Abstract
Global loss of DNA methylation has been known for decades as an epigenomic aberration associated with carcinogenesis and cancer progression. Loss of DNA methylation affects predominantly repetitive elements, which encompass >50% of the CpG dinucleotides present in the human genome. Because of the lack of an effective approach, no studies have been conducted to reveal such genome-wide methylation changes at a single-base resolution. To precisely determine the CpG sites with methylation loss during progression of pediatric intracranial ependymomas, we exploited a high-throughput bisulfite sequencing approach that simultaneously generates methylation profiles for thousands of Alu elements and their flanking sequences. Comparison of the methylation profiles of normal and tumor tissues revealed that the methylation status of the majority of CpG sites adjacent to or within Alu repeats remain unaltered, while a small set of CpG sites gain or lose methylation in ependymomas. Compared to the CpG sites with stable methylation level between normal control and ependymomas, the differentially methylated CpG sites are enriched in the sequences with low CpG density in the flanking regions of Alu repeats, rather than within the Alu sequences themselves. In addition, the CpG sites that are hypermethylated in ependymomas are proximal to CpG islands, whereas those that are hypomethylated are overrepresented in intergenic regions. Lastly, aberrant methylation of several genomic loci was confirmed to be associated with the aggressive primary tumors and the relapsed ependymomas.
Keywords: DNA methylation, bisulfite sequencing, pediatric brain tumor
Ependymomas are glial tumors, derived from radial glial cells, that develop from the neuroepithelial lining of the ventricles, and from the central canal of the spinal cord (1). They are the third most prevalent brain tumor in childhood accounting for ≈10% of neuroepithelial tumors (2, 3). Clinical behavior of ependymomas varies over a wide range, from prolonged remissions, through multiple relapses at shorter intervals, to tumors that grow relentlessly resembling the aggressiveness of high-grade gliomas. Sadly, nearly 50% of children with ependymoma succumb to the disease within 5 years of diagnosis. The outcome of relapsing patients is even worse, with a 5-year survival of 24% and 27% for children younger and older, respectively, than 3 years of age at initial diagnosis (4). Unfortunately, the association between clinical outcome of ependymomas and histological grades—i.e., WHO grades II and III—could not be firmly established (5–7). The biological basis for the significantly poorer outcome of patients remains totally elusive.
Both genetic and epigenetic analyses have been conducted in an effort to uncover the molecular mechanisms underlying clinical behavior and outcome of ependymomas (8–10). In the human genome, most CpG islands are hypomethylated to allow an open chromatin organization and facilitate nearby gene expression. By contrast, repetitive elements, such as LINEs, SINEs, and LTRs, are heavily methylated, thus preventing their transcription and retrotransposition and maintaining genomic stability (11, 12). During tumor development and progression, two distinct but concurrent epigenetic abnormalities are commonly observed: localized hypermethylation, typically affecting CpG islands, and global hypomethylation, which predominantly occurs in repeats. Localized epigenetic alterations have been reported to occur in ependymomas, such as promoter hypermethylation of RASSF1A and of TRAIL pathway-related genes (10, 13). Although global hypomethylation has not been associated yet with tumorigenesis and progression of ependymomas, it has been recognized as a hallmark of many tumors. Importantly, it has been shown to be an early event in the development of chronic lymphocytic leukemia, colon cancer, and breast cancer (14–17). Furthermore, the degree of methylation loss was associated with disease stage in hepatocellular carcinoma, ovarian cancer, and glioma, and may serve as an important prognostic factor (11, 18, 19). Interestingly, remethylation of DNA repeats via folate supplementation was shown to limit the aggressiveness of gliomas (20). However, in spite of the well established significance of global loss of methylation in cancer development and progression, to date no genome-wide analysis has been conducted to precisely localize such hypomethylated CpG sites in any tumor.
As the most abundant repetitive elements in the human genome, Alu elements comprise >10% of the human genome sequence. Of the 30 million CpG dinucleotides in the human genome, ≈7 million are contributed by >1 million Alu elements. In this study, we generated single-base-resolution epigenomic data for a large number of Alu elements with the aim of understanding global loss of DNA methylation patterns and the identification of epigenetic marks underlying the wide range of clinical behavior observed in pediatric ependymomas. This study was initiated to investigate: (i) whether global loss of DNA methylation occurs during development and progression of ependymomas; (ii) whether both gain and loss of methylation may occur within Alu elements and in their flanking sequences during disease progression; (iii) whether such epigenetic alterations are restricted to a nonrandom subset of CpG sites in the genome; (iv) whether there may be genomic features associated with global gain and loss of DNA methylation; and (v) whether severity of clinical behavior in ependymomas may be explained by qualitative and/or quantitative differences in their epigenomes.
Results
Global Hypomethylation Is Associated with Ependymoma Progression.
To ascertain epigenomic alterations occurring in ependymomas, 10 normal tissue samples were microdissected from the fourth ventricle lining and from the lateral ventricle lining of five individuals to be used as controls for infratentorial and supratentorial ependymomas, respectively. In addition, a total of 52 infratentorial or supratentorial ependymoma samples derived from 36 patients were included in the study (Table S1). These samples were classified into three major groups according to disease stage and outcome: primary nonaggressive (PN), primary aggressive (PA), and relapsed ependymomas (RL).
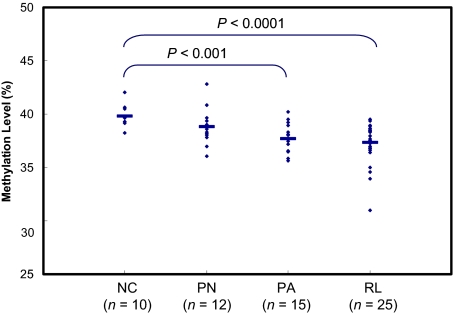
Because Alu elements encompass ≈25% of all CpG dinucleotides present in the human genome, collectively their methylation levels serve as representations of the epigenomic alterations that may be occurring genome-wide (21, 22). To maximize both the sensitivity and the specificity of these assays, we designed a set of primers targeting Alu elements rich in CpG dinucleotides (23). We determined bioinformatically that after bisulfite conversion, there are 45,810 distinct Alu elements with sequences 100% identical to the primers that we designed. Using this set of primers, the methylation levels of ≈180,000 CpG sites can be determined simultaneously in a single pyrosequencing reaction. In this study, we used this primer set to determine the Alu methylation levels in genomic DNA derived from ependymomas and normal tissue controls. The average methylation values were 39.8, 38.8, 37.7, and 37.4 for the normal ventricle linings, the PN tumor samples, the PA tumor samples, and the RL tumor samples, respectively (Fig. 1). When compared to methylation levels exhibited by the normal tissue controls, we found that there was a significant loss of methylation in PA and RL tumor samples. By contrast, there is only a minor loss of methylation in the PN tumor samples.
Fig. 1.
Global Alu methylation profile in ependymomas. Each dot represents the methylation level of an individual sample. The x axis represents the sample groups. The y axis represents the global Alu methylation levels determined by pyrosequencing of intra-Alu PCR products. Group comparisons were conducted for normal controls and tumor tissues with P values determined by t test (two-sample assuming unequal variances).
Bisulfite Sequencing of Alu Epigenomes in Ependymomas.
To further determine the specific CpG sites undergoing gain or loss of methylation in ependymomas, we exploited the high-throughput bisulfite sequencing strategy that we described in ref. 24. Methylation profiles from thousands of Alu elements and their flanking sequences were generated for one normal tissue control (fourth ventricle lining, microdissected) and six ependymomas (two PN, two PA, and two RL). We obtained data for >456 million nucleotides from 2.6 million sequence reads that were derived from the aforementioned Alu amplicon libraries. After the removal of primer and adaptor sequences, 2.1 million sequence reads encompassing 319 million nucleotides were unambiguously mapped to the human genome (Table S2). A total of 12 million methylation data points were generated for 264,304 distinct CpG sites that are widely distributed in the human genome (Fig. S1A). For each chromosome, methylation data were obtained for ≈0.9% of all CpGs. More than 91.6% of these CpG sites were derived from Alu elements (Fig. S1B), as predicted. Because the Alu-specific primer used for construction of the Alu amplicon libraries was designed to amplify a subset of Alu elements rich in CpG dinucleotides, AluY elements were particularly overrepresented as predicted. Indeed, a total of 193,270 CpG sites, or 73.1% of 264,304, were found to localize to AluY elements.
Methylation Variation of CpG Sites in Alu Elements and Flanking Sequences.
To examine the extent to which our genome is under pressure to keep Alu elements stably methylated, we first examined the methylation stability of CpG sites in Alu elements and their flanking sequences in our samples. We identified a set of 12,890 CpG sites that were sequenced at least 10 times in all samples, and ascertained the maximum methylation difference observed for each CpG site in the seven samples. In the set of 12,890 CpG sites, 9,817 or 76.2% of the CpG sites exhibited <20% methylation alterations—i.e., the highest methylation difference observed in any pairwise comparison was <20%. At the other end of the spectrum, 0.8% of CpG sites were found to exhibit methylation differences exceeding 80% (Fig. S2A). This indicated that the majority of CpG sites are relatively stable with respect to their methylation status in all seven samples.
An important question to be addressed is whether the CpG sites for which ≥20% difference in methylation was observed are randomly distributed in the genome. For the set of 12,890 CpG sites, the CpG sites that were <500 bp apart were clustered. A total of 2,149 clusters—with six CpG sites on average—were obtained. We found that in 623 clusters, all of the CpG sites had a maximum methylation difference of <20%, 120 clusters had all CpG sites with a maximum methylation difference of ≥20%, and there were 1,406 clusters in which some of the CpG sites had ≥20% methylation alterations. We interpret the fact that the majority of genomic loci contained some CpG sites that exhibited ≥20% methylation differences and others that did not as indicative that the methylation stability of CpG sites may vary in spite of their colocalization.
Of these 12,890 CpG sites, 3,073 exhibited ≥20% methylation differences among the seven samples and were widely distributed into 1,526 genomic loci. To determine whether the distribution of CpG sites exhibiting ≥20% methylation difference is random, a simulation analysis was performed 10,000 times. For each simulation, a set of 3,073 CpG sites were randomly selected from the 12,890 CpG sites and clustered based on their genomic coordinates. For all 10,000 simulations, the randomly selected CpG sites were always distributed in >1,526 clusters (Fig. S2B). This simulation result indicated that the CpG sites with ≥20% methylation alterations tend to cluster in the genome (P < 0.0001).
Genomic Features Associated with Methylation Gain and Loss in Alu Elements and Flanking Sequences.
In the previous section, we described the analysis performed for the CpG sites that were represented in 10 or more reads in each of the seven samples. Such analysis was adequate to reveal the magnitude of methylation variation (but not the direction of methylation change) exhibited by CpG sites among all samples. Next, we performed all possible pairwise comparisons with the seven samples. A total of 85,900 distinct CpG sites were represented in 10 or more reads in at least two samples. The number of such CpG sites in the different pairwise comparisons ranged from 24,208 to 46,065 (Table S3). Compared to normal control, ependymoma tissues exhibited various levels of loss of DNA methylation. Approximately 3.4–8.6% of the CpG sites in the ependymomas had a loss of methylation that was ≥20%. In addition, a trend was observed in that the more aggressive ependymomas exhibited the most loss of methylation. Interestingly, although the number of CpG sites that exhibited loss of methylation far exceeded that of CpG sites in which gain of methylation was observed, 1.8–2.6% of CpG sites did exhibit a methylation gain of ≥20% in the tumor tissues. This result suggests that the Alu elements and their flanking sequences may gain or lose methylation during development and progression of ependymomas.
Previously, it was shown that the methylation statuses of CpG islands are associated with their genomic sequence features (25, 26). Thus, we hypothesized that the differentially methylated CpG sites identified in this study may also show association with specific genomic sequence features. To better understand the patterns of methylation loss and gain observed in ependymomas, we focused our analysis on the six pairwise comparisons between normal and tumor tissues. Using the same criteria as those discussed above, three sets of CpG sites were identified: (i) 10,414 CpG sites with ≥20% loss of methylation in the ependymomas, (ii) 2,110 CpG sites with ≥20% gain of methylation in the ependymomas, and (iii) a reference set of 45,010 CpG sites with methylation changes of <10% in any of the six pairwise comparisons. In the human genome, >60% of SNPs are associated with the spontaneous deamination of methylated CpGs in germ cells (27). To eliminate a potential bias, a total of 1,829 CpG sites (89.3% in SINEs) with methylation-associated SNPs were excluded from this study.
Based on the previous association studies (25, 26), we focused on 285 genomic DNA attributes, including 15 genomic features and 270 sequence characteristics (Table S4). For all of the attributes compiled, statistical comparisons against the reference set were conducted for the CpG sites with ≥20% methylation gain or loss. After family-wise error rate justification, a number of genomic features were found to be associated with a methylation gain or loss (Table 1 and 2). Compared to the reference set, the CpG sites with either ≥20% gain or loss of methylation in the ependymomas are underrepresented in the Alu sequences. Approximately 90.7% of the CpG sites in the reference set (defined above) are located in SINEs, which are predominately Alu elements. For the CpG sites with significant methylation gain or loss, the ratios decreased to 81.7% and 83.2%, respectively. This suggests that there is a greater constraint to maintain the methylation status of the CpG sites within Alu elements than of those residing in their flanking sequences. Although the CpG sites with either methylation gain or loss are sometimes within the same genomic features, there are occasions when this is not the case. The CpG sites hypermethylated in ependymomas are closer to the most adjacent CpG islands, whereas the CpG sites that are hypomethylated in ependymomas are overrepresented in the intergenic regions and farther from the closest transcriptional factor binding sites, when compared to the reference set. Regarding DNA sequence characteristics, lower CpG ratios were observed in the sequences within 1-kb windows flanking the CpG sites that exhibit ≥20% methylation gain or loss. For methylation gain, GATC, TGGA, TTCC, and TTAG tetranucleotides were enriched within such 1-kb sequence windows. In contrast, for the CpG sites showing ≥20% methylation loss, enrichments for CCCC and TCTG tetranucleotides were observed. Full descriptions of these genomic features and the statistical results are provided in Table S4.
Table 1.
Top 10 genomic features and DNA-related attributes associated with the CpG sites with 20% or more methylation loss in ependymomas
| Attribute name | Direction of change* | Statistical test | Significance (not adjusted) | Significance (Bonferroni) | Significance threshold (FDR) |
| In SINE | Decrease | χ2 test | 0.00E+00 | 0.00E+00 | 3.48E-05 |
| In intron | Decrease | χ2 test | 4.07E-40 | 6.11E-39 | 6.97E-05 |
| In gene | Decrease | χ2 test | 2.11E-39 | 3.17E-38 | 1.05E-04 |
| Length contribution by SINE | Decrease | Wilcoxon rank-sum | 2.68E-25 | 7.22E-23 | 1.39E-04 |
| In LTR | Increase | χ2 test | 3.50E-20 | 5.26E-19 | 1.74E-04 |
| In LINE | Increase | χ2 test | 8.54E-18 | 1.28E-16 | 2.09E-04 |
| Distance to the most adjacent TFBS† | Increase | Wilcoxon rank-sum | 2.53E-16 | 3.80E-15 | 2.44E-04 |
| tetraNT_CCCC | Increase | Wilcoxon rank-sum | 6.45E-15 | 1.74E-12 | 2.79E-04 |
| cgRatio | Decrease | Wilcoxon rank-sum | 8.00E-13 | 2.16E-10 | 3.14E-04 |
| tetraNT_TCTG | Increase | Wilcoxon rank-sum | 8.55E-13 | 2.31E-10 | 3.48E-04 |
*Direction of change (increase or decrease) indicated the presentation of the CpG sites in term of genomic features or DNA-related attributes examined.
†TFBS: 3,837,187 putative transcription factor binding sites conserved in the human/mouse/rat alignment for 398 transcriptional factors (28).
Table 2.
Top 10 genomic features and DNA-related attributes associated with the CpG sites with 20% or more methylation gain in ependymomas
| Attribute name | Direction of change* | Statistical test | Significance (not adjusted) | Significance (Bonferroni) | Significance threshold (FDR) |
| In SINE | Decrease | χ2 test | 2.35E-43 | 3.53E-42 | 3.48E-05 |
| In LINE | Increase | χ2 test | 4.63E-15 | 6.95E-14 | 6.97E-05 |
| cgRatio | Decrease | Wilcoxon rank-sum | 3.09E-11 | 8.35E-09 | 1.05E-04 |
| Length contribution by SINE | Decrease | Wilcoxon rank-sum | 1.78E-09 | 4.80E-07 | 1.39E-04 |
| tetraNT_GATC | Decrease | Wilcoxon rank-sum | 3.21E-08 | 8.65E-06 | 1.74E-04 |
| cgCount | Decrease | Wilcoxon rank-sum | 6.52E-08 | 1.76E-05 | 2.09E-04 |
| tetraNT_TGGA | Increase | Wilcoxon rank-sum | 3.88E-07 | 1.05E-04 | 2.44E-04 |
| tetraNT_TTCC | Increase | Wilcoxon rank-sum | 4.15E-07 | 1.12E-04 | 2.79E-04 |
| tetraNT_TTAG | Decrease | Wilcoxon rank-sum | 1.53E-06 | 4.14E-04 | 3.14E-04 |
| Distance to the most adjacent CGI | Decrease | Wilcoxon rank-sum | 5.83E-05 | 8.74E-04 | 3.48E-04 |
*Direction of change (increase or decrease) indicated the presentation of the CpG sites in terms of genomic features or DNA-related attributes examined.
To probe the reliability of the methylation differences ≥20% identified between normal (fourth ventricle lining) and tumor tissues, we included a published dataset derived from normal cerebellum in this comparative analysis (24). As expected because the roof of the fourth ventricle is formed by the cerebellum, similar methylation profiles were observed for the fourth ventricle lining and the normal cerebellum from the two human subjects. Approximately 96% of the CpG sites in these two tissues differed by <20% in methylation level. Thus, we conducted all six pairwise comparisons between normal cerebellum and ependymomas, and compared the results to the pairwise comparisons between normal fourth ventricle lining and ependymomas. A strong positive correlation (r = 0.77) of methylation differences between ependymomas and these two normal controls was observed for the CpG sites that exhibited at least 20% methylation difference between ependymomas and fourth ventricle lining (Fig. S3).
Methylation Status of Alu Elements May Serve as Prognostic Marker in Ependymoma.
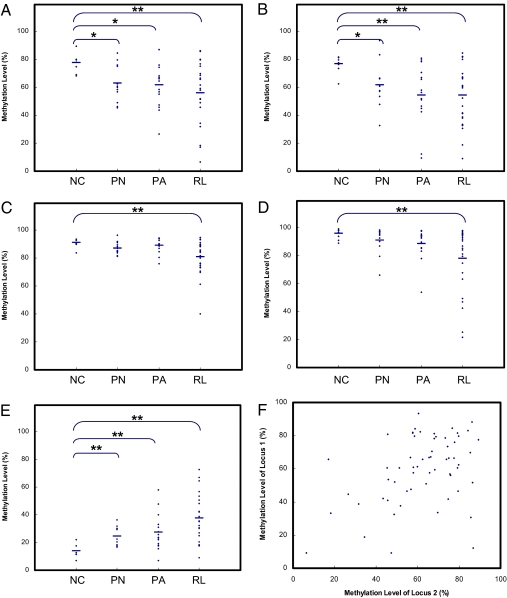
To further explore the association between the methylation aberrations observed on individual Alu elements and the aggressiveness and/or recurrence of ependymomas, we performed validation experiments on a panel of ependymoma samples. We chose the genomic loci that exhibited ≥20% methylation difference in ependymomas of different stages. Similarly to the global Alu methylation profiles observed in ependymomas, substantially different DNA methylation profiles were observed for five examined genomic loci in ependymomas of different stages (Fig. 2). Particularly for the relapsed ependymomas, striking differences in DNA methylation were observed for all five genomic loci. For loci 1–4, varying levels of DNA methylation loss were observed in ependymomas (Fig. 2 A–D), whereas locus 5 showed an increased level of methylation in a number of aggressive and relapsed ependymomas (Fig. 2E). Compared to normal control, the relapsed ependymomas exhibited significant methylation differences in all five loci (t test, P < 0.001).
Fig. 2.
Pyrosequencing validations. (A–E) Methylation profiles of five genomic loci in a panel of ependymoma tissues and normal controls. Each dot represents the methylation level of an individual sample. The x axis represents the sample groups. The y axis represents the average methylation level determined by pyrosequencing. NC, PN, PA, and RL represent normal control (n = 10), primary nonaggressive (n = 12), primary aggressive (n = 15), and relapsed ependymoma tissues (n = 25), respectively. Group comparisons were conducted for normal controls and tumor tissues with P values determined by t test (two-sample assuming unequal variances). *, P < 0.01 in comparison with the normal controls; **, P < 0.001 in comparison with the normal controls. (F) Correlation of methylation levels of genomic locus 1 and 2. Each dot represents the methylation levels of genomic locus 1 and 2 in an individual sample.
We further examined the correlations among the methylation levels of global Alu and the methylation levels of all five genomic loci examined (Table S5). The global Alu methylation levels were found to be moderately and positively correlated (Pearson's r from 0.32 to 0.44) with the methylation levels of all four genomic loci, which exhibited loss of methylation in ependymomas. Only little and negative correlation (Pearson's r = −0.1) was found between the global Alu methylation levels and the methylation levels of locus 5, which is hypermethylated in ependymomas. Interestingly, the methylation levels of genomic loci 1, 2, and 4 showed moderate positive correlation between each other. The correlation of the methylation levels of loci 1 and 2 is shown as an example in Fig. 2F. In addition, moderate negative correlations were found between the methylation levels of these three hypomethylated loci and those of the hypermethylated locus 5 (Pearson's r from −0.32 to −0.38).
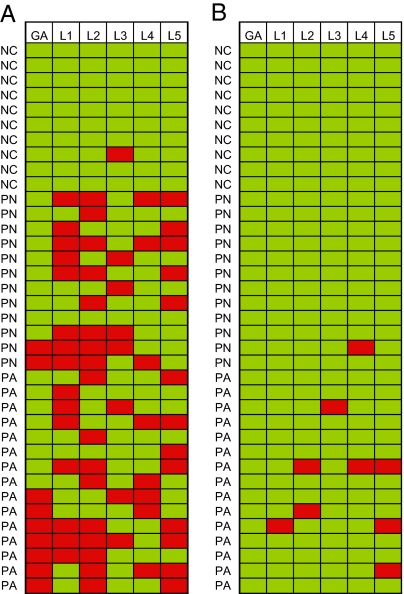
To explore whether the methylation levels of this subset of specific Alu elements could serve as indicators for severity of clinical behavior in ependymomas, we used two classifiers: the first to distinguish primary ependymomas from normal tissue, and the second to discriminate between aggressive and nonaggressive primary tumors. To maximize its potential clinical application, we limited these analyses to primary tumors and normal controls—i.e., recurrent ependymoma specimens were excluded. Using the cut-off values defined in Materials and Methods, only 33% of tumors exhibited altered global Alu methylation but 96% demonstrated methylation alteration in at least one of the five loci examined (Fig. 3A). Differences in global levels of Alu methylation alone failed to distinguish aggressive from nonaggressive primary ependymomas. In contrast, 5 of the 15 primary aggressive tumors investigated showed methylation alterations in at least one of the five loci examined (Fig. 3B). Thus, compared to the classifications based on global levels of Alu methylation, the sensitivity to discern between normal and tumor tissues was increased from 0.33 to 0.96 with a drop in the specificity from 1 to 0.9. Most promisingly, the sensitivity to detect a primary aggressive tumor was increased from 0 to 0.33, with a drop in specificity from 1 to 0.95, when using the methylation data from five specific loci. In summary, the combination of the methylation levels of multiple specific genomic loci greatly improved the classification sensitivity.
Fig. 3.
Methylation status of global Alu and five specific loci and the clinical behavior of ependymomas. (A) Classifier distinguishing the ependymomas from normal. Green squares = methylation level of the sample is within the cut-offs, two standard derivations from the average methylation levels of normal controls; red squares = methylation level of the sample is either below the cut-off for hypomethylated loci or above the cut-off for hypermethylated locus. (B) Classifier distinguishing the primary aggressive ependymomas from primary nonaggressive ones. Green squares = methylation level of the sample is within the cut-offs, two standard derivations from the average methylation levels of primary nonaggressive ependymomas; red squares = methylation level of the sample is either below the cut-off for hypomethylated loci or above the cut-off for hypermethylated locus. Each row represents a tissue sample, and each column represents a specific locus (L1 to L5) or global Alu methylation level (GA) indicated on top. NC, PN, and PA represent normal control, primary nonaggressive, and aggressive ependymoma tissues, respectively.
Last, to visualize the methylation data reported in this study, a genome browser has been developed and installed at http://cmbteg.childrensmemorial.org/cgi-bin/gbrowse/btech.
Discussion
One of the main challenges in the management of pediatric ependymomas is to cure patients with minimum damage to the developing brain. The identification of epigenetic marks that might enable tumor stratification according to clinical behavior would constitute a significant advancement in that it would provide the basis for the development of individually tailored treatment strategies. In this study, we generated large-scale sequence-based methylation data from primary (nonaggressive and aggressive) and recurrent ependymomas as well as from normal tissue control, to uncover alterations in genome-wide patterns of Alu methylation occurring during disease development and progression. A limitation that is typical of studies such as this is that, at least in part, the observed variations are likely to be contributed by a wide range of factors, from genomic and tumor heterogeneity to exposure to different therapeutic agents. Despite such limitations, we demonstrated that methylation losses in Alu elements are insignificant in primary nonaggressive ependymomas but that such losses increase in aggressive primary tumors and further yet in relapsed ependymomas. In particular, our data suggested that the methylation status of some Alu elements may serve as prognostic factors for a subset of aggressive ependymomas.
A few interesting observations were made upon analysis of the bisulfite sequence data derived from ependymomas and normal tissue control. First, an examination of the magnitude of the methylation gains and losses observed in ependymomas at different stages of disease progression revealed that in fact significant changes in methylation are restricted to a relatively small number of CpG sites in Alu repeats and in their flanking sequences. Furthermore, we found that both gain and loss of methylation may occur in Alu and flanking sequences. In addition, the observed gains and losses of DNA methylation in Alu repeats and flanking sequences are not randomly distributed throughout the genome. CpG sites within Alu elements seem to be more resistant to alterations in methylation than those mapping to their 5′ flanking sequences. By contrast, we found that genomic regions with a lower density of CpG dinucleotides are more vulnerable to alterations in methylation. Lastly, we observed that CpG sites in intergenic regions are more likely to exhibit losses in methylation, whereas those adjacent to CpG islands tend to gain methylation.
Materials and Methods
Genomic DNA Preparation.
Human snap-frozen tissues were obtained from the tissue bank of the Department of Pathology, Children's Memorial Hospital at Chicago under an IRB-approved protocol (#2007-12971). Genomic DNA was extracted with the Qiagen DNeasy tissue kit.
Construction and Sequencing of Alu Amplicon Libraries.
Seven Alu amplicon libraries were constructed with the genomic DNA extracted from one normal fourth ventricle lining and six ependymoma tissues. The Alu amplicon libraries were constructed as described in ref. 24. 454 sequencing was conducted with the Roche Genome Sequencer FLX System at the Sequencing Core Facility of Children's Memorial Research Center, Northwestern University Feinberg School of Medicine. Sequence data processing and determination of CpG methylation was conducted as described in ref. 24.
Statistical Analysis of the Association Between Methylation Aberrations and DNA-Related Attributes.
A comprehensive list of attributes that can be linked directly to CpG sites or to the sequence in a 1-kb window surrounding the CpG dinucleotide of interest (CpG ± 499 bp) was compiled. The data for most of these attributes were calculated based on the UCSC Genome Annotation Database (28). The attributes for DNA sequence features were directly calculated based on the DNA sequence extracted from the reference human genome sequence. All of the attributes are either in the numerical form or Boolean form (such as presence or absence in a gene). All of the CpG sites harboring a methylation-associated SNP (27) were excluded in the analysis. The nonparametric Wilcoxon rank-sum and χ2 statistical tests were performed for each attribute in numerical form and Boolean form, respectively. Using the CpG sites with stable methylation levels in normal and tumor samples as a reference set, statistical significance was determined for all attributes for differentially methylated CpG sites. Significance thresholds were adjusted for multiple testing by using the Bonferroni correction, and the family-wise error rate was set to be <1%.
PCR and Pyrosequencing.
Bisulfite pyrosequencing reactions were conducted as described in refs. 23 and 24. Except for genomic locus 5, nested PCR reactions were adopted to amplify the target regions from bisulfite modified genomic DNA. The primers used in the PCR runs and pyrosequencing reactions are shown in Table S6.
Classifiers for the Stratification of Nonaggressive and Aggressive Primary Ependymomas and Normal Tissues.
The cut-off values used to distinguish primary tumors from normal tissue were set at (±) two standard deviations of the average methylation levels (global Alu methylation level and those of each of the five selected loci) observed in normal controls. Similarly, the cut-off values used to distinguish aggressive from nonaggressive primary tumors were set at ±2 SD of the average methylation levels (global Alu methylation level and those of each of the five selected loci) observed in nonaggressive primary tumors. The sensitivity of the classifier was defined based on the calculated true positive rate—i.e., the percentage of true positives in all positive instances. Accordingly, the specificity of the classifier was defined based on the calculated true negative rate—i.e., the percentage of true negative in all negative instances.
Supplementary Material
Acknowledgments
We thank Dr. Elio Vanin (Children's Memorial Research Center, Chicago) for critical reading and valuable comments on the manuscript. This work was supported by the Everett/O'Connor Charitable Trust, the Dr. Ralph and Marian C. Falk Medical Research Trust, the Gus Foundation, the Maeve McNicholas Memorial Foundation, and the Medical Research Institute Council.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913836107/DCSupplemental.
References
- 1.Taylor MD, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 2.CBTRUS. Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004-2005. Hinsdale, IL: Central Brain Tumor Registry of the United States; 2009. [Google Scholar]
- 3.Agaoglu FY, et al. Ependymal tumors in childhood. Pediatr Blood Cancer. 2005;45:298–303. doi: 10.1002/pbc.20212. [DOI] [PubMed] [Google Scholar]
- 4.Messahel B, et al. Children's Cancer Leukaemia Group Brain Tumour Committee. Relapsed intracranial ependymoma in children in the UK: patterns of relapse, survival and therapeutic outcome. Eur J Cancer. 2009;45:1815–1823. doi: 10.1016/j.ejca.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Kano H, Niranjan A, Kondziolka D, Flickinger JC, Lunsford LD. Outcome predictors for intracranial ependymoma radiosurgery. Neurosurgery. 2009;64:279–287. doi: 10.1227/01.NEU.0000338257.16220.F7. [DOI] [PubMed] [Google Scholar]
- 6.Ernestus RI, Schröder R, Stützer H, Klug N. The clinical and prognostic relevance of grading in intracranial ependymomas. Br J Neurosurg. 1997;11:421–428. doi: 10.1080/02688699745925. [DOI] [PubMed] [Google Scholar]
- 7.Bouffet E, Perilongo G, Canete A, Massimino M. Intracranial ependymomas in children: a critical review of prognostic factors and a plea for cooperation. Med Pediatr Oncol. 1998;30:319–329. doi: 10.1002/(sici)1096-911x(199806)30:6<319::aid-mpo1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 8.Palm T, et al. Expression profiling of ependymomas unravels localization and tumor grade-specific tumorigenesis. Cancer. 2009;115:3955–3968. doi: 10.1002/cncr.24476. [DOI] [PubMed] [Google Scholar]
- 9.Mendrzyk F, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 10.Michalowski MB, et al. Methylation of RASSF1A and TRAIL pathway-related genes is frequent in childhood intracranial ependymomas and benign choroid plexus papilloma. Cancer Genet Cytogenet. 2006;166:74–81. doi: 10.1016/j.cancergencyto.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 12.Roman-Gomez J, et al. Repetitive DNA hypomethylation in the advanced phase of chronic myeloid leukemia. Leuk Res. 2008;32:487–490. doi: 10.1016/j.leukres.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton DW, Lusher ME, Lindsey JC, Ellison DW, Clifford SC. Epigenetic inactivation of the RASSF1A tumour suppressor gene in ependymoma. Cancer Lett. 2005;227:75–81. doi: 10.1016/j.canlet.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 14.Paz MF, et al. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in normal tissues and human primary tumors. Cancer Res. 2002;62:4519–4524. [PubMed] [Google Scholar]
- 15.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 16.Stach D, et al. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res. 2003;31:E2. doi: 10.1093/nar/gng002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 18.Lin CH, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001;61:4238–4243. [PubMed] [Google Scholar]
- 19.Watts GS, et al. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hervouet E, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clin Cancer Res. 2009;15:3519–3529. doi: 10.1158/1078-0432.CCR-08-2062. [DOI] [PubMed] [Google Scholar]
- 21.Weisenberger DJ, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang AS, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie H, Wang M, Tange Y, Bonaldo MD, Soares MB. A primer design algorithm for global analysis of CpG methylation. In: Zhang J, Li G, Yang JY, editors. 2009 International Joint Conference on Bioinformatics, Systems Biology and Intelligent Computing. Los Alamitos, CA: IEEE Computer Society; 2009. pp. 126–130. [Google Scholar]
- 24.Xie H, et al. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 2009;37:4331–4340. doi: 10.1093/nar/gkp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das R, et al. Computational prediction of methylation status in human genomic sequences. Proc Natl Acad Sci USA. 2006;103:10713–10716. doi: 10.1073/pnas.0602949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bock C, et al. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet. 2006;2:e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie H, Wang M, Bischof J, Bonaldo MdeF, Soares MB. SNP-based prediction of the human germ cell methylation landscape. Genomics. 2009;93:434–440. doi: 10.1016/j.ygeno.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.