Abstract
Karrikins are a class of seed germination stimulants identified in smoke from wildfires. Microarray analysis of imbibed Arabidopsis thaliana seeds was performed to identify transcriptional responses to KAR1 before germination. A small set of genes that are regulated by KAR1, even when germination is prevented by the absence of gibberellin biosynthesis or light, were identified. Light-induced genes, putative HY5-binding targets, and ABRE-like promoter motifs were overrepresented among KAR1-up-regulated genes. KAR1 transiently induced the light signal transduction transcription factor genes HY5 and HYH. Germination of afterripened Arabidopsis seed was triggered at lower fluences of red light when treated with KAR1. Light-dependent cotyledon expansion and inhibition of hypocotyl elongation were enhanced in the presence of germination-active karrikins. HY5 is important for the Arabidopsis hypocotyl elongation, but not seed germination, response to karrikins. These results reveal a role for karrikins in priming light responses in the emerging seedling, and suggest that the influence of karrikins on postfire ecology may not be limited to germination recruitment.
Keywords: smoke, photomorphogenesis
Germination and seedling establishment are critical phases of the plant life cycle and are consequently sensitive to a variety of external cues, such as light, temperature, and nutrients. Fire events have a dramatic environmental impact that extends beyond immediate biotic destruction. The reduced competition for resources in the postfire environment creates an opportunity for seedling establishment that is taken advantage of by many species whose germination is recruited by either smoke or heat stimulation (1).
The reduction of vegetative canopy and ground cover that accompanies fire also gives rise to changes in the intensity and spectrum of solar radiation at the soil surface. Both light properties are highly influential on seed germination and plant development. The light-regulated control of germination is mediated by the phytochrome family of photoreceptors, primarily by PhyA and PhyB (2). Phytochrome proteins are converted between two isoforms by light: an inactive red (R) light absorbing form (Pr) and an active far-red (FR) light absorbing form (Pfr). The ratio of R:FR is influenced by canopy filtering, and soil type, moisture, and depth, which in turn affects the equilibrium of Pfr:Pr (3, 4).
Substantial work has begun to unravel the complex signal transduction networks that regulate specific plant developmental processes after light is perceived (5, 6). As the hy5 mutant exhibits photomorphogenesis defects under a wide range of light spectra, the bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) is considered a transducer of light signals from multiple photoreceptors (7–10). Its closest homolog, HYH, has a weaker, partially overlapping role that is evident primarily under blue light conditions (11). HY5 and HYH are both targeted for degradation in the dark by COP1 (11–13). ChIP-chip analysis recently revealed that 14.5% of Arabidopsis gene promoters and an estimated 60% of early phytochrome-induced gene promoters are putatively bound by HY5 (14). HY5 does not have a single role in transcription; its targets may be either activated or repressed in the hy5 mutant (14). HY5 acts with additional transcriptional regulators. For example, the double B-box zinc finger proteins STH2 and STH3/LZF1 physically interact with HY5 and act as positive regulators of light signaling during early seedling establishment (15–17). HY5 may serve as an important integration point for light and hormone signaling because it has been linked to auxin, cytokinin, and abscisic acid (ABA) responses; and gibberellin (GA) represses photomorphogenesis in darkness via COP1 by decreasing HY5 protein accumulation (18–22).
Karrikins (KAR) have been established as a new class of signaling molecules in smoke from burning vegetation that trigger seed germination for many angiosperms (23–26). Although karrikins can overcome primary dormancy (PD) in Arabidopsis thaliana seed, the effectiveness of KAR1 on germination varies by ecotype and seed dormancy state: highly dormant seed has reduced response, whereas the rapid germination rates of nondormant seed can mask any positive influence from karrikin (25). KAR1 does not recover germination of the ga1-3 mutant, which has no detectable GA biosynthesis, or enhance its sensitivity to exogenous GAs. Active karrikins induce transcript levels of GA biosynthetic enzymes GA3ox1 and GA3ox2, but there is no corresponding influence on either GA4 or ABA levels during the first 48 h of imbibition. Unlike GAs, karrikins cannot replace a light requirement for germination (25). Thus, although karrikins require GA biosynthesis to influence germination, it is unlikely that karrikin responses are exclusively GA-mediated. Here, we report an investigation into molecular responses to KAR1 that ultimately revealed crosstalk with light-dependent processes during germination and seedling establishment.
Results
Identification of KAR1-Responsive Genes.
To identify molecular responses to karrikin before germination, we used Affymetrix ATH1 microarrays to examine transcriptional profiles of the PD seed of the Landsberg erecta (Ler) ecotype imbibed for 24 h in water or 1 μM KAR1. This time point is well before completion of germination, because radicle protrusion was not observed in KAR1-treated seeds at 48 h imbibition and had reached only ∼13% in KAR1-treated seeds by 72 h (Fig. S1). It was previously demonstrated that germination of the GA-deficient ga1-3 mutant, which cannot germinate in the absence of exogenous GAs or other hormone perturbations, is not recovered by KAR1 (25). To identify transcriptional changes specific to KAR1 treatment rather than general germination processes, we also performed an analysis of ga1-3 seed under the same conditions.
We first identified transcriptional changes that were consistent between both genotypes (Criteria I; Table S1) and then, out of the remaining genes, those that were significantly responsive to KAR1 in only one genotype (Criteria II; Table S2). A few general observations can be made from the Criteria I probesets. First, KAR1 induced ∼6-fold more transcripts at 24 h than it repressed (Table 1). Second, although the KAR1 treatment substantially enhanced PD seed germinability, few “strong” fold changes were observed. Only 30 transcripts were induced at least 2-fold by KAR1, and only one was repressed at least 2-fold (Table 1). Third, the magnitude of transcript induction in response to KAR1 was largely consistent in PD Ler and ga1-3 seed, despite the disparity in germination outcome between these genotypes. Of the 118 probesets induced > 1.5-fold by KAR1 in PD Ler, 85 (75%) also had FC > 1.5 in ga1-3. Of the 10 most up-regulated transcripts in PD Ler seed, 7 were also among the 10 most up-regulated in ga1-3. This trend was less notable for down-regulated transcripts, because only 4 probesets (20%) were repressed at least 1.5-fold by KAR1 in both genotypes.
Table 1.
Summary of transcript changes induced in imbibed seed by KAR1
| Genotype | Criteria I | Criteria II | |
| Induced | PD Ler | 118 (30) | 13 (2) |
| KAR1/H2O ≥ 1.5 (2.0) | ga1-3 | 109 (29) | 18 (3) |
| Repressed | PD Ler | 15 (1) | 11 (0) |
| KAR1/H2O ≤ −1.5 (−2.0) | ga1-3 | 23 (1) | 7 (0) |
K/W, KAR1/water.
Thus, despite the lack of an overt ga1-3 germination response to KAR1, a common set of putative KAR1-responsive genes was identified. Because many more transcripts were significantly induced than repressed, and there was more consistency between induced gene sets in PD Ler and ga1-3, we chose to focus further analysis on the 118 Criteria I up-regulated probesets in PD Ler, corresponding to 121 genes, hereafter referred to as the KAR-UP set.
KAR-UP Gene Set Is Enriched for Light-Responsive Transcripts.
To determine whether KAR1 induces similar transcriptional effects as other treatments influencing germination, we performed multiple comparisons to previously published microarray datasets examining light-, dormancy-, and hormone-responsive genes (14, 27–39). These results are summarized in Table S3 and S4, and the most striking findings are discussed here.
We noted that HY5 and its homolog HYH were significantly up-regulated by KAR1. Comparison of the KAR-UP genes to the Lee et al. (14) ChIP-chip results for HY5 revealed 64 (54%) putative HY5 targets. This represents a significant, >3-fold enrichment of potential HY5 targets among KAR1-induced genes.
We next examined whether early light-responsive genes were overrepresented among the KAR-UP transcripts. Tepperman et al. (36) identified sets of genes responding to continuous red light (Rc) within 1 h of light exposure in 4-d-etiolated seedlings. 16 KAR-UP genes were present among the 206 robustly up-regulated light-responsive transcripts (∼1% of ATH1 array). Inclusion of the less strongly light-responsive genes in the comparison indicated that 31% of KAR-UP genes were induced by red light (vs. 5%). Among the red light-induced genes, 29 of the 37 (78%) promoters were putatively bound by HY5. In contrast, only 5% of KAR-UP genes were present in the Rc down-regulated list, which was not significantly different from the 2% prevalence of those genes. Consistent with the predominance of PhyA in early light responses, the majority of the Rc-induced KAR-UP genes were classified as PhyA-responsive. A comparison with 405 genes (2% of ATH1 array) induced in light-grown seedlings 1 h after UV-B exposure revealed 31 genes (26% of KAR-UP) in common (35). Thus, there is significant enrichment of both early red and UV-B light-induced genes among KAR-UP transcripts.
PIL5 mediates repression of germination in the dark. ChIP-chip was recently used to identify PIL5 binding sites (34). Among KAR-UP genes, 12% had PIL5 binding sites (vs. 3%of genome). Corresponding with this, 21% of KAR-UP genes were classified as repressed by PIL5 (vs. 6%). Thus a >3.5-fold enrichment of PIL5-regulated genes was observed among those induced by KAR1.
Finally, we performed an analysis for enriched transcription factor binding sites among promoters (maximum 1 kb) of KAR-UP genes using Athena (40). An Abscisic Acid Response Element (ABRE)-like binding site motif, (C/G/T)ACGTG(G/T)(A/C), was present in the promoters of 52 (43%) of the 121 genes represented by the 118 probesets. This motif is significantly enriched among the KAR-UP genes when compared against the genome-wide distribution of 18%. The ABRE-like binding site motif includes motifs such as ABA-responsive element (ABRE), (C/T)ACGTGGC, and the light-responsive element G-box, CACGTG. HY5 has been demonstrated to bind G-box motifs in the promoters of some light-regulated genes (7). Among KAR-UP genes, 35 of the 52 (67%) promoters containing ABRE-like binding site motifs were previously classified as HY5-binding promoters (14).
Expression Patterns of KAR1-Responsive Transcripts.
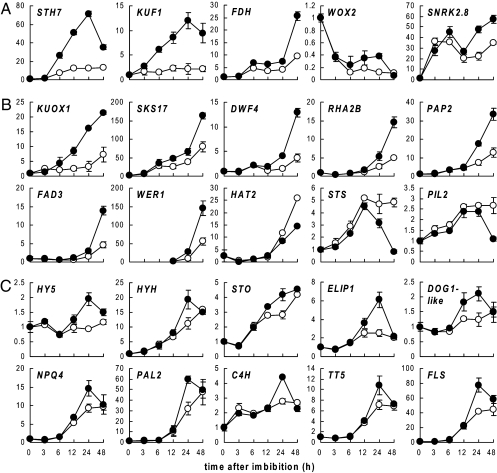
Transcript abundance of 23 Criteria I and two Criteria II (DWF4, WER1) genes was reexamined by qRT-PCR during a 48-h time course of seed imbibition. Two KAR-UP genes encoding a putative F-box protein (At1g31350) and an oxidoreductase (At5g07480) are previously uncharacterized and here referred to as KUF1 and KUOX1. Three general transcriptional response patterns to KAR1 were identified: early, late, and transient. Early-responsive genes (STH7, KUF1, FDH, WOX2, SNRK2.8) were induced within 6 h of imbibition (Fig. 1A). Late-responsive genes (KUOX1, SKS17, DWF4, RHA2b, PAP2, FAD3, WER1, HAT2, STS, PIL2) feature a sustained KAR1 response that begins around 12–24 h and continues through 48 h (Fig. 1B). Finally, transient response genes (HY5, HYH, STO, ELIP1, DOG1-like, NPQ4, PAL2, C4H, TT5, FLS) exhibit only a temporary induction by KAR1 that peaks at 24 h imbibition (Fig. 1C). The last four genes in this group are involved in phenylpropanoid or flavonoid biosynthesis.
Fig. 1.
Expression of KAR1-responsive genes during seed imbibition. Three general transcriptional responses to KAR1 were observed: early (A), late (B), and transient (C). Relative expression was assayed over a 48-h time course by qRT-PCR on total RNA from PD Ler seed imbibed in water-agar (open circles) or 1 μM KAR1 (filled circles) in continuous white light at 20 °C. Mean ± SEM, n = 2.
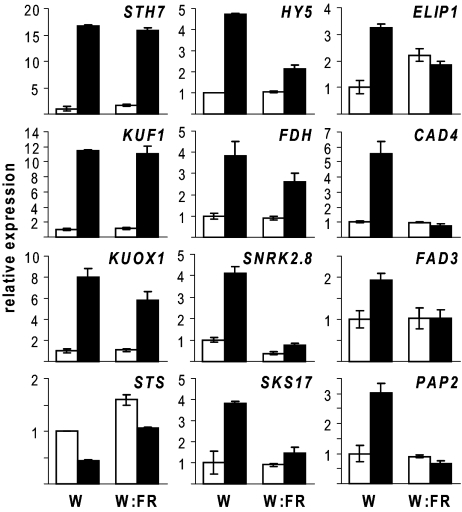
Because karrikins cannot replace a light requirement for germination in Arabidopsis, we next examined whether light was required for transcriptional changes of 12 KAR1-responsive genes in PD Ler seed at 24 h (Fig. 2). STH7 and KUF1 were strongly up-regulated by KAR1 independently of light. KUOX1, STS, HY5, FDH, SNRK2.8, and SKS17 transcripts had enhanced KAR1 responsiveness in seeds given a white light treatment. A FR light pulse abolished transcript induction by KAR1 for ELIP1, CAD4, FAD3, and PAP2. Notably, ELIP1, CAD4, and FAD3 are described as HY5 binding targets (14). Thus, although light is not required for all transcriptional responses to KAR1, it can have a synergistic relationship.
Fig. 2.
Influence of light on KAR1-responsive transcripts. PD Ler seed were imbibed for 2 h under white light (W) on water-agar (open bars) or 1 μM KAR1 (filled bars), then placed into darkness for 22 h. A subset (W:FR) were treated with 5 min of FR (6 μE) light to deactivate Pfr before dark transfer. Relative expression was assayed by qRT-PCR after 24 h imbibition. W water control expression for each gene was set to 1, and other values were scaled accordingly. Mean ± SEM, n = 2.
KAR1 Enhances Arabidopsis Seed Germination Under Low Light Fluences.
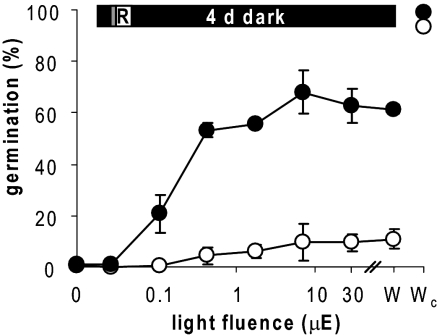
The enrichment of light-responsive genes among KAR1-induced transcripts suggested that karrikin may affect developmental responses to light. We examined the Arabidopsis seed germination response at a range of fluences in the presence and absence of KAR1. We used nondormant seed to minimize the KAR1 influence on germination via dormancy and focus on the effect of karrikin on light sensitivity. Seeds were imbibed for 1 h in darkness, given a 5′ pulse of FR to deactivate any Pfr, treated with 1 h of R light at various fluences, and germination recorded after a further 96 h of darkness. Germination was markedly increased by KAR1 at low R fluences. In keeping with prior observations (25), KAR1 could not induce germination in the absence of a R light pulse, and germination was similar to the control for seeds kept in continuous white light instead of the extended dark incubation (Fig. 3). PhyB is present in the seed, whereas PhyA is synthesized during the 24- to 96-h period of imbibition (41–43). To consider the contribution of both phytochromes to light responsiveness, we also performed the experiment with a 48-h dark period between the FR and R pulses (Fig. S2). Similar results were obtained, although with increased germination across all fluences of R light.
Fig. 3.
KAR1 enhances light-dependent germination of Arabidopsis seed. Afterripened (7 months) seed was imbibed on water-agar (open circles) or 1 μM KAR1 (filled circles) in the dark for 1 h, exposed to 5 min FR (6 μE), then given 1 h R light of the indicated intensity. Germination was assessed after a further 4-d dark incubation. Seeds treated with a white light (100 μE) pulse (W) or continuous white light for 4 d (Wc) are shown for comparison. Mean ± SD, n = 3, >50 seeds per sample.
Karrikins Positively Influence Photomorphogenesis in Seedlings.
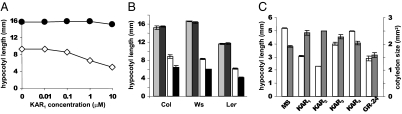
To determine whether karrikins can influence other light-responsive processes, we investigated the effect on KAR1 on Arabidopsis hypocotyl elongation. To reduce any potential growth contribution from KAR1 effects on seed dormancy, we used afterripened seed that was stratified after plating on media supplemented with nitrates. At 100 nM and higher concentrations, KAR1 inhibited hypocotyl elongation under continuous red (Rc) light (Fig. 4A). In darkness, however, KAR1 had no effect on hypocotyl elongation. It is notable that seedlings of the Col and Ws ecotypes responded as strongly to KAR1 as Ler, because in our hands Col seeds have little to no dormancy and hence only a minimal KAR1 germination response (Fig. 4B). Altogether, this suggested that the seedling growth response was independent of KAR1 effects on germination rates.
Fig. 4.
Promotion of de-etiolation by active karrikins. (A) Ler seedlings grown on indicated concentrations of KAR1 in Rc (open diamonds) or darkness (filled circles). (B) Hypocotyl elongation of three Arabidopsis ecotypes grown on 0.5× MS or 1 μM KAR1 in darkness (light and dark gray bars, respectively) or Rc (white and black bars, respectively). (C) Ler seedlings grown in Rc on 0.5× MS media supplemented with 1 μM KAR1, KAR2, KAR3, KAR4, or GR-24. Hypocotyl length (open bars) and total upper cotyledon surface area (filled bars). Mean ± SEM, n = 3, ≥14 hypocotyls per sample or ≥10 cotyledon pairs per sample. Where not visible, error bars are smaller than the symbol.
Further tests revealed that all active karrikins inhibit Arabidopsis hypocotyl elongation in a light-dependent manner, and the degree of inhibition corresponds with the relative effectiveness of each karrikin as a germination stimulant: KAR2 > KAR1 > KAR3 (Fig. 4C). KAR4, which was previously found to be inactive in promotion of Arabidopsis germination (25), did not significantly affect hypocotyl length. At the concentrations tested, no growth inhibition in darkness was observed for any of the karrikins. The active karrikins also promoted expansion of cotyledon surface area by ∼20–30% under Rc, whereas KAR4 did not. Interestingly, the synthetic strigolactone GR-24 was also an effective inhibitor of hypocotyl elongation, although it was previously reported as a weaker germination stimulant than active karrikins in Arabidopsis (Fig. 4C) (25). However, the effect of GR-24 on seedlings was distinct from karrikins, because cotyledon expansion was inhibited. We examined the hypocotyl elongation response in the presence of 1 μM KAR1 at a range of R and B+FR light fluences (Fig. S3 A and B). KAR1 reduced hypocotyl elongation under all light fluences tested.
To determine whether the light-dependent inhibition of hypocotyl elongation by karrikins occurs in other species, we tested the responses of lettuce (Lactuca sativa L. cv Grand Rapids) and wild turnip (Brassica tournefortii). Germination of these species is strongly activated by KAR1 but is oppositely influenced by light; lettuce requires only a very low light exposure to induce germination, whereas wild turnip germination is strongly inhibited by light. Both species displayed a similar enhancement of postgerminative light response by KAR1 as Arabidopsis (Fig. S3 C and D). However, hypocotyl elongation in the dark was inhibited at 10 μM KAR1 in lettuce and at all tested concentrations in the dark for wild turnip. This suggests that karrikin may induce growth effects even in the absence of light in some highly sensitive species.
Karrikins enhanced visible greening in the apical portion of the Arabidopsis hypocotyl (Fig. S3 E and F). Chlorophyll a and b content was increased by ∼15–20% in KAR1-treated seedlings of three tested ecotypes, which may be a direct consequence of increased cotyledon expansion (Fig. S3G).
HY5 Has a Role in the Hypocotyl Elongation Response to Karrikins.
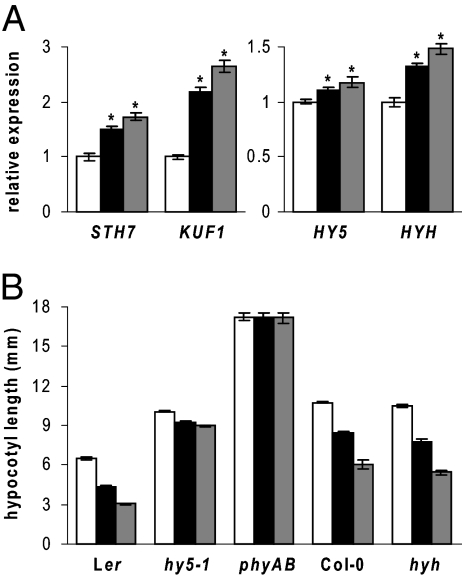
As HY5 targets are enriched among genes induced by karrikins after 24 h imbibition, and HY5 transcript is also transiently up-regulated by KAR1 in a light-enhanced manner (Figs. 1C and 2), we investigated the role of HY5 in seed responses to karrikins. Although hy5-1 seed exhibited reduced dormancy, consistent with a recent report (44), its germination was significantly responsive to karrikins (Fig. S4 A and B). The early karrikin-responsive transcripts STH7 and KUF1 were still inducible by KAR1 and KAR2 in the hy5-1 mutant, regardless of light treatment (Fig. S4 C and D). However, ELIP1, a putative HY5 target gene that requires light for significant induction by karrikins, was no longer positively responsive to karrikins in 24-h-imbibed hy5-1 seed (Fig. S4E). Thus, HY5 is not required for all karrikin responses in seed.
Some markers of karrikin response in 24-h-imbibed seed maintained expression changes postgermination. In seedlings grown 3 d under Rc, STH7 and KUF1 transcripts were still up-regulated by karrikins, and expression was weakly enhanced 11–17% for HY5 and 33–48% for HYH by KAR1 and KAR2, respectively (Fig. 5A). Because HY5 has been well established as an important regulator of deetiolation, we also examined light-dependent hypocotyl elongation responses to karrikin in hy5-1. In wild type, 1 μM KAR1 and KAR2 inhibited hypocotyl elongation under Rc by 33% and 53%, respectively. However, hypocotyl growth was only inhibited 10% by these karrikins in hy5-1 (Fig. 5B). A hyh mutant retained normal hypocotyl elongation responses to KAR1 and KAR2. Consistent with the absence of karrikin effects on dark-grown hypocotyls, the red light-insensitive phyA-201 phyB-5 mutant had no growth response to karrikins under Rc (Fig. 5B).
Fig. 5.
Hypocotyl elongation response to karrikins is reduced in hy5-1. (A) Relative expression assayed by qRT-PCR of STH7, KUF1, HY5, and HYH in Ler seedlings grown under Rc for 3 d as described for hypocotyl elongation assays. 0.5× MS control for each gene is scaled to 1. Mean ± SEM, n ≥ 4 biological replicates per treatment; *, P < 0.01, two-tailed t test. (B) Rc hypocotyl elongation response of Ler, hy5-1, phyA-201 phyB-5, Col, and hyh. Mean ± SEM, n = 3, ≥12 hypocotyls per sample. For (A) and (B), growth was on 0.5× MS (white), 1 μM KAR1 (black), or 1 μM KAR2 (gray).
Discussion
KAR1 is an effective germination stimulant for PD Ler seed in the presence of light. Despite producing a substantial difference in germination fate, our microarray analysis revealed that remarkably few genes (∼0.5%) were transcriptionally affected by KAR1 >1.5 fold after 24 h imbibition. Gene expression responses to KAR1 were detected for several genes as early as 6 h after the start of imbibition, consistent with the timing of a prior observation of KAR1 effects on GA3ox1 expression (Fig. 1). It is notable that many of the genes induced by KAR1 in PD Ler are comparably up-regulated in ga1-3, although ga1-3 does not complete germination. This suggests that although GA is ultimately required for KAR1 to stimulate germination, GA is not necessary for karrikin perception and early transcriptional responses during imbibition.
Although limited in size, the small set of genes we have identified with strong responses to KAR1 can be useful tools in future work. As markers of karrikin response, expression of these genes can be used to characterize putative karrikin-insensitive mutants identified through forward genetic screens. These markers can also be examined for interaction between transcriptional responses to karrikin and other stimuli, as in Fig. 2. Finally, genes that show an early and strong transcriptional response to karrikin are attractive candidates for reverse genetic investigations of potential roles in karrikin function. For example, STH7 was the most highly KAR1-induced transcript in all microarray experiments and is up-regulated by KAR1 within 6 h of imbibition. STH7 is a member of a small subfamily of eight double B-box zinc finger genes that are structurally related to the CONSTANS-LIKE family of transcription factors but lack the C-terminal CCT motif. Although STH7 has not been characterized, other subfamily members, including STH2, STH3/LZF1/DBB3, STO, and STH, have been implicated in light-dependent developmental processes (15–17, 45). Furthermore, STH2 and LZF1, the closest homologs of STH7, physically interact with HY5 and play positive roles in light-induced postgerminative processes.
In our analysis of karrikin up-regulated transcripts, we found a strong connection to light signaling. Putative HY5 binding sites and genes induced during early red light responses were overrepresented among genes responding positively to KAR1. These results led us to investigate the influence of karrikin on light-dependent processes. KAR1 enhanced light-dependent germination of Arabidopsis thaliana at a range of low fluences (Fig. 3). With longer imbibition times before the red light pulse, seeds became more sensitive to light treatment and the KAR1 treatment was even more effective (Fig. S2). One interpretation of these results is that karrikin promotes light sensitivity in seed germination. This leads to the attractive hypothesis that karrikins recruit germination in the seed bank by enhancing light perception; that is, seeds at greater depths in the soil will respond to the very low fluences of light they are receiving, or seeds will be more responsive to brief light exposures during soil disturbance. Although this may indeed be the case for some light-requiring species, this is not likely to be a broad karrikin mechanism. First, light levels attenuate very rapidly within millimeters of soil depth, and seeds in the upper soil layer where light can penetrate will also be most susceptible to fire damage. Second, not all KAR-responsive species are light-requiring, and germination of some species is even inhibited by light. It could be argued that karrikin response is a result of convergent evolution and has been integrated into germination pathways in different ways across angiosperms. However, Arabidopsis thaliana and Brassica tournefortii, both members of the Brassicacaeae, respond positively to karrikin and yet oppositely to light in germination. Thus, at least for these species, a more parsimonious hypothesis calls for a common karrikin response mechanism in which karrikins increase germination potential and light acts independently to modulate the karrikin effect.
Our examination of karrikin effects on postgerminative growth revealed that karrikins enhance light-dependent processes such as inhibition of hypocotyl elongation and expansion of cotyledons. The influence of KAR1 on Arabidopsis hypocotyl elongation required light and was effective across a range of fluences, consistent with enhanced light sensitivity rather than nonspecific hypocotyl growth inhibition. As in germination, KAR2 was the most effective karrikin. Hypocotyl elongation of lettuce and wild turnip was inhibited by KAR1, suggesting that, like germination, this response to karrikins may also be prevalent. It will be interesting to determine whether karrikins confer a selective advantage by preparing seedlings for emergence under the light conditions of the postfire environment.
Light is required for overt germination and hypocotyl growth responses to karrikins in Arabidopsis, but not for all transcriptional effects (Figs. 2, 3, and 4B). Because HY5 is a major component of light signal transduction, putatively binds 54% of KAR-UP genes, and is itself transcriptionally induced by karrikins, we propose that it has an important role in carrying out some aspects of the karrikin response. Indeed, inhibition of hypocotyl elongation under red light by karrikins was strongly attenuated in hy5-1 (Fig. 5B). As was exemplified by ELIP1 misregulation in the hy5-1 mutant (Fig. S4E), we expect that the loss of this broadly active transcription factor will affect the inducibility of many karrikin-responsive transcripts after 24 h imbibition. However, it is also clear that HY5 is not required for all karrikin responses, because germination and induction of STH7 and KUF1 transcripts retained significant responses in hy5-1 seed (Fig. S4). We conclude that karrikin perception and early signal transduction can occur independently of light or HY5, but must ultimately integrate with light-activated signaling pathways to produce developmental changes.
The influence of karrikins on postgermination light responses that we have described here indicates a broader importance for karrikins in plant development and suggests an intriguing ecological role beyond germination recruitment in the postfire environment.
Methods
Growth of Arabidopsis thaliana, seed storage, and surface sterilization were performed as described in ref. 25. hyh mutant was isolated from the WiscDsLox253D10 insertion line. No expression was detectable by RT-PCR using primers for full-length HYH cDNA.
Expression Analysis.
cRNA targets were prepared from total RNA using the GeneChip One-Cycle Target Labeling Kit (Affymetrix) and hybridized to GeneChip Arabidopsis ATH1 Genome Arrays (Affymetrix) according to the manufacturer's instructions. Signal intensities were GC-RMA-normalized using Avadis software (Strand Life Sciences). Statistical analysis and identification of karrikin-responsive genes was performed as described in SI Methods. For seed analysis, RNA isolation, cDNA synthesis, and qRT-PCR were performed as described in ref. 25. Two independent seed batches were examined with at least two technical replicates of each PCR. The average relative expression (vs. At5g46630, a clathrin adaptor complex subunit) and SE for the two biological replicates are shown. Levels of relative expression in dry seeds or the water control were adjusted to 1 when detectable, and all other values were scaled accordingly. RNA was extracted from 3-d-Rc seedlings using Qiagen RNeasy Plant Mini Kit. Primer pairs (Table S5) were designed by AtRTPrimer (46).
Lighting.
Arrays of R and FR light-emitting diodes (L660-04U, L735-04AU; Epitex) supplied R/FR light treatments. White fluorescent light (NEC Cool White) passed through Lee #120 (deep blue) filter, provided blue and FR (B+FR) light of approximately equal intensities. Light spectrum and intensity was measured with a Warsash Scientific EPP2000 spectrometer with irradiance calibration. Neutral density filters (Lee #210, #211) were used to produce a range of light fluences.
Light Response Assays.
Arabidopsis seeds were surface-sterilized and plated onto solid 0.5× Murashige & Skoog (MS)/2.5 mM Mes (pH 5.7) media supplemented with indicated concentrations of karrikins. In experiments comparing multiple karrikins and GR-24, 1,000× MeOH stocks were used. Otherwise, an aqueous KAR1 or KAR2 stock was used. After plating, seeds were stratified (dark, 4 °C, 3 d), exposed to 3 h of white fluorescent light (100 μE) at 20 °C to initiate germination, and wrapped in foil for 21 h at 20 °C. Plates were transferred to indicated light condition for 4 d before seedling image capture and hypocotyl measurement using ImageJ (National Institutes of Health). Unless noted otherwise, Rc intensity was 28 μE. Total upper surface area of both cotyledons was measured in ImageJ. Chlorophyll was extracted from seedlings in 2.5 mM phosphate buffered 80% acetone (pH 8), and concentration was determined by absorption spectroscopy (47).
Supplementary Material
Acknowledgments
We thank Kristen Feher for statistics consultation and the Arabidopsis Biological Resource Center for Arabidopsis thaliana seed. This work was supported by Australian Research Council Grants FF0457721 and DP0667197 and the Centres of Excellence program of the Government of Western Australia.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE20556).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911635107/DCSupplemental.
References
- 1.Van Staden J, Brown NAC, Jäger AK, Johnson TA. Smoke as a germination cue. Plant Species Biol. 2000;15:167–178. [Google Scholar]
- 2.Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- 3.Mandoli DF, Ford GA, Waldron LJ, Nemson JA, Briggs WR. Some spectral properties of several soil types: implications for photomorphogenesis. Plant Cell Environ. 1990;13:287–294. [Google Scholar]
- 4.Pons TL. Seed responses to light. In: Fenner M, editor. Seeds, the Ecology of Regeneration in Plant Communities. 2nd ed. New York, NY: CABI; 2000. [Google Scholar]
- 5.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 6.Franklin KA, Larner VS, Whitelam GC. The signal transducing photoreceptors of plants. Int J Dev Biol. 2005;49:653–664. doi: 10.1387/ijdb.051989kf. [DOI] [PubMed] [Google Scholar]
- 7.Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang LH, et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 9.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 13.Hardtke CS, et al. HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta S, et al. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20:2324–2338. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19:3242–3255. doi: 10.1105/tpc.107.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CS, et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008;54:205–219. doi: 10.1111/j.1365-313X.2008.03401.x. [DOI] [PubMed] [Google Scholar]
- 18.Vandenbussche F, et al. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 2007;49:428–441. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- 19.Sibout R, et al. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2006;2:1898–1911. doi: 10.1371/journal.pgen.0020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, et al. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA. 2008;105:4495–4500. doi: 10.1073/pnas.0710778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alabadi D, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- 23.Chiwocha SDS, et al. Karrikins: a new family of plant growth regulators in smoke. Plant Sci. 2009;177:252–256. [Google Scholar]
- 24.Light ME, Daws MI, Van Staden J. Smoke-derived butenolide: towards understanding its biological effects. S Afr J Bot. 2009;75:1–7. [Google Scholar]
- 25.Nelson DC, et al. Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 2009;149:863–873. doi: 10.1104/pp.108.131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. A compound from smoke that promotes seed germination. Science. 2004;305:977. doi: 10.1126/science.1099944. [DOI] [PubMed] [Google Scholar]
- 27.Bassel GW, Zielinska E, Mullen RT, Bewley JD. Down-regulation of DELLA genes is not essential for germination of tomato, soybean, and Arabidopsis seeds. Plant Physiol. 2004;136:2782–2789. doi: 10.1104/pp.103.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 29.Cao D, Cheng H, Wu W, Soo HM, Peng J. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 2006;142:509–525. doi: 10.1104/pp.106.082289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrera E, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrera E, et al. Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol. 2007;143:1669–1679. doi: 10.1104/pp.107.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa M, et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tepperman JM, Hwang YS, Quail PH. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- 37.Yamauchi Y, et al. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;16:367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 39.Zentella R, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- 41.Hennig L, Buche C, Eichenberg K, Schafer E. Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol. 1999;121:571–577. doi: 10.1104/pp.121.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinomura T, Nagatani A, Chory J, Furuya M. The Induction of Seed Germination in Arabidopsis thaliana Is Regulated Principally by Phytochrome B and Secondarily by Phytochrome A. Plant Physiol. 1994;104:363–371. doi: 10.1104/pp.104.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinomura T, et al. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Xiong L. Role of HY5 in abscisic acid response in seeds and seedlings. Plant Signal Behav. 2008;3:986–988. doi: 10.4161/psb.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumagai T, et al. The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2008;72:1539–1549. doi: 10.1271/bbb.80041. [DOI] [PubMed] [Google Scholar]
- 46.Han S, Kim D. AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics. 2006;7:179. doi: 10.1186/1471-2105-7-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res. 2002;73:149–156. doi: 10.1023/A:1020470224740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.