitutes one such pathway. Typically, the ligand–receptor interaction triggers a series of regulated proteolytic cleavages of N releasing the receptor's intracellular domain (NICD). NICD subsequently translocates to the nucleus, where it interacts with DNA-binding transcriptional regulators, namely CSL [CBF1, Su(H), and LAG-1] and its coactivator Mastermind to activate transcription of specific target genes (reviewed in refs. 1 and 2).
The canonical N pathway is deployed in three types of developmental processes: lateral inhibition, lineage restriction, and generation of compartment boundaries (3). Lateral inhibition primarily relies on small differences in the levels of N and its ligands (Delta or Serrate) among otherwise equivalent neighboring cells. Such differences arise stochastically among the neighboring cells, and eventually get amplified and fixed by a feedback control loop, ultimately producing two alternative cell types. In the case of lineage restriction, different cell fates between two daughter cells are generated through asymmetric segregation of regulators of N signaling, so that the unequal inheritance of a critical regulator brings to differential activation of the N receptor. Finally, spatial differences in N signaling within a developing field can generate a boundary of differential cell adhesion that separates cells into nonintermingling populations or compartments. The N pathway appears to be exceptionally dose sensitive. As a matter of fact, the gene encoding the N receptor was initially discovered because of haploinsufficiency, which leads to notches at the fly wing margin.
N signaling is central to a wide variety of contexts throughout fly development. During development of imaginal discs (the primordia of most of the external structures), N signaling controls cell differentiation, survival, and proliferation and is required for specifying the identity of different imaginal discs (4 –6). In the wing imaginal disk, N signaling activity is restricted to the dorsal/ventral compartment boundary, where it is necessary for keeping the two fields separate and the establishment of an organizer regulating the growth and patterning of the developing wing (7, 8). In addition, N-mediated lateral inhibition directs the specification of the sense organ precursors in the wing margin (9) and subsequently plays a role in defining the vein boundary (7, 10, 11).
Phenotypic studies combined with subsequent genetic and molecular analysis of wing development have been instrumental in the identification of new components of N and other signaling pathways. However, one important limitation of standard loss-of-function genetic screens for previously uncharacterized factors affecting wing development is that mutations in relevant genes that have cell vital functions cannot be recovered because they cause embryonic or larval lethality. To avoid the embryonic lethality, we performed an in vivo RNAi-knockdown screen to identify key regulators/activities using two different postembryonic wing specific Gal4 drivers, namely scalloped (sd)-Gal4 and nubbin (nub)-Gal4 to drive the expression of candidate UAS-RNAi transgenes.
One of the genes identified in our RNAi knockdown screen was hrp48. hrp48 encodes an abundant, essential RNA binding protein that belongs to the hnRNP A/B family in Drosophila [reviewed in (12)]. Previous studies on Hrp48 have shown that it functions as a splicing factor in the nucleus (13, 14). Recently, however, Hrp48 was also shown to be involved in regulation of RNA localization and translation regulation (15 –18). Its function in translational regulation is context dependent, and, depending upon the target, it can repress or activate translation.
Consistent with a vital function, we found that only small and virtually featureless “wings” were present when strong drivers were used to express the UAS-hrp48 RNAi in the wing disk. However, when weaker Gal4 lines were used, phenotypes reminiscent of defects in the N signaling pathway were readily evident. Consistent with the “notch-like” nature of the observed phenotypes, we show that levels of N receptor itself are compromised upon loss of hrp48. Interestingly, we observe that the Hrp48-dependent effects on N expression and signaling were very severe in females, whereas males were only mildly affected. This observation is rather counterintuitive, as one would expect that major signaling pathways would operate similarly in both sexes during monomorphic organ development. One possible explanation is that Hrp48 is required for prevention of sex-specific effects during monomorphic organ development. Such a mechanism is needed in light of the fact that the master switch gene Sex-lethal (Sxl), the activity of which is essential in every female cell, has been implicated in regulation of both Hh and N signaling (19, 20). Indeed, we find that the influence of Hrp48 on N pathway is almost entirely mediated by Sxl. Our data uncover a unique function for Hrp48 and support the conclusion that Hrp48 functions to restrict Sxl expression in developing monomrphic tissues to allow for proper N expression and signaling.
Results
Knockdown of Hrp48 Results in Sex-Specific Aberrant Wing Development.
hrp48 was identified as a candidate gene in an RNAi knockdown screen using two Gal4 lines, sd-Gal4 and nub-Gal4, that drive a high level of expression in the developing wing disk. Because the severity of the defects evident with these drivers made analysis difficult, we used a weaker wing Gal4 driver (C765) to only partially knock down hrp48 function. This driver had two striking effects on wing development. First, we observed multiple aberrant wing phenotypes including thick veins with deltas at wing tip, and variable marginal notches (Fig. S1 A–D). These phenotypes are typically associated with reduced N activity. N signaling plays a pivotal role in Drosophila wing margin and veins formation, and regulates the expression of genes involved in the formation of the D/V boundary in the disk and in wing margin patterning (11, 21, 22). Second, the N-like wing defects were much stronger in females than in males. Thus in addition to having an impact on N signaling during wing development, Hrp48 appears to have a sex-specific function in this developmental pathway.
To test further the effects of Hrp48 on wing development, we examined transheterozygous mutant combinations of hrp48 [hrp4802647/hrp48377 and hrp4810280/hrp48377 (15)], which produce viable flies of normal size (Fig. S1 E and F). Significantly, flies carrying both mutant combinations display precisely the same sort of N-like wing defects that are evident when hrp48 is knockdown using the weak Gal4 driver (Fig. S1 E–K). Moreover, also like the RNAi knockdown, the defects were much more severe in females than in males (Fig. S1 E–K).
N Signaling in Wing Development Is Hrp48 Dependent.
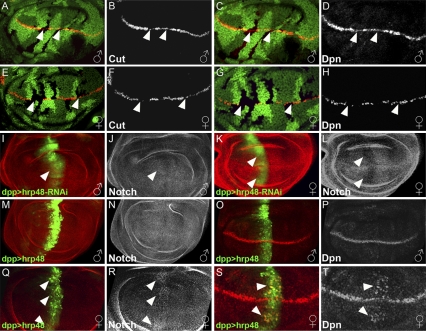
To further examine the role of Hrp48 in wing development, we first sought to confirm that it functions specifically in the N signaling pathway. For this purpose, we generated homozygous hrp4802647 mutant clones in the wing discs and analyzed the expression of two target genes of the N signaling pathway, namely cut and deadpan (dpn) as a readout. Expression levels of these two genes have been used as a reliable indicator of the strength of the N-dependent signaling (8) (Fig. S2). As expected, in the clones mutant for hrp48, the expression levels of Cut and Dpn were either substantially reduced or completely absent (Fig. 1 A–H). Furthermore, consistent with the adult phenotype, the reduction in the levels of Cut and Dpn proteins was significantly stronger in females than in males (compare Fig. 1 B and D with F and H).
Fig. 1.
Hrp48 is required for N expression and signaling in the developing wing. (A–H) Loss of Hrp48 reduces the expression of N target genes more significantly in females than in males. hrp48 LOF clones are marked by loss of GFP (A, C, E, and G) in wing discs (anterior to left, dorsal up). (A–D) Cut and Dpn expression is only partially lost in male discs (B and D, gray), whereas in females discs (E–H) it is abolished (F and H, gray). (I–L) Reduction of hrp48 results in a stronger reduction of N expression in females than in males. Knocking-down hrp48 expression using UAS-RNAi-hrp48 driven by dpp-Gal4 (marked by UAS-GFP, green) results in only a slight reduction in N protein (J) in male wing discs (I–J). (K–L) N protein (L) is clearly reduced in hrp48 knockdown female wing discs. (M–T) Hrp48 overexpression enhances N expression and signaling more significantly in females than in males. dpp-Gal4 driven hrp48 overexpression (M and Q, green) results in pronounced N up-regulation in females wing discs (R, gray) and no effect is observed in males (N, gray). dpp-Gal4 driven hrp48 overexpression (O and S, green) results in pronounced Dpn up-regulation in females wing discs (T, gray) while only a slight effect is observed in males (P, gray).
Similar results were also obtained using an alternative approach. hrp48-specific RNAi-mediated knockdown also leads to repression of the N targets, leading in turn to female-specific adult wing abnormalities (Fig. S3 A–J and O). Taken together, we conclude that hrp48 is required for the proper execution of N signaling pathway during wing development in both sexes. In addition, as indicated by our phenotypic analysis on the adult wings, Hrp48 also performs a female-specific function. As a result, loss of hrp48 function leads to a more pronounced reduction in N target gene expression in female wing discs.
Hrp48 Is Required for N Expression in the Developing Wing.
Reduction in the levels of N targets could result from the ability of Hrp48 to regulate the levels of N protein itself. To test this, we examined the effect of hrp48 knockdown on N protein levels and found that, upon loss of Hrp48, N expression was reduced. Interestingly, N protein levels were strongly reduced in female wing discs and only mildly affected in males (compare Fig. 1 J and L and Fig. S3O). In a complementary approach, we tested whether overexpression of Hrp48 is sufficient to augment the expression of N protein and its target Dpn in wing discs (Fig. 1 M–T). Again, such an enhancement was readily apparent only in the female wing discs (Fig. 1 Q–T), whereas in male discs there were only very mild, if any, effects (Fig. 1 M–P). Taken together, these results indicate that Hrp48 positively influences N expression levels in the wing discs, and this influence is vastly more pronounced in females, consistent with the sex-specific nature of Hrp48 function.
Ligand–receptor interactions trigger the proteolytic cleavage of N, releasing the intracellular domain of the receptor (NICD). NICD translocates to the nucleus, where it activates transcription of target genes (reviewed in refs. 2 and 23). The data presented in the previous sections support the conclusion that Hrp48 likely acts upstream of the activating cleavage event. If reduction of N protein is indeed the primary cause of the observed reduction in N transcriptional output, it should be possible to rescue the effect of hrp48 loss of function (LOF) by simultaneously expressing NICD. We used the dpp-Gal4 driver in combination with a UAS-hrp48-RNAi and a UAS-NICD to simultaneously knock down hrp48 expression and overexpress NICD. We found that, in this experimental setup, N target genes were activated (Fig. S3 K–N), suggesting that loss of Hrp48 affects N protein levels and therefore the pathway's transcriptional output.
Hrp48 Negatively Regulates Sxl Expression in Female's Developing Wing.
The data presented so far demonstrate that the phenotypes associated with the partial LOF hrp48 mutations resemble that of N. Moreover, mutant females display more severe phenotypes than males. Consistently, Hrp48 seems to control N protein levels in a sex-specific manner. In females, loss of Hrp48 leads to reduction in N protein levels, and overexpression of Hrp48 leads to enhancement. By contrast, in males, loss of hrp48 does not lead to a dramatic decrease in the N protein levels, and elevated Hrp48 activity is unable to boost N protein levels or to up-regulate N signaling.
Recently, we have shown that the master control gene for sex determination in Drosophila, Sxl, negatively regulates the N signaling pathway in females, and that this plays an important role in sexually dimorphic tissue development (20). Sxl appears to negatively regulate the pathway by reducing N protein accumulation, as higher levels of N are found in Sxl LOF clones compared with adjacent wild-type cells. This inhibition of N expression does not depend on the known downstream targets of Sxl such as transformer and double sex; rather, Sxl protein was shown to bind N mRNAs and likely down-regulates N translation. These data support the understanding that down-regulation of the N pathway by Sxl contributes to sex-specific differences in morphology.
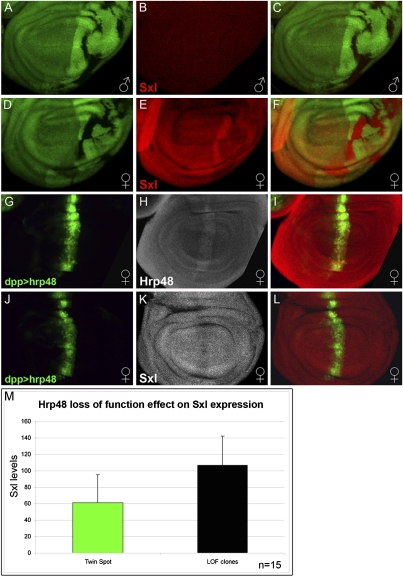
The direct connection between Sxl and N raised the possibility that the sex-specific effects of Hrp48 arise because it is required to attenuate Sxl activity in female wing primordium to maintain levels of N expression appropriate for normal development. In the simplest version of this model, Hrp48 would negatively regulate the levels of Sxl protein. In accordance, we found that Sxl protein levels are indeed significantly elevated in hrp48 LOF clones generated in female wing discs (Fig. 2 D–F and 2 M). Critically, however, loss of hrp48 is not able to induce Sxl protein expression in males (Fig. 2 A–C). Further supporting the antagonistic regulatory relationship between Hrp48 and Sxl, overexpression of Hrp48 in the wing disk led to a concomitant reduction in Sxl protein levels (Fig. 2 J–L).
Fig. 2.
Hrp48 negatively regulates Sxl expression in female developing wings. (A–F) hrp48 loss of function clones marked by the loss of GFP (A and D, green), results in Sxl up-regulation in females (E and F, red) but not in males (B and C, red). (C) Merge of A and B. (F) Merge of D and E. (G–L) hrp48 overexpression reduces Sxl protein levels. dpp-Gal4 driven UAS-hrp48 (G and J, marked by GFP, green) increases the levels of Hrp48 protein (H, gray) and reduces the levels of Sxl protein (K, gray) in females. (I) Merge of G and H. (L) Merge of J and K. (M) Quantification of Sxl expression levels in both hrp4802647/hrp4802647 mutant clones and sister wild-type twin spot clones (clonal average pixel intensity, n = 15).
Sxl Negatively Regulates Both N Expression and Signaling During Wing Development.
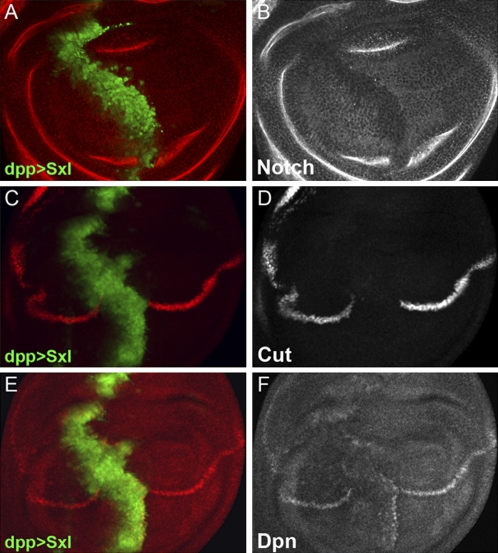
These data suggest that the negative effects of loss of hrp48 function on N signaling in female wings could, in fact, be mediated through the elevation of Sxl protein. If this is indeed the case, elevated levels of Sxl protein should be sufficient to repress N mRNA translation irrespective of Hrp48. Indeed, we found that N expression was abolished where Sxl was overexpressed in female discs (Fig. 3 A and B). Conversely, when we knocked down Sxl expression in female wing disks by RNAi (Fig. 4 A and B), we observed an increase in N protein levels (Fig. 4 C and D), confirming that Sxl negatively regulates N expression in wing discs.
Fig. 3.
Sxl negatively regulates both Notch expression and signaling. (A and B) Sxl overexpressed under the regulation of the dpp-Gal4 driver (A, marked by GFP, green) down-regulates N protein levels (B, gray). (C–F) Sxl overexpression (C and E, marked by GFP, green) down-regulates the expression of N signaling target genes Cut (D) and Dpn (F).
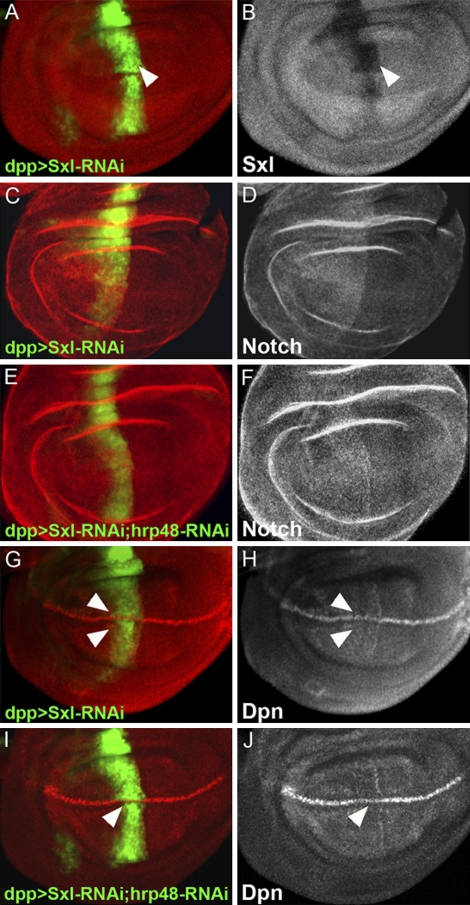
Fig. 4.
Effect of Hrp48 knockdown on Notch protein and signaling is rescued by Sxl knockdown. Sxl levels are reduced (A, red; B, gray) by dpp-Gal4–driven UAS-RNAi-Sxl (A, marked by GFP, green). (C and D) Sxl knockdown on the A–P boundary (C, marked by GFP, green) results in elevated levels of N protein (C, red; D, gray). (E and F) hrp48 and Sxl simultaneous knockdown (E, marked by GFP, green) results in elevated levels of N protein on the D/V boundary (E, red; F, gray). (G and H) Sxl knockdown on the A/P boundary (G, marked by GFP, green) results in Dpn up-regulation on the D/V boundary (G, red; H, gray). hrp48 and Sxl simultaneous knockdown on the A/P boundary (I, marked by GFP, green) results in Dpn up-regulation on the D/V boundary (I, red; J, gray).
We then examined the effect of Sxl on N signaling in the developing wing. We found that overexpression of Sxl abolished the expression of N target genes, cut and dpn (Fig. 3 D and F). Furthermore, knocking down Sxl expression led to mild ectopic expression of Dpn (Fig. 4 G and H). In accordance with our previous findings (20), these results demonstrate that Sxl attenuates N expression and therefore N signaling in female wing development.
Reducing Sxl Relieves the Negative Effects of Loss-of hrp48 on N Expression and Signaling.
The results presented so far show that knocking down Hrp48 expression (either by generating LOF clones of hrp48 or by using a relevant UAS-RNAi in combination with a dpp-Gal4 driver) abolished the expression of N and its target genes, namely Cut and Dpn in female wing discs (Fig. 1 F, H, and L). On the other hand, elevated expression of Sxl protein alone is sufficient to mimic the attenuation in N levels. Combined together, these two observations support the idea that Sxl is a critical target of Hrp48. Furthermore, N expression and signaling in females is maintained primarily by controlling Sxl expression. If so, LOF phenotype of hrp48 should be ameliorated by concomitant reduction in Sxl levels. To examine this idea, we performed a “rescue” experiment in which we simultaneously knocked down Sxl and hrp48 expression using RNAi and monitored the expression levels of N and its target Dpn. Indeed, we found that reduction in Sxl prevented the characteristic reduction in both N expression and signaling associated with reduction in Hrp48 (Fig. 4 F and J). Combined together, these two observations demonstrate that Sxl is a critical target of Hrp48-dependent repression and that Hrp48 functions in a homeostatic control mechanism that does not allow Sxl levels to exceed a given threshold in female wings.
Hrp48 Negatively Regulates Sxl Expression in Eye and Antenna Imaginal Discs.
Next, we wondered whether such a homeostatic mechanism is in place in other tissues and, if so, whether it could be assayed using similar experimental conditions. We therefore sought to determine whether, like in the wing, Hrp48 is differentially required in males and females for the development of other monomorphic organs. Using eyeless (ey)-Gal4 in combination with UAS-hrp48-RNAi, we knock down Hrp48 expression in the eye. Although a moderate reduction in eye size was observed in males, female eyes were severely reduced in size and completely lost pigmentation (Fig. S4 B and C). Similar results were reported by Hammond et al., who used semilethal homozygous hrp48 mutant flies (hrp48k16203/hrp48k16203) (14).
These results prompted us to determine the regulatory relationship between hrp48 and Sxl in the developing eye. To this end, we used the flip-recombinase under the control of eyeless promoter (ey-Flp) to generate homozygous hrp4802647 mutant clones. Importantly, loss of hrp48 function resulted in up-regulation of Sxl expression in both eye and antenna discs of females (Fig. S4H), whereas it remained unexpressed in males (Fig. S4E). Thus, Hrp48 appears to negatively regulate Sxl expression in the development of various monomorphic organs.
Discussion
In Drosophila, the primary sex determination switch is controlled by the X chromosome-to-autosome ratio, which in females (2×/2A) leads to the early expression of the master switch gene, Sex lethal (Sxl). Once activated, Sxl promotes its own synthesis by directing the female-specific splicing of Sxl premRNAs. Although this autoregulatory feedback loop maintains the female-determined state, it is not activated in males (1×/2A). As a result, Sxl transcripts in males are spliced in the nonproductive default mode. Most aspects of somatic sexual development are controlled by Sxl via several gene cascades. These include the transformer (tra)→doublesex(dsx) and tra→fruitless (fru) sexual differentiation pathways. Sxl controls the dsx and fru pathways by regulating tra splicing. Furthermore, Sxl turns off the dosage compensation system in females by blocking the translation of msl-2. It also functions in an msl-2-independent dosage compensation system by directly downregulating the translation of X-linked genes the transcripts of which contain Sxl protein binding sites in their 3′ UTRs (24, 25). In addition, Sxl is thought to control some of the sexually dimorphic traits, such as body size, via mechanisms that are independent of the Sxl→tra→dsx-fru regulatory cascade (26). Consistent with this idea, Sxl was recently shown to down-regulate the activity of the N-signaling pathway (20). It was proposed that repression of N translation by Sxl generates some sexually dimorphic traits, such as the bristles on abdominal segments A5–A6, and functions to promote female development in specific tissues such as the follicle cells of the ovary.
The effects of Sxl on N signaling raised an important question regarding monomorphic developmental contexts. With the exception of genital discs, most organ primordia follow identical developmental routes and develop in similar fashion in both sexes. Moreover, irrespective of the sex, N signaling plays a central role during imaginal disk development. It is therefore critical that a mechanism be in place in females that senses and maintains the Sxl protein level within homeostatic limits. This mechanism must ensure that the amount of Sxl protein in female cells is sufficient to promote sexual differentiation and to repress dosage compensation, but does not exceed a critical threshold to avoid abrogation of N signaling. Assuming that such sensor indeed exists, it needs to fulfill the following criteria: (i) loss of its activity ought to result in a phenotype that resembles that of N; (ii) the effects of loss of this sensor should be sex-specific; (iii) upon loss of the sensor, there should be a corresponding change in N protein levels and/or activity; and (iv) the deleterious phenotypic consequences that result from the loss of the sensor should be ameliorated by reducing Sxl levels.
In the studies reported here, we have uncovered a regulatory link between Hrp48, Sxl and N signaling and have demonstrated that Hrp48 qualifies for such a sensor. We have found that reducing Hrp48 activity results in sex-specific defects. Adult females compromised for Hrp48 function display strong phenotypes reminiscent of hypomorphic N mutations, whereas males are only mildly affected. These phenotypes arise as a consequence of sex-specific reduction in total amount of N protein, which in turn is a result of increase in Sxl levels. We further show that Hrp48 negatively regulates Sxl expression in different monomorphic contexts such as eye and antenna discs. Finally, we show that Sxl is the critical mediator of this effect. Our conclusion that the effect of Hrp48 on the N signaling is mediated through repression of Sxl is based on several lines of evidence. First, the “gain” and “loss” of hrp48 expression influences Sxl and N levels in a reciprocal manner. Second, reducing Sxl expression in females rescues the strong defects induced by the loss of hrp48 on both N expression and signaling. Finally, in males, where Sxl is absent, adverse effects caused by the absence of Hrp48 on N signaling are observed but they are relatively minor. The fact that there are effects in males indicates that hrp48 can have an impact on N signaling by a mechanism that is independent of Sxl. In this regard, it is also noteworthy that the hypomorphic, semiviable alleles of hrp48 induce lethality in both sexes; however, the female viability is considerably lower (14).
Although the sex-specific expression of Sxl proteins is controlled at the level of alternative splicing, recent studies have shown that the levels of Sxl protein are regulated in females via translational control that uses the Sxl 3′ and 5′ UTRs (25). Interestingly, in different developmental contexts, Hrp48 activity has been implicated in both of these regulatory mechanisms namely alternative premRNA splicing and translational control. We reasoned that if Hrp48 negatively affects the efficiency of female splicing, then the amount of female-specific mRNAs should be increased in hrp48 LOF background compared with control females. Using northern and semiquantitative RT-PCR analysis, we found that the level and pattern of Sxl sex-specific transcripts appeared to be unaffected in both males and females mutant for hrp48 (Fig. S5), suggesting that hrp48 does not influence either the efficiency or the specificity of the alternative splicing. Likewise there were no alterations in the pattern of polyA addition of Sxl mRNAs. The general stability of the Sxl message also seemed unaffected, suggesting very little, if any, contribution of a mechanism based on RNA stability. It is therefore likely that hrp48 exerts its effects via translational control of Sxl message, possibly by physically interacting with components of the translational machinery. Further experiments will focus on understanding the precise biochemical details.
Taken together, our data elucidate a unique mechanism that results in restricting the amounts of Sxl protein in female tissues by an hnRNP. This sex-specific function of a general RNA binding protein seems to play a critical role during proper development of sexually monomorphic organs. More generally, our data bring into focus the essential nature of the homeostatic regulatory controls that ensure simultaneous development of sexually monomorphic as well as dimorphic organs.
Experimental Procedures
Fly Strains and Genetic Analysis.
The P element insertion line hrp4802647and the EMS allele- hrp48377 were generously provided by Trudi Schüpbach (15). The following transgenic stocks were used in this study: UAS-Sxl, UAS-flp, UAS-RNAi-hrp48 (NIG #10377R1), UAS-RNAi-Sxl (NIG #18350R-3, VDRC #3131), UAS-NINT, UAS-NExd-DN, UAS-Ser, UAS-Hrp48-HA, UAS-GFP. Transgenes were expressed using the Gal4/UAS binary system with the following drivers: nubbin-Gal4, hh-Gal4, dpp-Gal4, ey-Gal4, and C765-Gal4.
Generation of LOF Clones.
Mutant clones were generated using Flp-mediated mitotic recombination. Clones were induced either with hh-Gal4/UAS-flp or by heat shock (60 min at 37 °C). Genotypes of dissected larvae were as follows. Hrp4802647 LOF clones in the wing: yw hsp70-flp; Hrp4802647 FRT40/ubi-GFP FRT40; UAS-flp, hh-Gal4 or yw hsp70-flp; Hrp4802647 FRT40/ubi-GFP FRT40. Hrp4802647 LOF clones in the eye: yw,ey-flp ; Hrp4802647 FRT40/FRT40.
Immunohistochemistry
Imaginal discs from third instar larvae were fixed and stained by standard techniques. The specific primary antibodies used were: rabbit anti-Hrp48 (1:500, a gift from D.C. Rio), mouse anti-Cut (1:150; DSHB #2B10), rabbit anti-Dpn (1:500; a gift from Y.N. Jan), mouse anti-Nint (1:300; DSHB # C17.9C6), rabbit anti-Nicd (1:300; a gift from S. Atravanis-Tsakonas), mouse anti-Sxl (1:500). We used secondary antibodies conjugated with rhodamine red-X or Cy5 (1:400; Jackson Labs). Images were taken on a TE2000-E confocal microscope (Nikon).
Quantification of Hrp48 LOF and Knockdown Effects
Sxl expression levels (intensity) were sampled (using Adobe PhotoShop) in Hrp48 mutant clones and sister wild-type twin spot. Sampled field size was equal in both the mutant and wild-type sibling clones. Clonal average pixel intensity for mutants and twin spots are shown in a histogram (Fig. 2M). In both males and females, Hrp48 was knocked down on the AP boundary using the Dpp-Gal4 driver. Intensity levels (for Notch Cut and Dpn) were measured (using Adobe PhotoShop) on the AP boundary and adjacent tissue for each wing disk. Sampled field size was equal in both experiment and control. Average pixel intensities are shown in a histogram (Fig. S3O).
Plasmid Construction.
Construct containing full-length hrp48 cDNA was used, and hrp48-HA was prepared by standard PCR amplification. Following sequencing, this was inserted in-frame into the pUAST vector.
Supplementary Material
Acknowledgments
We thank Y. N. Jan, D. C. Rio, T. Schüpbach, S. Artavanis-Tsakonas, National Institute of Genetics Fly Stock Center (NIG), The Vienna Drosophila RNAi Center, The Bloomington Drosophila Stock Center, Developmental Studies Hybridoma Bank, Drosophila Genomics Resource Center (DGRC), for fly stocks, antibodies and DNA clones. We thank E. Ordan and O. Ziv for insightful discussions and comments on the manuscript and A. Fiensod and Y. Grosman for technical assistance. The authors declare that they have no competing financial interests. This research was supported by grants from the F.I.R.S.T. (Bikura) Individual Grant Application 1450/09, Israel.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910570107/DCSupplemental.
References
- 1.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006:883–885. doi: 10.1126/stke.3642006cm7. [DOI] [PubMed] [Google Scholar]
- 2.Kadesch T. Notch signaling: The demise of elegant simplicity. Curr Opin Genet Dev. 2004;14:506–512. doi: 10.1016/j.gde.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Bray S. Notch signalling in Drosophila: Three ways to use a pathway. Semin Cell Dev Biol. 1998;9:591–597. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- 4.Giraldez AJ, Cohen SM. Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development. 2003;130:6533–6543. doi: 10.1242/dev.00904. [DOI] [PubMed] [Google Scholar]
- 5.Kurata S, Go MJ, Artavanis-Tsakonas S, Gehring WJ. Notch signaling and the determination of appendage identity. Proc Natl Acad Sci USA. 2000;97:2117–2122. doi: 10.1073/pnas.040556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafel N, Milán M. Notch signalling coordinates tissue growth and wing fate specification in Drosophila. Development. 2008;135:3995–4001. doi: 10.1242/dev.027789. [DOI] [PubMed] [Google Scholar]
- 7.de Celis JF, Bray S, Garcia-Bellido A. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development. 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 8.Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Benjumea FJ, Cohen SM. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development. 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- 10.Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- 11.Sturtevant MA, Bier E. Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development. 1995;121:785–801. doi: 10.1242/dev.121.3.785. [DOI] [PubMed] [Google Scholar]
- 12.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 13.Burnette JM, Hatton AR, Lopez AJ. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics. 1999;151:1517–1529. doi: 10.1093/genetics/151.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond LE, Rudner DZ, Kanaar R, Rio DC. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrich JS, Clouse KN, Schüpbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–1958. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- 16.Huynh JR, Munro TP, Smith-Litière K, Lepesant JA, St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell. 2004;6:625–635. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- 17.Nelson MR, et al. A multiprotein complex that mediates translational enhancement in Drosophila. J Biol Chem. 2007;282:34031–34038. doi: 10.1074/jbc.M706363200. [DOI] [PubMed] [Google Scholar]
- 18.Yano T, et al. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- 19.Horabin JI. Splitting the Hedgehog signal: Sex and patterning in Drosophila. Development. 2005;132:4801–4810. doi: 10.1242/dev.02054. [DOI] [PubMed] [Google Scholar]
- 20.Penn JK, Schedl P. The master switch gene sex-lethal promotes female development by negatively regulating the N-signaling pathway. Dev Cell. 2007;12:275–286. doi: 10.1016/j.devcel.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Celis JF, García-Bellido A. Roles of the Notch gene in Drosophila wing morphogenesis. Mech Dev. 1994;46:109–122. doi: 10.1016/0925-4773(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 22.Xu T, Rebay I, Fleming RJ, Scottgale TN, Artavanis-Tsakonas S. The Notch locus and the genetic circuitry involved in early Drosophila neurogenesis. Genes Dev. 1990;4:464–475. doi: 10.1101/gad.4.3.464. [DOI] [PubMed] [Google Scholar]
- 23.Weng AP, Aster JC. Multiple niches for Notch in cancer: Context is everything. Curr Opin Genet Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Kelley RL, Wang J, Bell L, Kuroda MI. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 25.Yanowitz JL, Deshpande G, Calhoun G, Schedl PD. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of sex-lethal. Mol Cell Biol. 1999;19:3018–3028. doi: 10.1128/mcb.19.4.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.