Abstract
Aneuploidy is a characteristic feature of established cancers and can promote tumor development. Aneuploidy may arise directly, through unequal distribution of chromosomes into daughter cells, or indirectly, through a tetraploid intermediate. The polo family kinase Plk4/Sak is required for late mitotic progression and is haploinsufficient for tumor suppression in mice. Here we show that loss of heterozygosity (LOH) occurs at the Plk4 locus in 50% of human hepatocellular carcinomas (HCC) and is present even in preneoplastic cirrhotic liver nodules. LOH at Plk4 is associated with reduced Plk4 expression in HCC tumors but not with mutations in the remaining allele. Plk4+/− murine embryonic fibroblasts (MEFs) at early passage show a high incidence of multinucleation, supernumerary centrosomes, and a near-tetraploid karyotype. Underlying these phenotypes is a high rate of primary cytokinesis failure, associated with aberrant actomyosin ring formation, reduced RhoA activation, and failure to localize the RhoA guanine nucleotide exchange factor Ect2 to the spindle midbody. We further show that Plk4 normally localizes to the midbody and binds to and phosphorylates Ect2 in vitro. With serial passaging Plk4+/− MEFs rapidly immortalize, acquiring an increasing burden of nonclonal and clonal gross chromosomal irregularities, and form tumors in vivo. Our results indicate that haploid levels of Plk4 disrupt RhoGTPase function during cytokinesis, resulting in aneuploidy and tumorigenesis, thus implicating early LOH at Plk4 as one of the drivers of human hepatocellular carcinogenesis. These findings represent an advance in our understanding of genetic predisposition to HCC, which continues to increase in incidence globally and particularly in North America.
Keywords: aneuploidy, hepatocellular carcinoma, Polo-like kinase, RhoA, Ect2
The role of aneuploidy as a cause rather than a consequence of cancer development has recently been demonstrated for genes encoding the mitotic checkpoint machinery (1 –3). Heterozygosity for Mad1, Mad2, or CENP-E allows anaphase to proceed despite the presence of lagging chromosomes, increasing the rates of aneuploidy, chromosomal instability, and spontaneous tumorigenesis. CENP-E in particular is not known to have a function apart from its role as a mitotic motor and checkpoint protein, suggesting that the development of spontaneous tumors in CENP-E+/− mice is a result of chromosomal instability per se (1). In murine mammary epithelial cells lacking p53 function, tetraploidy precedes aneuploidy and predisposes to tumor development (4). Defects in cytokinesis result in both tetraploidy and multiple centrosomes, which can form multipolar spindle intermediates, resulting in unequal segregation of chromosomes (5). Thus mutations that impair cytokinesis and induce tolerance of the tetraploid state are likely to be carcinogenic. However, our knowledge of the roles played in cancer progression by pathways controlling telophase and by genetic mutations leading to tetraploidy is incomplete.
The polo family member Plk4 localizes to centrioles throughout the cell cycle and is essential for centriole duplication (6, 7). Plk4 null embryos arrest at day 7.5 post coitus and display dumbbell-shaped cells with anaphase bridges, indicating that Plk4 is also required in late mitosis, possibly at telophase (8). Plk4+/− embryos develop normally, but adult mice display a 16-fold increase in the incidence of spontaneous hepatomas compared with Plk4+/+ littermates. The wild-type Plk4 allele was retained in the liver tumors, indicating that Plk4 is a haploinsufficient tumor suppressor (9). Cell cycle progression was impaired in regenerating Plk4+/− liver with a ≈6-h delay in M phase completion, whereas multipolar spindle formation was increased 6-fold and p53 activation was impaired. These defects culminated in widespread disorganization of liver architecture and eventually generalized hepatic dysplasia in 100% of Plk4+/− animals after hepatectomy.
To investigate the mechanisms underlying Plk4 haploinsufficiency for tumorigenesis, we examined the cellular phenotypes of murine embryonic fibroblasts (MEFs) isolated from Plk4+/− embryos. Here we report a primary defect in cytokinesis in Plk4+/− MEFs associated with failure to appropriately localize the actomyosin contractile ring and failure to complete cleavage furrow ingression. We show that RhoGTPase activation in the Plk4+/− cells is impaired and identify a potential substrate for Plk4, the mitotic Rho GEF (guanine nucleotide exchange factor) Ect2. Defective cytokinesis resulted in multiple centrosomes, multinucleation, and a near-tetraploid karyotype, and with serial passaging immortalization in vitro, aneuploidy, and tumorigenesis in vivo. These Plk4+/− phenotypes were exaggerated in the absence of p53 function, a circumstance known to favor survival of tetraploid cells.
Results and Discussion
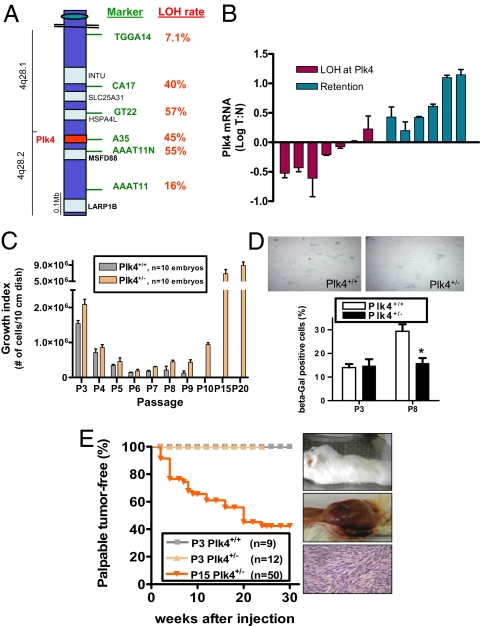
To determine the Plk4 status in human hepatocellular carcinoma (HCC), we queried six microsatellite markers within/near the Plk4 locus in 32 sequential cases resected and banked at our institution, looking for loss of heterozygosity (LOH) in microdissected tumor tissue compared with adjacent nonneoplastic liver. For the four markers closest to the Plk4 locus, the LOH rate averaged 50%, whereas at more distal sites the rate fell to background levels of 10–15% (Fig. 1A and Fig. S1A). By comparison, the majority of unrelated informative microsatellite markers show an LOH rate of <20% in human HCC (10). The present results, taken together with those of a small initial series (9), indicate that there is loss of one Plk4 allele in a significant proportion of established HCC. In five patients with multifocal hepatomas, each of the foci displayed LOH, in each case affecting the same allele (Fig. S1B) and therefore consistent with a field effect that confers predisposition to carcinogenesis. Furthermore, in three livers removed from transplant recipients, we observed LOH at the Plk4 locus in preneoplastic cirrhotic nodules, comparing these with adjacent nonnodular liver tissue. By contrast with the findings in HCC, the LOH rate at the Plk4 locus in pancreatic and colorectal adenocarcinomas (11–17%) was similar to background levels typical for unrelated microsatellite markers on 4q, which are ≈17% and ≈29%, respectively (11, 12) (Fig. S1 C and D). Reduced expression of Plk4 in microdissected liver tumor compared with adjacent nonneoplastic liver correlated with LOH (Fig. 1B). In addition, sequencing revealed no mutations in the remaining Plk4 allele in cases with LOH. Of note, in hepatomas with retention of heterozygosity at Plk4, Plk4 expression was increased on average 3-fold in tumor vs. adjacent nontumorous liver. This is in keeping with the increased Plk4 expression we previously observed in neoplastic compared with paired nonneoplastic tissues (13), further highlighting the significance of the decreased Plk4 expression in hepatoma cases with LOH. Thus, LOH at the Plk4 locus and reduced Plk4 expression may occur early in the process of human hepatocellular carcinogenesis and play a causative role in tumor initiation, consistent with the high rate of spontaneous hepatocellular cancer development in Plk4+/− mice.
Fig. 1.
Haploid levels of Plk4 are common in human hepatoma and drive cancer formation by murine cells. (A) Schematic representation of a region of the q arm of human chromosome 4, showing high rates of LOH at microsatellite markers closest to Plk4 in microdissected human hepatomas (n = 32 cases). All provisional, validated, and reviewed (the latter are bolded) genes are shown in black (University of California, Santa Cruz Gene Browser); none are recognized as or suggested to be tumor suppressors. (B) Decreased Plk4 mRNA levels in patients with LOH at the Plk4 locus, comparing tumor (T) in microdissected hepatoma samples with adjacent nonneoplastic liver (N) from the same patient. (C) Spontaneous immortalization of Plk4+/− MEFs (mean ± SEM). y axis indicates the total number of live cells present on a 10-cm dish 5 days after plating 3 × 105 cells. (D) Beta galactosidase staining (blue) in MEFs at passage 3 (P3) and P8 (Top), showing reduced senescence in P8 Plk4+/− cells (n = 3 lines/genotype, *P = 0.001 vs. P8 Plk4+/+). (E) P15 Plk4+/− MEFs formed tumors at the site of s.c. injection into NOD-SCID mice, whereas P3 wild-type and P3 Plk4+/− cells did not. A representative example of a high-grade tumor with spindle morphology is shown.
Primary MEFs were used to investigate the mechanism of Plk4 haploinsufficiency for tumor suppression. Plk4+/− MEFs became immortalized within 10–12 routine passages (19 of 20 lines; Fig. 1C). Under the same culture conditions, Plk4+/+ MEF lines (15 of 15) became senescent by passage 9 without the emergence of immortal colonies (Fig. 1D). Six of ten P15 Plk4+/− lines gave rise to palpable tumors when injected s.c. into nonobese diabetic/severe combined immunodeficient (NOD-SICD) mice (Fig. 1E), whereas P3 Plk4+/+ and P3 Plk4+/− MEFs were not tumorigenic (observed to 8 months). The P15 Plk4+/− derived tumors were grossly palpable after a latency period of 2–20 weeks and on histologic evaluation showed a high mitotic index, consistent with rapid proliferation (Fig. 1E).
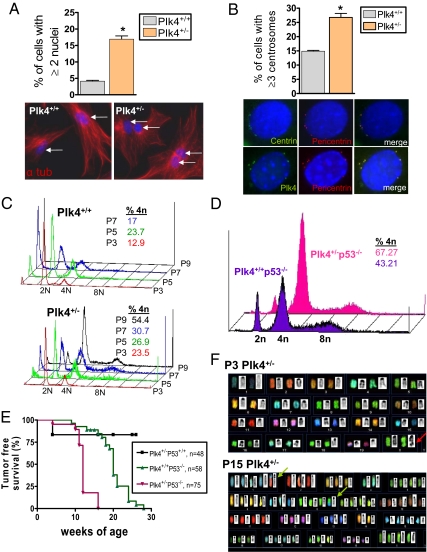
Even at early passage (P3), the incidence of multinucleation was higher in Plk4+/− than Plk4+/+ MEFs (Fig. 2A). In addition, a significantly greater fraction of P3 Plk4+/− than P3 Plk4+/+ MEFs contained three or more centrosomes, as identified by staining for centrin, pericentrin, γ-tubulin, and Plk4 itself (Fig. 2B and Fig. S2A). The supernumerary centrosomes were structurally intact by electron microscopy, and the majority stained positive for the mother centriole marker Cep-170 (14) (Fig. S2 A and B). FACS analysis showed increasing DNA content with serial passage of Plk4+/− MEFs, whereas Plk4+/+ MEFs remained largely diploid (Fig. 2C).
Fig. 2.
Plk4+/− MEFs show multinucleation, increased centrosome number, and aneuploidy. (A) Increased incidence of multinucleation in P3 Plk4+/− interphase MEFs (n = 400 cells per genotype; *P = 0.007). Representative photomicrographs of MEFs stained for DNA (blue) and α-tubulin (red). (B) Supernumerary centrosomes in P3 Plk4+/− interphase MEFs (n = 200 cells per genotype, *P = 0.014). Centrosomes stain positive for centrin (green), pericentrin (red), and Plk4 (green). (C) Increasing ploidy with increasing passage (P) number in Plk4+/− compared with Plk4+/+ MEFs. Plk4+/+ MEFs could not be examined past P7 because of senescence. (D) Increased ploidy in P3 Plk4+/− MEFs is exaggerated in p53 null background. (E) Plk4+/−p53−/− mice show accelerated development of spontaneous lymphomas and sarcomas compared with Plk4+/+p53−/− mice (P < 0.0001). (F) Representative spectral karyotype analyses showing diploidy and nonclonal structural change (red arrow) in P3 Plk4+/− MEFs and near tetraploidy and development of multiple clonal chromosomal rearrangements (green arrows), gains, and losses in late-passage Plk4+/− MEFs; 10 spreads were evaluated per embryo.
The checkpoint protein P53 plays a role in regulating centrosome duplication, and P53 haploinsufficiency in mice is accompanied by early conversion to aneuploidy and tumorigenesis (15, 16). As expected for the combination of haploid levels of two gene products that regulate a common end point, the incidence of supernumerary centrosomes and tetraploidy was exaggerated in Plk4+/−p53−/− compared with Plk4+/+p53−/− MEFS and with Plk4+/−p53+/+ MEFs, all at P3 (Fig. 2D and Fig. S2C). Furthermore, the development of palpable tumors (lymphomas and sarcomas) was accelerated in Plk4+/−p53−/− compared with Plk4+/+p53−/− mice (Fig. 2E). Spectral karyotype (SKY) analysis was performed to directly assess chromosomal instability. At P3, Plk4+/+ cells had the expected normal 2n karyotype, whereas Plk4+/− cells showed a mix of 2n and 4n ploidy, as well as a few nonclonal structural chromosomal aberrations (Fig. 2F and Table S1). By P15, Plk4+/− MEFs were predominantly near-tetraploid with clonal numeric and structural changes, consistent with the development of chromosomal instability, aneuploidy, and clonal selection (Fig. 2F and Tables S1 and S2). Interestingly, two of eight Plk4+/− MEF lines at P15 displayed a translocation or deletion at 4C4 that was associ-ated with loss of the cyclin-dependent kinase inhibitor/tumor suppressor p16 by FISH analysis (Figs. S3 A and B and S4A). After formation of palpable tumors by late-passage Plk4+/− MEFs, tumor cells that had been exposed to the in vivo environment revealed further clonal chromosomal rearrangements (Fig. S4B and Table S2). The 4C4 breakpoint was retained in vivo where it had been observed in cell culture.
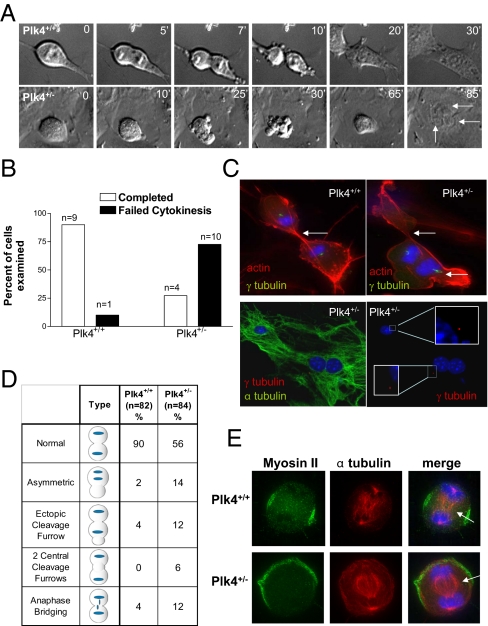
Nine of 10 P3 Plk4+/+ MEFs underwent normal cytokinesis, a process that took on average 15 min, starting from the time of cell rounding and ending at the time of flattening and loss of refractility (Fig. 3 A and B and Movie S1). By contrast, 10 of 14 mononuclear P3 Plk4+/− MEFs that initially established a bipolar spindle displayed a failure of cytokinesis, often with vigorous cortical membrane blebbing near the cellular equator (Fig. 3 A and B, Fig. S5 A–C, and Movie S2). Plk4+/− MEFs that underwent abortive cytokinesis subsequently reattached to the culture plate after ≈65 min, forming bi- and trinucleate cells (Fig. 3A and Fig. S5 B and C). The delay may reflect an ongoing attempt to correctly position the cleavage furrow, and could also represent enforcement of a post anaphase/pretelophase checkpoint, previously suggested to be an F-actin–dependent sensor of spindle and cleavage ring alignment (17). In transgenic P3 Plk4+/− MEFs expressing YFP-α-tubulin to label the mitotic spindle, more than half of mitotic cells that set up a bipolar spindle failed to complete cytokinesis (Fig. S5 A and B). Clustering of centrosomes at the spindle pole can allow establishment of a bipolar spindle even in cells with multiple centrosomes such as P3 Plk4+/− MEFs (9). However, we found that of 50 Plk4+/− MEFs undergoing aberrant cytokinesis, 23 (46%) had one centrosome at each pole (as shown in Fig. 3C, Bottom), ruling out the possibility that the observed defect in cytokinesis was secondary to the presence of supernumerary centrosomes.
Fig. 3.
Plk4 is required for myosin II localization and completion of cytokinesis. (A) Time-lapse DIC microscopy of mitotic P3 MEFs, showing failure of cytokinesis in a bipolar Plk4+/− cell, resulting in multinucleation (arrows indicate nuclei, time points are in minutes). (B) Proportion of bipolar mitotic P3 Plk4+/+ and Plk4+/− MEFs completing and failing cytokinesis. (C) (Upper) P3 Plk4+/+ and Plk4+/− MEFs in late mitosis stained for F-actin (red), γ-tubulin (green), and DNA (blue). White arrows indicate cleavage furrow formation. (Lower) P3 Plk4+/− MEF in late mitosis stained for DNA (blue) and α-tubulin (green) and/or γ-tubulin (red). (Insets) A single γ-tubulin–positive centrosome is seen at each pole. (D) Summary of mitotic phenotypes in P3 Plk4 MEFs. Plk4+/− MEFs frequently undergo asymmetric division and ectopic cleavage furrow formation. (E) Representative immunofluorescent images of MEFs in cytokinesis showing mislocalized myosin II in Plk4+/− cells (arrows indicate midbody location).
Transient placement of the primary cleavage furrow was typically asymmetric to the spindle midbody, an event that was followed by extensive cortical membrane blebbing (Fig. 3A and Fig. S5B). The cleavage furrow appeared to be rapidly forming, dissolving and reforming elsewhere, indicating an inability to establish the location of the cellular equator. Cleavage furrow positioning requires activation of RhoA at a specified cortical site, which promotes the formin-directed generation of F-actin filaments and the Rock/Citron-dependent phosphorylation and recruitment of myosin II (18). In logarithmically growing Plk4+/+ and Plk4+/− MEFs, phalloidin staining showed that Plk4 insufficiency disrupts the proper positioning of actin microfilaments at the cellular equator (Fig. 3C). Only cells with a bipolar spindle were examined in this and subsequent experiments, to avoid confounding effects of multiple centrosomes on actinomyosin organization. Whereas 90% of Plk4+/+ cells had a strong, equatorial distribution of cortical actin, 32% of bipolar Plk4+/− MEFs displayed an abnormal distribution of actin during cytokinesis (Fig. 3D). An increased fraction of Plk4+/− MEFs also demonstrated persistent intercellular anaphase bridging, indicating a failure to pinch-off and complete cleavage (Fig. 3D).
During cytokinesis, myosin II contracts against the equatorial F-actin microfilaments and generates the force required for membrane invagination at the cleavage furrow; localization of myosin II must be precisely maintained to effect complete cleavage. Plk4+/+ MEFs in anaphase and telophase consistently show a tight persistent equatorial localization of cortical myosin II, whereas in Plk4+/− cells myosin II is frequently poorly localized and labile (8 of 12 and 7 of 20 Plk4+/− cells examined in anaphase and telophase, respectively; Fig. S6 A and B), suggesting impaired RhoA-Rock/Citron-mediated localization. The observed blebbing phenotype was also consistent with an inability to retain myosin II at the equator, similar to Diaphanous deficiency (18). To confirm that the asymmetric division of bipolar P3 Plk4+/− MEFs was due to improper cleavage furrow placement rather than aberrant chromosome segregation, monastrol-synchronized MEFs were released into blebbistatin, a potent inhibitor of myosin II that blocks the contractile ring. Asymmetric DNA segregation was not observed, but myosin II localization was eccentric or ectopic in 40% of Plk4+/− MEFs compared with 10% of Plk4+/+ MEFs (n = 60 cells per genotype; Fig. 3E), indicating that asymmetric division is due to improper cleavage furrow formation. These results place Plk4 upstream of the RhoA-dependent regulation of cytokinesis.
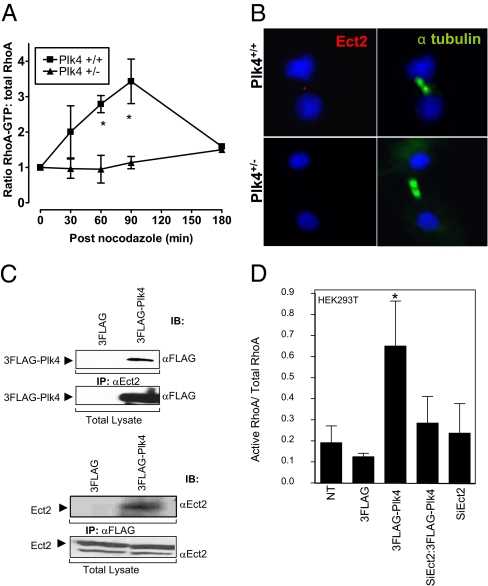
The centralspindlin complex (CSC) is composed of MKLP1 and CYK-4/MgcRacGAP, and mislocalization of these components phenocopies aspects of the defective actin and myosin II localization we observed here. Another polo family member, Plk1, localizes to the midbody and is required for proper CSC localization and subsequent execution of cytokinesis (19 –22). Plk1 localized normally in Plk4+/− cells, to the centrosomes in metaphase, the central spindle in anaphase, and the midbody in telophase, even in asymmetrically dividing cells (100% of cells examined, n = 11 cells per phase of mitosis; Fig. S7 A and B). Localization of the CSC component MKLP1 was also normal in nine of nine Plk4+/− MEFs examined in cytokinesis. Plk4 itself was observed at the midbody in 82% of Plk4+/+ MEFs but in only 10% of Plk4+/− cells (n = 11 and 20 cells respectively; P < 0.001; Fig. S7C). Haploid levels of Plk4 may be present at the midbody in Plk4+/− MEFs but below the limit of detection in this assay. Although CSC localization is appropriate, signaling from the central spindle to the cortex via the Ect2-RhoA-Rock/Citron cascade seems to be impaired in Plk4 heterozygous cells. Indeed, activated RhoA levels after release from a nocodazole-induced M-phase arrest remained low in Plk4+/− MEFs, whereas in wild-type cells they rose with the formation of the contractile ring (Fig. 4A). This failure of RhoA activation by 180 min was not attributable to a delay in establishing a bipolar spindle after release from nocodazole, because the fraction of cells that had done so was similar in wild-type and heterozygous cells examined at 30 min (53% vs. 47%, respectively), 60 min (75% vs. 67%), and 90 min (68% vs. 68%) after release (n = 40 cells per time point per genotype).
Fig. 4.
Plk4 is required for RhoA activation and Ect2 localization in cytokinesis. (A) Reduced levels of activated RhoA in Plk4+/− MEFs after release from nocodazole (mean ± SEM, n = 3, *P < 0.001 vs. heterozygous). (B) Representative images of MEFs in cytokinesis. Staining for Ect2 (red), α-tubulin (green), and DNA (blue) reveals Ect2 localization to the spindle midbody in Plk4+/+ but not Plk4+/− cells. (C) Reciprocal coimmunoprecipitation assay on HEK293T cells shows physical interaction of Flag-Plk4 with endogenous Ect2. (D) Ratio of GTP-bound/total RhoA in unsynchronized HEK293T cells transfected with 3Flag-Plk4, with or without prior depletion of Ect2 (siEct2), showing dependence of Plk4-induced RhoA activation on Ect2 (mean ± SEM, n = 2).
The RhoA GEF Ect2/Pebble undergoes a dramatic redistribution to the spindle in prometaphase and to the midbody in anaphase and telophase (23, 24). Ect2 localizes to the central spindle through direct association with CYK-4/MgcRacGAP (25, 26), in a Plk1-dependent manner (27, 28). Ect2 localized precisely to the midbody in 18 of 19 P3 Plk4+/+ MEFs in telophase but was absent from the midbody in 14 of 20 P3 Plk4+/− MEFs at the same stage (P < 0.001 vs. Plk4+/+; Fig. 4B). Thus, localization of Ect2 to the central spindle and subsequent RhoA activation to effect placement and stabilization of the cleavage furrow require Plk4. The defect in Ect2 localization in Plk4 heterozygous cells occurs despite normal Plk1 localization, suggesting that Plk4 acts downstream of Plk1 in the signaling cascade that culminates in RhoA activation.
Flag-Plk4 expressed in HEK293T cells coimmunoprecipitated with endogenous Ect2 (Fig. 4C). Ect2 contains four putative consensus sequence sites for phosphorylation by the Plk4 kinase domain (Fig. S8A) (29). We examined the ability of Plk4 kinase domain to phosphorylate Ect2 in an in vitro kinase assay (Fig. S8B). For this experiment, full-length Ect2 was incubated with Plk4 kinase domain in the presence of 33P-labeled γ-ATP. The resulting phospho blots revealed the presence of several phosphorylated proteins, including the Plk4 kinase domain itself. Four of the proteins were analyzed by MS and proved to be near full-length N-terminal fragments of Ect2, each of which contained the four consensus sequence sites. GTP-RhoA levels in unsynchronized P3 Plk4+/− MEFs were notably reduced compared with wild-type cells (Fig. S9A), and transfection of HEK293T cells with Flag-Plk4 increased activated RhoA levels (Fig. S9B), demonstrating that Plk4 is indeed an activator of RhoA. Furthermore, the activation of RhoA by Flag-Plk4 in HEK293T cells was Ect2 dependent (Fig. 4D), suggesting that the physical interaction between Plk4 and Ect2 is functionally relevant.
Here we have shown that Plk4 is required in a dose-dependent manner for Ect2 localization to the cleavage furrow, activation of RhoA, stable positioning of the actomyosin ring, and completion of cytokinesis. The error-prone events of mitosis in Plk4+/− cells correlate with high rates of tumorigenesis in vivo, similar in this regard to other haploinsufficient mitotic proteins such as MAD2 and 14-3-3sigma (30). An insufficiency in 14-3-3sigma disrupts the mTor-mediated switch from cap-dependent to cap-independent translation at G2/M, resulting in insufficient translation of mitotic proteins (30, 31). As with Plk4 haploinsufficiency, this leads to cytokinesis failure and tumorigenesis. Failure of cytokinesis yields tetraploidy, which predisposes to aneuploidy, reflecting an unstable genome (4). Early passage Plk4+/− MEFs are near-tetraploid and by P15 develop aneuploidy characterized by multiple large-scale deletions and translocations. In human hepatomas, LOH at the Plk4 locus occurs in 50% of cases, is seen in multiple separate early hepatomas from the same patient, and is accompanied by reduced Plk4 expression; this loss is not associated with mutation in the remaining Plk4 allele. These findings indicate that haploid levels of Plk4 cause chromosomal instability and constitute a significant factor in human hepatocellular carcinogenesis.
Materials and Methods
Human Tumor LOH and Expression Analysis.
Human hepatoma and nonneoplastic liver, gallbladder, pancreatic adenocarcinoma, and normal pancreas specimens were processed and microdissected as described by Ko et al. (9). The primer sequences for the additional microsatellite markers used here were as follows: AAAT11N, ACA GAG CGA AAC TCT GTC TC (forward) and GAA GAT CTT CAA AGC TGA AAG (reverse); A35, AGT CTA GGT GAT AGA GC (forward) and CAA ATA TTG TAA CAG AGC (reverse). The concentration of magnesium per reaction was 1.5 mM for AAAT11N and 1 mM for A35. The annealing temperature was 54 °C for AAAT11N and 53 °C for A35. The expected amplicon sizes of AAAT11N and A35 were 95 and 93 bp, respectively. Thirty-two cases were informative for TGGA14, GT22, AC17, and AAAT11, 15 for A35, and 12 for AAAT11N. This study was approved by the Mount Sinai Hospital Research Ethics Board.
For real-time RT-PCR analysis, microdissected hepatoma tissue was incubated with Proteinase K solution for 16 h and RNA isolated using standard TRIzol and chloroform methods. RNA pellet was stained using 2 μL of pellet paint (Novagen; 70748-3) to facilitate visualization. Random primed RT-PCR was performed with SuperScript II (Invitrogen). Reverse transcription was done for 60 min at 42 °C and terminated by heating to 70 °C for 15 min. Each sample was diluted to a volume of 12.5 μL and added to an equal volume of 2× SYBR green I Master Mix reagents (Applied Biosystems). The following primers were used for human Plk4: AAT CAA GCA CTC TCC AATC (forward) and TGT GTC CTT CTG CAA ATC (reverse). Forty-five cycles of denaturing (95 °C) and annealing (60 °C) were performed in an ABI-PRISM 7200 HT. Relative quantification was done using PBDG as a housekeeping gene. The 2-ΔΔCt method was used to calculate the relative expression in the tumor tissue compared with the paired nonneoplastic tissue.
Plk4 Mice and MEFs.
Plk4 mutant mice were generated on a 129Sv/CD1 background as described by Hudson et al. (8), backcrossed onto a CD1 background. Plk4+/− p53+/+ female mice (129Sv/CD1) were crossed with commercially available Plk4+/+ p53−/− male mice (JAX Mice, B6;129S2-Trp53tm1Tyj/J; Jackson Laboratory) to generate the Plk4-p53 compound mutant mice. All animal procedures were reviewed and approved by the Animal Care Committee, Samuel Lunenfeld Institute, Mount Sinai Hospital. MEFs were generated from 13.5-day-old embryos and cultured in DMEM plus 10% FCS at 37 °C in 5% CO2. To measure growth, passage-2 MEFs were thawed, plated, cultured overnight, washed, trypsinized, and counted using a hemocytometer. Cells (3 × 105) were plated in a 10-cm dish, cultured for 5 days, then washed, trypsinized, and counted; the total number of cells on the plate was calculated and recorded as the growth index at P3. The growth index for subsequent passages was similarly derived.
Tumorigenicity in Vivo.
Eight-week-old NOD-SCID mice were anesthetized with 0.5 mL of 1.25% Avertin (Sigma) by i.p. injection, injected in the scruff of the neck with 2.5 × 106 cells mixed with 600 μL of Matrigel (BD Basement Membrane Matrix, High Concentration), and observed for tumor development. Each embryo line was injected into a total of six mice.
Indirect Immunofluorescence.
Cells were washed and fixed with 0.5 mL of 4% paraformaldehyde for 20 min at room temperature and permeabilized with 0.5 mL of 0.5% Nonidet-P40 (Bioshop) for 15 min. Primary antibody was used for 2 h at room temperature or overnight at 4 °C. One hundred microliters of 3% BSA/PBS were used with the following primary antibody dilutions: α-tubulin, 1:200 (ab15426; Abcam); Plk4, 1:100 (ab2642; Abcam); Plk1, 1:100 (06-813; Millipore); Pericentrin, 1:200 (ab4448; Abcam); γ-tubulin, 1:200 (T6557; Sigma); nonmuscle myosin IIA, 1:200 (M8064; Sigma), Ect2 1:100 (H300; Santa Cruz Biotechnology). Corresponding conjugated secondary antibodies were added for 1 h at room temperature, along with Hoechst (1:400) and TRITC-phalloidin or FITC-phalloidin (1:800).
FACS Analysis.
At each passage, 2 × 105 cells were fixed for 24 h in ice-cold 80% ethanol for FACS analysis. Staining with propidium iodide (0.05 mg/mL) was done at room temperature for 1h. Fluorescence was measured using the FASCalibur cell sorting system (BD Biosciences). DNA content was analyzed using CellQuest (BD Biosciences) software. A gate for propidium iodide width was set to exclude cell doublets and triplets that could be confused with polyploid cells. Gated cells (200,000) were counted for each genotype, and percentage of cells with 2N, 4N, and 8N DNA content was expressed as a percentage of total gated cells. DNA content histograms are representative of two independent experiments using three separate lines of MEFs.
Spectral Karyotyping.
Plk4+/− MEF cultures were harvested after 4-h treatment with colcemid (0.05 μg/mL). Subsequently, cells were exposed to 0.075 M KCl hypotonic treatment for 20 min at 37 °C and then fixed in three changes of methanol/acetic acid in ratio 3:1. Slides were dropped from fixed cell suspension. The SKY mouse probe from Applied Spectral Imaging was hybridized to the slides according to the manufacturer's instructions. The SKY-metaphase images were captured using an SD 200 spectral bioimaging system (Applied Spectral Imaging) attached to an Axioplan 2 microscope (Zeiss). Captured metaphase images were analyzed using SKYView software version 1.6.2 (Applied Spectral Imaging). For each MEF line, 10 SKY-metaphase spreads were karyotyped using both spectral and inverted DAPI images.
Live Cell Imaging.
Plk4+/+ and Plk4+/− MEFs were grown overnight on delta T cell culture dishes (Bioptechs) at a density of 5 × 104 cells. The next day, dishes were rinsed with fresh DMEM plus 10% FCS and attached to the Delta T4 temperature controller (Bioptechs) set at 37 °C. Approximately 70% of mitotic Plk4+/− MEFs established a bipolar spindle; only bipolar cells were assessed in subsequent assays of cytokinesis. The cells were visualized by differential interference phase contrast (DIC) imaging using an Olympia Deconvolution Microscope. Rounded and refractile (t = 0) mitotic Plk4+/+ and Plk4+/− MEFs were observed as they proceeded through mitosis. DIC images were captured at 30-s intervals until cytokinesis was completed and the two daughter cells separated, or up to 90 min.
To observe in detail the process of cytokinesis, MEFs were synchronized by monstastrol-blebbistatin block, as described by Straight et al. (32). Briefly, Plk4+/+ and Plk4+/− MEFs were synchronized at G1/S by double thymidine block, washed with PBS, and released into 100 mM monastrol (Sigma) for 6 h to arrest cells in prometaphase. Cells were then released into 100 mM blebbistatin (Sigma) for 60 min to arrest in late mitosis, washed with PBS, fixed, and permeabilized.
Rho-GTP Binding Assays and Western Blotting.
Plk4+/+ and Plk4+/− cells at passage 3 were synchronized at the G1/S boundary by double thymidine block. Cells were released into 100 nM nocodazole for 24 h to synchronize in mitosis. Cells were harvested at 0, 30, 60, and 90 min after release from nocodazole block, and lysates were assayed for levels of activated Rho-GTP using Rhotekin-GST agarose according to the manufacturer's directions (Upstate). Four hundred micrograms of total protein lysate was incubated with 30 μg of Rhotekin Rho Binding Domain (RBD)-GST agarose for 1 h at 4 οC. Beads were washed with lysis buffer, and total protein lysate or RBD-GST pulldown was separated by SDS/PAGE. Membranes were probed with anti-RhoA (06-770; Upstate).
To assay the levels of activated Rho-GTP in unsynchronized (interphase) MEFs and HEK293T cells, cells were cultured for 24 h to 40% confluence. HEK293T cells were transfected with either 3Flag, RFP, 3Flag-Plk4, or RFP-Ect2 for 24 h. To determine the effect of Ect2 depletion, unsynchronized HEK293T cells were incubated for 48 h with or without an SiEct2 construct (Dharmacon), then transfected for an additional 24 h with 3Flag-Plk4. Rho GTPase activity was determined by RBD pulldown, as described above. Densitometry was performed using Fluor-S Max multiImager (BioRad Quantity One). Serum starvation and RhoA activation with 10 μM nocodazole were conducted as negative and positive controls, respectively, according to the manufacturer's recommendations (Cytoskeleton). Cells were harvested and lysed, and GTP-bound RhoA was extracted from the lysates by RBD pulldown, as described above. GTP-bound RhoA and total RhoA in whole-cell lysates were assayed using anti-RhoA antibody (Cytoskeleton). Blots were scanned and quantified using Fluor-S Max multiImager (BioRad Quantity One).
Coimmunoprecipitation of Flag-Plk4 and Endogenous Ect2.
HEK293T cells were transfected with pCMV:3Flag-Plk4 construct. Cells were harvested in lysis buffer [50 mM Tris-Cl (pH 7.5), 100 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 5 mM NaF, 10% glycerol, and 0.1% (wt/vol) Na deoxycholate] supplemented with mixture of protease and phosphatase inhibitors (Roche). Two micrograms of either Flag agarose beads (Sigma) or rabbit Ect2 antibody were added to 40 μg cleared total lysates and incubated for 1 h at 4°C. Complexes generated with Ect2 antibody were incubated with protein G Sepharose 4 Fast Flow (Amersham Pharmacia Biotech). Beads were washed, resolved through 10% SDS/PAGE gel, and electroblotted. Blots were incubated either with the mouse anti-Flag antibody (Sigma) 1:2,000 or rabbit Ect2 antibody (H300; Santa Cruz Biotechnology) 1:2,000.
In Vitro Kinase Assay.
GST-tagged kinase domain of human Plk4 (residues 1–390) and full-length Ect2 (residues 1–884) were each subcloned into modified pGEX-2T vectors (GE Healthcare), expressed in bacteria, and purified by affinity chromatography using Glutathione Sepharose (GE Healthcare). After cleavage and removal of the GST-tag with His-tagged TEV protease, the isolated Plk4 kinase domain and full-length Ect2 were subjected to subtractive Nickel column (Hi Trap Chelating HP column; GE Healthcare). Flow-through containing Plk4 and Ect2 proteins was concentrated and applied to a Superdex 75 gel filtration column, and both proteins were purified to homogenicity by elution on a sizing column. Five micrograms of Plk4 kinase domain were mixed with either 5 μg myelin basic protein or increasing amounts of Ect2 in kinase reaction buffer [20 mM Hepes (pH 7.5), 250 mM NaCl, 2 mM DTT, 20 mM MnCl2, 20 mM MgCl2, 50 mM NaF, 25 mM β-glycerophosphate, and 0.1 mM Na3VO3] and incubated with 33P-labeled γ-ATP at 30 °C for 15 min. Kinase reaction samples were resolved through 12% SDS/PAGE gel. Film was exposed to the dried gel and subsequently scanned by Storm phosphoimager (Molecular Dynamics). Cold reactions were run in parallel without 33P-labeled γ-ATP.
Supplementary Material
Acknowledgments
Supported by grants from the Canadian Institutes of Health Research (to C.J.S.) and the National Cancer Institute of Canada (to J.W.D.). Cep170 rabbit polyclonal antibodies were generously donated by Erich Nigg. We thank Peter E. Wu, Karina Pacholczyk, Catherine O'Brien, and Jorge Cabezas for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910941107/DCSupplemental.
References
- 1.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 2.King RW. When 2+2=5: The origins and fates of aneuploid and tetraploid cells. Biochim Biophys Acta. 2008;1786:4–14. doi: 10.1016/j.bbcan.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver BA, Cleveland DW. The role of aneuploidy in promoting and suppressing tumors. J Cell Biol. 2009;185:935–937. doi: 10.1083/jcb.200905098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 5.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 7.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Hudson JW, et al. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr Biol. 2001;11:441–446. doi: 10.1016/s0960-9822(01)00117-8. [DOI] [PubMed] [Google Scholar]
- 9.Ko MA, et al. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 10.Li SP, et al. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J Hepatol. 2001;34:840–849. doi: 10.1016/s0168-8278(01)00047-2. [DOI] [PubMed] [Google Scholar]
- 11.Jiang LX, et al. Tumor suppress genes screening analysis on 4q in sporadic colorectal carcinoma. World J Gastroenterol. 2008;14:5606–5611. doi: 10.3748/wjg.14.5606. discussion 5609–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seymour AB, et al. Allelotype of pancreatic adenocarcinoma. Cancer Res. 1994;54:2761–2764. [PubMed] [Google Scholar]
- 13.Macmillan JC, Hudson JW, Bull S, Dennis JW, Swallow CJ. Comparative expression of the mitotic regulators SAK and PLK in colorectal cancer. Ann Surg Oncol. 2001;8:729–740. doi: 10.1007/s10434-001-0729-6. [DOI] [PubMed] [Google Scholar]
- 14.Guarguaglini G, et al. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatachalam S, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: Reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey M, et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 17.Pardo M, Nurse P. Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science. 2003;300:1569–1574. doi: 10.1126/science.1084671. [DOI] [PubMed] [Google Scholar]
- 18.Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci USA. 2005;102:13473–13478. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Zhou T, Kuriyama R, Erikson RL. Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J Cell Sci. 2004;117:3233–3246. doi: 10.1242/jcs.01173. [DOI] [PubMed] [Google Scholar]
- 20.Neef R, et al. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- 21.Burkard ME, et al. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci USA. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santamaria A, et al. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol Biol Cell. 2007;18:4024–4036. doi: 10.1091/mbc.E07-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalamalasetty RB, Hummer S, Nigg EA, Sillje HH. Influence of human Ect2 depletion and overexpression on cleavage furrow formation and abscission. J Cell Sci. 2006;119:3008–3019. doi: 10.1242/jcs.03032. [DOI] [PubMed] [Google Scholar]
- 25.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon GC, et al. Sequential Cyk-4 binding to ECT2 and FIP3 regulates cleavage furrow ingression and abscission during cytokinesis. EMBO J. 2008;27:1791–1803. doi: 10.1038/emboj.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkard ME, et al. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 2009;7:e1000111. doi: 10.1371/journal.pbio.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe BA, Takaki T, Petronczki M, Glotzer M. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009;7:e1000110. doi: 10.1371/journal.pbio.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung GC, Ho CS, Blasutig IM, Murphy JM, Sicheri F. Determination of the Plk4/Sak consensus phosphorylation motif using peptide spots arrays. FEBS Lett. 2007;581:77–83. doi: 10.1016/j.febslet.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 30.Wilker EW, et al. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 31.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.