Abstract
Glioblastoma multiforme (GBM) is the most common and lethal primary brain cancer that is driven by aberrant signaling of growth factor receptors, particularly the epidermal growth factor receptor (EGFR). EGFR signaling is tightly regulated by receptor endocytosis and lysosome-mediated degradation, although the molecular mechanisms governing such regulation, particularly in the context of cancer, remain poorly delineated. Here, high-resolution genomic profiles of GBM identified a highly recurrent focal 1p36 deletion encompassing the putative tumor suppressor gene, Mig-6. We show that Mig-6 quells the malignant potential of GBM cells and dampens EGFR signaling by driving EGFR into late endosomes and lysosome-mediated degradation upon ligand stimulation. Mechanistically, this effect is mediated by the binding of Mig-6 to a SNARE protein STX8, a protein known to be required for late endosome trafficking. Thus, Mig-6 functions to ensure recruitment of internalized receptor to late endosomes and subsequently the lysosomal degradation compartment through its ability to specifically link EGFR and STX8 during ligand-stimulated EGFR trafficking. In GBM, the highly frequent loss of Mig-6 would therefore serve to sustain aberrant EGFR-mediated oncogenic signaling. Together, these data uncover a unique tumor suppression mechanism involving the regulation of receptor trafficking.
Keywords: glioblastoma, vesicle, STX8
Glioblastoma multiforme (GBM) is the most aggressive form of malignant glioma and stands as one of the most lethal cancers with median survival of ≈12–15 months (1). Extensive molecular and genomic studies of human glioma have identified numerous genetic and genomic alterations resulting in activation of multiple receptor tyrosine kinases, most notably epidermal growth factor (EGFR), which is found to be amplified and overexpressed in ∼45% of primary GBMs, although much less frequently in low-grade gliomas (2, 3). Clinically, overexpression of EGFR has been correlated with poor prognosis in GBM patients (4), and the precise wiring of the EGFR network and the regulation of its signaling pathway in GBM have always been an area of active investigation. It is well known that multiple mechanisms are engaged in the activation of the EGFR pathway during tumor initiation and progression, including receptor amplification and activating receptor mutations (5). Intriguingly, EGFR mutations occurring in GBM often involve the deletions in the extracellular domain or cytoplasmic tails, such as the EGFRvIII mutant missing the extracellular ligand binding domain (5), whereas EGFR kinase domain mutations commonly found in nonsmall cell lung cancer (NSCLC) are rare in GBM, suggesting distinctive oncogenic EGFR networks in different tumor types.
A hallmark feature of malignant glioma is its rampant genomic instability accompanied by numerous recurrent chromosomal structural aberrations that serve as a key pathological driving force for tumor progression and many of them remain to be characterized (6, 7). GBM possesses a highly rearranged genome and high-resolution genome analysis has uncovered myriad somatic alterations on the genomic and epigenetic levels (2, 3). Here, using an integrated genomic and functional analysis, we have identified Mig-6 as a candidate tumor suppressor that regulates EGFR trafficking and turnover in GBM cells. Mig-6 was originally identified as a mitogen-inducible gene and has been implicated in the feedback regulation of a variety of signaling processes, including the EGFR pathway (8–11). Ablation of Mig-6 was shown to induce tumor formation in various tissues, supporting the tumor suppressor function of Mig-6 (12–14). However, the role of Mig-6 during gliomagenesis is largely unknown. We report that Mig-6 functions to suppress the malignant potential of GBM cells by enhancing EGFR trafficking into late endosomes/lysosomes and promoting its degradation. Further molecular and cell biology studies identified STX8, a SNARE protein required for late-endosome fusion (15–17), as a Mig-6-binding protein to form a complex with EGFR during receptor trafficking. The strong interaction between Mig-6 and STX8 upon ligand activation therefore ensures recruitment of internalized EGFR to late endosomes and subsequently the lysosomal degradation compartment.
Results
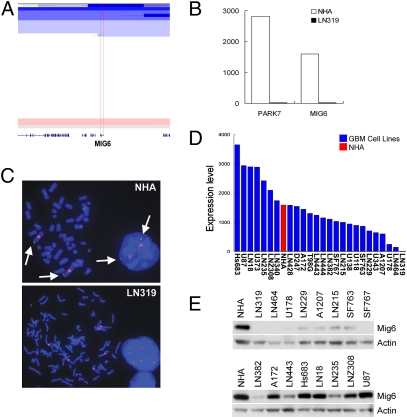
Genomic analysis of GBM has revealed myriad alterations with uncertain pathogenetic significance. Recurrent deletion of chromosome 1p36 is among the most common genomic events in multiple tumor types (18–23), although the structural complexity of these deletions and the uncertain definition of the commonly targeted region have hampered definitive identification of potential tumor suppressor(s) (24). In recent high-resolution array comparative genomic hybridization (CGH) analysis of GBM (18 tumors, 20 cell lines) that showed recurrent 1p36 deletion (Fig. 1A, 5/38; 13.2%), we delineated a unique 270-kb minimal common region (MCR) of deletion containing only two known genes, PARK7 and Mig-6 (ERRFI1) (Fig. 1A), and showing corresponding decrease in the expression of these genes in samples with genomic deletion (Fig. 1B). Consistent with our data, The Cancer Genome Atlas (TCGA) dataset confirmed recurrent deletion of this region (62/339; 18.3%) that defined an MCR containing seven known genes including PARK7 and Mig-6 (Fig. S1). Most notably, Mig-6 expression is down-regulated at both mRNA and protein levels in ∼50% of primary tumor samples and GBM cell lines, some of which do not show genomic deletion of Mig-6, indicating that additional mechanisms ensure Mig-6 down-regulation in human GBM (Fig. 1 D and E). Whereas PARK7 is an oncogene that inhibits the PTEN tumor suppressor (25–27), Mig-6 shows many properties consistent with a tumor suppressor role including cancer-prone phenotypes in knockout mice (12, 13, 28). In the context of this study, it is notable that Mig-6 was identified as a mitogen-inducible gene and is a presumed adaptor protein linked to the regulation of a variety of sig-naling pathways, including the key GBM oncogene EGFR (8–11). This profile prompted us to assess the functional relevance of Mig-6 and determine its mechanism of action in GBM.
Fig. 1.
Mig-6 is deleted in GBM cell lines and tumor samples. (A) Array-CGH heat map detailing Mig-6 deletion at chromosome 1p36 in primary GBM tumor specimens. Regions of amplification and deletion are denoted in red and blue, respectively. (B) mRNA expression levels of PARK7 and Mig-6 in normal human astrocytes (NHA) and LN319 cells. (C) FISH analysis of Mig-6 deletion in the GBM cell line, LN319. The green signals (arrows) indicate hybridization using a Mig-6-specific BAC probe, whereas the red signals indicate hybridization using a chromosome 1-specific centrosome reference probe. (D) mRNA level of Mig-6 in NHA and GBM cell lines. (E) Mig-6 protein level analyzed by Western blot in NHA and GBM cell lines.
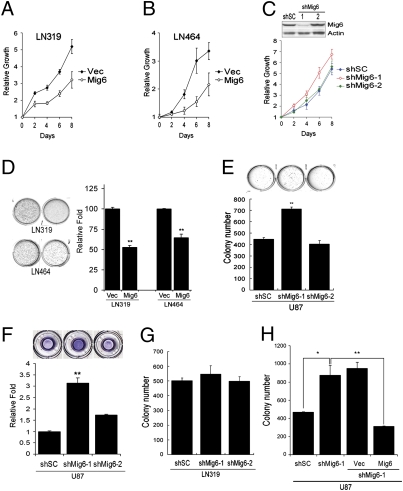
Reconstituting Mig-6 in LN319 and LN464 cells, two GBM cell lines that lack Mig-6 protein expression (Fig. 1 C–E), inhibits cellular proliferation as well as anchorage-independent growth (Fig. 2 A, B, and D), a hallmark for cellular transformation. Conversely, the shRNA-mediated knockdown of Mig-6 (shMig-6-1, 90% suppression) in U87 cells results in increased cell proliferation and anchorage-independent growth and invasion relative to control and ineffective Mig-6 shRNAs (e.g., shMig-6-2) (Fig. 2 C, E, and F). Mig-6 knockdown in additional GBM cells also promotes anchorage-independent growth (Fig. S2 A and B). Consistently, these shMig-6-1 cellular phenotypes are not elicited in LN319 cells carrying a homozygous 1p36 deletion (Fig. 2G), indicating that the effects of Mig-6 depletion are specific and not due to off-target effects. In addition, shMig-6-1-induced anchorage-independent growth is suppressed in U87 cells by expression of shRNA-resistant Mig-6 (Fig. S2C, Fig. 2H). Together, these gain- and loss-of-function studies demonstrate that Mig-6 functions as a potent tumor suppressor and is a key target of the 1p36 deletion event in GBM.
Fig. 2.
Mig-6 functions as a tumor suppressor. Reconstiting Mig-6 expression in (A) LN319 or (B) LN464 cells inhibits cell proliferation. Vec, empty vector control. (C) Knockdown of Mig-6 expression in U87 cells (Inset) accelerates cellular proliferation. shSC, hairpin control. (D) (Left) Reconstitution of Mig-6 expression in LN319 or LN464 cells attenuates anchorage-independent growth in soft agar. (Right) Histogram quantification. Error bars indicate ±SD [**, P = 0.002 (LN319) and 0.008 (LN464), n = 3]. (E) (Upper) Knockdown of Mig-6 expression in U87 cells promotes anchorage-independent growth in soft agar. (Lower) Histogram quantification. Error bars indicate ±SD (**, P = 0.001, n = 3). (F) Knockdown of Mig-6 expression in U87 cells promoted cell invasion in a Boyden chamber assay. (Upper) Representative images show the invasion of U87 cells expressing control (shSC and shMig-6-2) and targeting shRNA (shMig-6-1) after 16-h induction with 10% FBS followed by crystal violet (0.2%) staining. (Lower) Histogram quantification. Error bars indicate ±SD (**, P = 0.003, n = 3). (G) shRNA targeting Mig-6 has no effect on soft agar growth of LN319 cells. (H) Reconstitution of Mig-6 expression inhibits soft agar growth of U87 cells expressing Mig-6 targeting shRNA (shMig-6-1). Error bars indicate ±SD (*, P = 0.03; **, P = 0.01; n = 3).
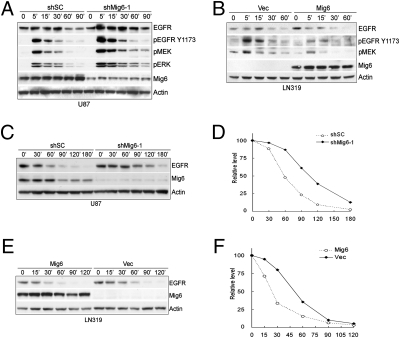
As a signature event in GBM, EGFR is amplified and mutated in 50% of GBM, highlighting its oncogenic role during GBM progression. Mig-6 has been shown to function as a feedback inhibitor of EGFR signaling (28, 29). Moreover, studies in human cancer cells as well as mouse models implicate that Mig-6 is a tumor suppressor of ErbB receptor-dependent carcinogenesis (12, 30). Thus, we examined the impact of Mig-6 on EGFR signaling in GBM cells. Consistent with previous reports (29, 31), Mig-6 knockdown in U87 cells enhances EGFR phosphorylation in response to EGF and downstream activation of MEK-ERK signaling (Fig. 3A). Similarly, Mig-6 reconstitution in LN319 cells inhibits the activation of the EGFR pathway (Fig. 3B), indicating that Mig-6 functions to limit the activation of EGFR signaling in these cells. Interestingly, Mig-6 overexpression fails to inhibit EGFRvIII signaling, a constitutively active EGFR mutant commonly found in GBM (32), in both murine astrocytes and the human GBM cell line, even though Mig-6 binds to EGFRvIII independent of ligand induction (Fig. S3), suggesting direct interaction of Mig-6 may not be sufficient to suppress EGFR signaling. In agreement with this notion, expression of Mig-6 showed a diminished inhibitory effect on the soft-agar growth of LN319 cells expressing EGFRvIII whereas the anchorage-independent growth of cells expressing wild-type EGFR was dramatically suppressed by Mig-6 (Fig. S4).
Fig. 3.
Mig-6 suppresses EGFR signaling and promotes ligand-induced receptor degradation. (A) Knockdown of Mig-6 expression in U87 cells enhances the activation of EGFR and the downstream signaling pathway in response to EGF treatment. Cells were treated with EGF (20 ng/mL) for the indicated times and cell lysates were immunoblotted with the indicated antibodies. (B) Reconstitution of Mig-6 expression in LN319 cells attenuates the activation of EGFR and the downstream signaling pathway in response to EGF treatment. Cells were treated with EGF (20 ng/mL) for the indicated times and cell lysates were immunoblotted with the indicated antibodies. (C) Knockdown of Mig-6 expression in U87 cells delayed EGFR degradation induced by EGF stimulation. Cells were pretreated with cycloheximide (CHX) (10 μg/mL) for 1 h before being treated with EGF (20 ng/mL) in the presence of CHX for the indicated times and cell lysates were subjected to immunoblotting with the indicated antibodies. (D) Histogram quantification of EGFR level (normalized with actin level) in C. (E) Reconstitution of Mig-6 expression in LN319 cells promotes EGFR degradation induced by EGF stimulation. Cells were treated as described in C and lysates were subjected to immunoblotting with the indicated antibodies. (F) Histogram quantification of EGFR level (normalized with actin level) in E.
Rapid EGFR internalization and degradation in response to EGF serves as a major negative feedback regulatory mechanism to control the duration and intensity of EGFR signaling (33, 34). Importantly, in contrast to wild-type EGFR, the EGFRvIII mutant does not undergo ligand-induced internalization and the subsequent lysosome-mediated degradation (35, 36), suggesting that Mig-6 may regulate EGFR endo/lysosomal trafficking and/or degradation. Indeed, shRNA depletion of Mig-6 in U87 cells results in delayed EGFR turnover upon EGF treatment whereas expression of Mig-6 in LN319 cells reduces the half-life of EGF-activated EGFR (Fig. 3 C–F). Consistent with our finding that Mig-6 failed to regulate EGFRvIII signaling, the half-life of EGFR vIII is not affected in the presence of Mig-6 (Fig. S5). Together, these data indicate that Mig-6 functions to desensitize EGFR signaling, possibly by promoting ligand-induced degradation of EGFR in GBM cells.
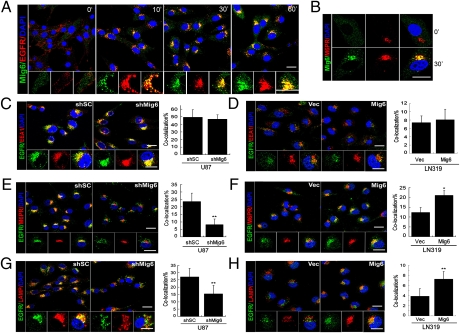
Given the evidence that Mig-6 enhances EGFR degradation, we next used confocal microscopy to examine whether Mig-6 controls receptor trafficking through endo-lysosomal compartments in LN319 cells expressing ectopic Mig-6 and U87 depleted of Mig-6. Indeed, the ligand stimulation by EGF strongly promotes the colocalization of endogenous Mig-6 with EGFR in vesicle structures (Fig. 4A) in U87 cells. Under these conditions, increased colocalization of Mig-6 with the late endosomal marker CI-M6PR (37) was noted in U87 cells treated with EGF for 30 min (Fig. 4B). In contrast, PDGFRβ did not colocalize with Mig-6 in PDGF-BB-treated U87 cells (Fig. S6), indicating that Mig-6 does not play a general role for receptor degradation. Interestingly, the colocalization of EGFR with early endosome antigen (EEA1), a marker for early endosomes, in response to EGF is comparable in Mig-6-depleted 87 cells, LN319 cells overexpressing Mig-6, and their respective control cells, indicating that Mig-6 does not regulate receptor traffic through early endosomes (Fig. 4 C and D). To determine whether Mig-6 controls subsequent EGFR trafficking to late endosomes, we performed receptor colocalization studies in Mig-6-depleted U87 cells and LN319 cells overexpressing Mig-6. As expected, we showed reduced colocalization of the EGFR with the CI-M6PR in Mig-6-depleted U87 cells relative to cells transduced with control siRNA (8.6% vs. 23.6%) (Fig. 4E). Similarly, reconstitution of Mig-6 in LN319 cells promoted EGFR trafficking to late endosomes compared to control cells (21% vs. 12.3%) (Fig. 4F). Consistent with a role for Mig-6 in late endosomal EGFR trafficking, receptor colocalization with the lysosomal marker LAMP1 is reduced in Mig-6-depleted U87 cells and increased in LN319 cells expressing Mig-6 (Fig. 4 G and H). Together, these data indicate a requirement for Mig-6 in the lysosomal degradation of EGFR and termination of receptor signaling by ensuring the trafficking of activated EGFR into late endosomes.
Fig. 4.
Mig-6 promotes EGFR localization into late endosome and lysosome compartments upon ligand induction. (A) Mig-6 colocalizes with EGFR in vesicle structures upon EGF stimulation. U87 cells were treated with EGF (20 ng/mL) for the indicated times and subjected to immunofluorescence staining with anti-Mig-6 (green), anti-EGFR (red), and DAPI (blue). (Scale bar: 20 μm.) (B) Mig-6 localizes in late endosomes upon EGF treatment. U87 cells were treated with EGF for 30 min and subjected to immunofluorescence staining with anti-Mig-6 (green), anti-M6PR (red), and DAPI (blue). (Scale bar: 20 μm.) (C) Knockdown of Mig-6 expression in U87 cells or (D) reconstitution of Mig-6 expression in LN319 cells shows a limited effect on the recruitment of EGFR to early endosomes. Cells were treated with EGF for 10 min and subjected to immunofluorescence staining with anti-EGFR (green), anti-EEA1 (red), and DAPI (blue). Quantification of colocalization between EGFR and EEA1 signals is shown in the histograms. Error bars indicate ±SD. (Scale bar: 20 μm.) (E) Knockdown of Mig-6 expression in U87 cells attenuates the recruitment of EGFR to late endosomes. Cells were treated with EGF for 30 min and subjected to immunofluorescence staining with anti-EGFR (green), anti-M6PR (red), and DAPI (blue). Quantification of colocalization between EGFR and CI-M6PR signals is shown in the histograms. Error bars indicate ±SD (**, P = 0.001; n = 5). (Scale bar: 20 μm.) (F) Reconstitution of Mig-6 expression in LN319 cells promotes the recruitment of EGFR to late endosomes. Cells were treated and stained as described in E. Quantification of colocalization between EGFR and CI-M6PR signals is shown in the histograms. Error bars indicate ±SD (*, P = 0.015; n = 5). (Scale bar: 20 μm.) (G) Knockdown of Mig-6 expression in U87 cells attenuates the recruitment of EGFR to lysosomes. Cells were treated with EGF for 60 min and subjected to immunofluorescence staining with anti-EGFR (green), anti-LAMP1 (red), and DAPI (blue). Quantification of colocalization between EGFR and LAMP1 signals is shown in the histograms. Error bars indicate ±SD (**, P = 0.006; n = 5). (Scale bar: 20 μm.) (H) Reconstitution of Mig-6 expression in LN319 cells promotes the recruitment of EGFR to lysosomes. Cells were treated and stained as described in G. Quantification of colocalization between EGFR and LAMP1 signals is shown in the histograms. Error bars indicate ±SD (**, P = 0.006; n = 5). (Scale bar: 20 μm.)
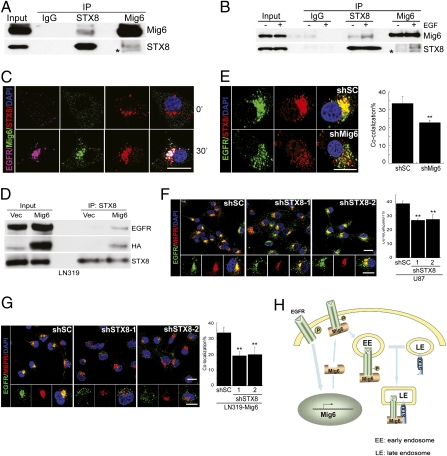
Regulation of receptor transit through endocytic compartments is under tight spatial and temporal control and remains an area of active investigation. To gain mechanistic insight into Mig-6-regulated EGFR degradation, we screened for Mig-6-interacting proteins using high-throughput yeast two-hybrid technology (Table S1). Most notable among the recurrent Mig-6-interacting proteins (7 of 74 positive colonies) was the t-SNARE Syntaxin 8 (STX8). STX8 is localized in the endosomal compartments and regulates the trafficking of EGFR from early to late endosomes (15–17). We first confirmed the interaction between endogenous Mig-6 and STX8 by coimmunoprecipitation using U87 cell lysates (Fig. 5A), an interaction enhanced by EGF treatment (Fig. 5B). Consistently, we detected colocalization of Mig-6 and STX8 in vesicular compartments 30 min after EGF treatment (Fig. 5C). Moreover, the binding of EGFR to STX8 is greatly enhanced in the presence of Mig-6 (Fig. 5D), indicative of a trimeric complex containing EGFR, Mig-6, and STX8. Consistent with this tenet, EGF-induced EGFR and STX8 colocalization decreased in Mig-6-depleted U87 cells (Fig. 5E). Together, these data strongly indicate that Mig-6 promotes the formation of a complex containing ligand-activated EGFR and STX8. Because STX8 is important for the fusion and trafficking of late endosomes (15, 17), we next examined whether STX8 is required for Mig-6-mediated EGFR trafficking to late endosomes. Knockdown of STX8 in U87 cells (Fig. S7A) significantly decreased the transit of EGFR into late endosomes (Fig. 5F). More importantly, the enhanced recruitment of EGFR to late endosomes by Mig-6 in LN319 cells was blocked by shRNA depletion of STX8 (Fig. S7B, Fig. 5G), indicating that STX8 is critical for Mig-6-mediated EGFR trafficking to late endosomes. Thus, Mig-6 functions via STX8 to enhance EGF-induced receptor degradation by regulating the trafficking of internalized EGFR into late endosomes and subsequent lysosomes (Fig. 5H).
Fig. 5.
Mig-6 interacts with STX8 to control EGFR trafficking through late endosome. (A) Mig-6 interacts with STX8 in vivo. U87 cell lysates were immunoprecipitated (IP) with normal mouse IgG or with an antibody against STX8 or Mig-6 and immunoblotted with the indicated antibodies. *, nonspecific band. (B) The interaction between Mig-6 and STX8 is enhanced by EGF stimulation. U87 cells were treated with EGF (20 ng/mL) for 30 min. Cell lysates were subjected to IP and immunoblotting as in A. (C) Complex formation between EGFR, Mig-6, and STX8. U87 cells were treated with EGF (20 ng/mL) for 30 min and subjected to immunofluorescence staining with anti-EGFR (purple), anti-Mig-6 (green), anti-STX8 (red), and DAPI (blue). (Scale bar: 20 μm.) (D) Mig-6 mediates the complex formation between EGFR and STX8. LN319 cells expressing vector (Vec) or Mig-6 were treated with EGF (20 ng/mL) for 30 min. Cell lysates were subjected to IP with anti-STX8 and immunoblotted with the indicated antibodies. (E) Knockdown of Mig-6 expression in U87 cells inhibits colocalization between EGFR and STX8. U87 cells were treated with EGF (20 ng/mL) for 30 min and subjected to immunofluorescence staining with anti-EGFR (green), anti-STX8 (red), and DAPI (blue). Quantification of colocalization between EGFR and STX8 signals is shown in the histograms. Error bars indicate ±SD (**, P = 0.001; n = 5). (Scale bar: 20 μm.) Knockdown of STX8 expression in (F) U87 cells and in (G) LN319 cells expressing Mig-6 (LN319-Mig-6) inhibits the recruitment of EGFR to late endosomes. Cells were treated with EGF (20 ng/mL) for 30 min and subjected to immunofluorescence staining with anti-EGFR (green), anti-M6PR (red), and DAPI (blue). Quantification of colocalization between EGFR and CI-M6PR signals is shown in the histograms. Error bars indicate ±SD (**, P < 0.001; n = 5). (Scale bar: 20 μm.) (H) Mig-6 regulates EGFR trafficking. Upon EGF stimulation, Mig-6 is recruited to activated EGFR and suppresses EGFR downstream signaling. During the ligand-stimulated EGFR trafficking, the interaction of Mig-6 and STX8 in the late endosomes recruits STX8 to the Mig-6-EGFR complex, which ensures EGFR trafficking from early endosomes to late endosomes and subsequent degradation. Note that the continuous presence of ligand also induces the expression of Mig-6 at the transcriptional level, which further keeps EGFR signaling in check.
Discussion
Many molecular genetic and histopathological studies, together with recent large-scale cancer genome sequencing projects, have identified EGFR and its downstream signaling networks as one of the most deregulated components of human GBM and other cancers (1–3). Here, by using unbiased genome-scale analysis, we have identified Mig-6 as a key tumor suppressor in GBM that functions as a negative regulator for EGFR signaling.
Multiple mechanisms can result in activation of the EGFR pathway during the initiation and development of GBM, including increased EGFR expression, enhanced autocrine signaling, and EGFR mutations (5). EGFR mutations commonly found in glioma typically involve the extracellular domains, such as the EGFRvIII mutant, as well as C-terminal deletion of the EGFR distal to the kinase domain (2). Those mutations also inhibit the receptor endocytosis and trafficking and thus stabilize the mutant receptor (35, 36, 38), underscoring the importance of the deregulation of the vesicle trafficking pathway as a mechanism for EGFR-mediated gliomagenesis. Although several regulators of the receptor trafficking pathway, such as Cbl, have been implicated in a number of human cancer types (39), the role of additional trafficking components during glioma development remains largely unexplored. Our data indicate Mig-6 promotes EGFR turnover through its regulation of the vesicle trafficking pathway. The identification of complex formation with STX8, a SNARE family protein important for the fusion of late endosomes, is unique and provides a mechanistic insight into how Mig-6 modulates sorting of EGFR to late endosome–lysosome compartments upon ligand activation. Our findings that endocytic trafficking and degradation of EGFR are closely controlled by Mig-6 indicate that the antagonistic effects of Mig-6 are exerted through perturbations in EGFR localization that alter the duration of EGFR signaling. As a consequence, Mig-6 deficiency in tumor cells further contributes to the “amplification” of EGFR signals during the development of GBM. Consistent with this notion, in the analysis of a total of 337 GBM samples from the TCGA dataset, we observed a positive correlation between the genomic alterations of Mig-6 and EGFR amplification (Fig. S8A). Surprisingly, in a limited analysis using patient samples with confirmed mutant EGFRvIII status, we also found that 47.4% (9/19) of samples coexist in both the EGFRvIII mutant and Mig-6 deletion/loss (Fig. S8B), suggesting a positive correlation between Mig-6 genomic alteration and EGFRvIII. Notably, in the human GBM, the mutant EGFRvIII has been found exclusively in samples with concurrent high-amplitude wild-type EGFR focal amplification. In addition, EGFRvIII is known to dimerize and activate wild-type EGFR during tumor pathogenesis (40). Therefore, whereas there is no impact of Mig-6 on EGFRvIII degradation, the loss of Mig-6 in the context of the EGFRvIII mutant could presumably serve to boost EGFR signaling by stabilizing of wild-type EGFR that is coexpressed with EGFRvIII. Moreover, due to the fact that multiple receptor tyrosine kinases (RTKs) are coactivated in human GBM (41) and the capacity of Mig-6 to regulate their signaling (8, 10), it is also conceivable that the loss of Mig-6 may function to enable activation of RTKs other than EGFR. Phase III trials for glioma treatment using monoclonal antibodies (mAbs) against EGFR are currently underway. Interestingly, one potential mechanism of the action of EGFR mAbs may involve their ability to target the EGFR for degradation in the lysosome (42). Hence deregulation of the EGFR trafficking components, such as by Mig-6 deletion, in gliomas may also contribute toward sensitivity to these and other future EGFR-targeted therapies. Such mechanistic insights into EGFR signaling may provide a framework for the selection of patients suitable for such trials.
Materials and Methods
Cell Lines.
The human glioma cell lines LN235, LN319, LN464, LNZ308, and U87 and the human embryonic kidney cell line HEK293T were purchased from American Type Culture Collection (ATCC). Primary murine astrocytes were isolated from 5-day-old pups with indicated genotypes and maintained in DMEM containing 10% FBS as previously described (43, 44).
Antibodies for Immunofluorescence and Western Blot Analysis.
The following antibodies were used: Mig-6 (8); EGFR (Upstate); PDGFRβ, pAkt, pMEK, and pERK (Cell Signaling); CI-M6RP, EEA1, and LAMP1 (Abcam); STX8 (BD Biosciences); HA (Roche); and pEGFR Y1173 and β-actin (Santa Cruz Biotechnology).
Proliferation Assays.
Proliferation assays were performed on 12-well plates in triplicate with 10,000 cells per well. Cells were fixed in 10% formalin in PBS and stained with crystal violet at 2-day increments starting after cell adherence (day 0). At the conclusion of the assay, crystal violet was extracted with 10% acetic acid and absorbance at 595 nm was measured with a 96-well plate reader.
Anchorage-Independent Growth Assay.
Between 5,000 and 10,000 cells per well were seeded in medium containing 0.4% low-melting agarose on top of bottom agar containing 1% low-melting agarose in regular medium. After 14–21 days, colonies were stained with iodonitrotetrazoliumchloride (Sigma) and counted with Totallab TL100 software.
Cell Invasion Assay.
Cell invasion assays were performed in Boyden chambers with matrix proteins as per manufacturer's protocol (BD Biosciences). A total of 200,000 cells were washed and then seeded in serum-free medium. The chemoattractant was medium plus 10% FBS.
Lentiviral-Mediated shRNA Targeting.
Lentiviral shRNA clones targeting Mig-6, STX8, and nontargeting control construct shGFP were obtained from the RNAi Consortium at the Dana-Farber/Broad Institute (sequences available upon request). Lentiviruses were produced in 293T cells with packing mix (ViraPower Lentiviral Expression System; Invitrogen) as per manufacturer's instruction.
Yeast Two-Hybrid Interaction Screening.
A pretransformed human fetal brain cDNA library (Clontech) was screened (1 × 106 clones) using the AH109 yeast reporter strain and the Matchmaker Two-Hybrid System 3 (Clontech), according to the manufacturer's instructions. Plasmid DNA from 200 potential positive clones was isolated after transformation into Escherichia coli strain DH5, followed by DNA sequencing using the provided prey vector-specific primers. Informative sequencing data were obtained for 109 of the 200 clones, 74 of which contained partial to full-length coding sequence and were further considered for downstream analysis.
Coimmunoprecipitation Analysis.
Cells were harvested in lysis buffer consisting of 20 mM Tris (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM EGTA, 1 mM EDTA, 5 mM sodium pyrophosphate, 50 mM NaF, 10 mM β-glycerophosphate, 1 mM sodium vanadate, 0.5 mM DTT, 1 mM PMSF, and 1× Protease Inhibitor Mixture (Roche). One to 1.5 mg of total protein was incubated with 1 μg of indicated antibodies and Protein A agarose (RepliGen) at 4 °C overnight with rocking. Immunoprecipitation complexes were eluted by boiling in SDS loading buffer and resolved on NuPAGE 4–12% Bis-Tris gels (Invitrogen) for immunoblotting analysis.
Immunofluorescence Analysis.
Cells were cultured on coverslips, followed by fixation for 15 min at room temperature in 4% paraformaldehyde in PBS, permeabilization for 5 min at room temperature in 0.1% Triton X-100 in PBS, and blocking for 1 h at room temperature in 1% BSA in PBS. Slides were then incubated overnight at 4 °C with indicated antibodies. Slides were stained for 1 h at room temperature with the corresponding Alexa Fluor secondary antibodies (Invitrogen) and mounted with mounting medium with DAPI (Vector). Microscopic images were obtained with a Zeiss LSM 510 confocal microscope in the Harvard NeuroDiscovery Center (HNDC) optical imaging core, using constant exposure times for each channel in individual experiment. Signal intensity and colocalization were measured with ImageJ software. Magnification was ×630 unless otherwise indicated.
FISH.
Mig-6 DNA probe was extracted from BAC clone CTD-2289F6 (Invitrogen) and labeled by nick translation mix (Roche). The centromere-specific CEP1 probe (Abbott Laboratories) served as a ploidy reference. FISH signal evaluation and acquisition were performed manually using filter sets and software developed by Applied Spectral Imaging.
Statistical Analysis.
Statistical analysis was performed using the unpaired Student's t test. For all experiments with error bars, standard deviation was calculated to indicate the variation within each experiment, and values represent mean ± SD.
Supplementary Material
Acknowledgments
H. Ying is a recipient of the Marsha Mae Moeslein Fellowship from the American Brain Tumor Association. H. Zheng was supported by Helen Hay Whitney Foundation. K.L.S. is supported by a Postdoctoral Fellowship from the American Cancer Society (PF-07-039-01-CSM). R.W. is supported by a Mildred Scheel Fellowship (Deutsche Krebshilfe). J.M.S. is supported by a Ruth L. Kirschstein National Research Service Award Fellowship. J.-H.P. was supported by the Damon Runyon Cancer Research Foundation. Grant support comes from the Goldhirsh Foundation (R.A.D.) and from National Institutes of Health Grants RO1CA99041 (to L.C.), 5P01CA95616 (to L.C., and R.A.D), and CA119075 (to L.A.E.). R.A.D. is an American Cancer Society Research Professor supported by the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914930107/DCSupplemental.
References
- 1.Furnari FB, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinojima N, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 5.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 6.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 8.Fiorentino L, et al. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makkinje A, et al. Gene 33/Mig-6, a transcriptionally inducible adapter protein that binds GTP-Cdc42 and activates SAPK/JNK. A potential marker transcript for chronic pathologic conditions, such as diabetic nephropathy. Possible role in the response to persistent stress. J Biol Chem. 2000;275:17838–17847. doi: 10.1074/jbc.M909735199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pante G, et al. Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J Cell Biol. 2005;171:337–348. doi: 10.1083/jcb.200502013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick M, Bürger C, Funk M, Müller R. Identification of a novel mitogen-inducible gene (mig-6): Regulation during G1 progression and differentiation. Exp Cell Res. 1995;219:527–535. doi: 10.1006/excr.1995.1261. [DOI] [PubMed] [Google Scholar]
- 12.Ferby I, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 13.Jeong JW, et al. Mig-6 modulates uterine steroid hormone responsiveness and exhibits altered expression in endometrial disease. Proc Natl Acad Sci USA. 2009;106:8677–8682. doi: 10.1073/pnas.0903632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YW, et al. Evidence that MIG-6 is a tumor-suppressor gene. Oncogene. 2007;26:269–276. doi: 10.1038/sj.onc.1209790. [DOI] [PubMed] [Google Scholar]
- 15.Prekeris R, Yang B, Oorschot V, Klumperman J, Scheller RH. Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol Biol Cell. 1999;10:3891–3908. doi: 10.1091/mbc.10.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramaniam VN, et al. Preferential association of syntaxin 8 with the early endosome. J Cell Sci. 2000;113:997–1008. doi: 10.1242/jcs.113.6.997. [DOI] [PubMed] [Google Scholar]
- 17.Antonin W, et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bièche I, et al. Two distinct regions involved in 1p deletion in human primary breast cancer. Cancer Res. 1993;53:1990–1994. [PubMed] [Google Scholar]
- 19.Brodeur GM, Sekhon G, Goldstein MN. Chromosomal aberrations in human neuroblastomas. Cancer. 1977;40:2256–2263. doi: 10.1002/1097-0142(197711)40:5<2256::aid-cncr2820400536>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Asano T, et al. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 21.Kleer CG, Bryant BR, Giordano TJ, Sobel M, Merino MJ. Genetic changes in chromosomes 1p and 17p in thyroid cancer progression. Endocr Pathol. 2000;11:137–143. doi: 10.1385/ep:11:2:137. [DOI] [PubMed] [Google Scholar]
- 22.Moley JF, et al. Consistent association of 1p loss of heterozygosity with pheochromocytomas from patients with multiple endocrine neoplasia type 2 syndromes. Cancer Res. 1992;52:770–774. [PubMed] [Google Scholar]
- 23.Mori N, et al. Chromosome band 1p36 contains a putative tumor suppressor gene important in the evolution of chronic myelocytic leukemia. Blood. 1998;92:3405–3409. [PubMed] [Google Scholar]
- 24.Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim RH, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Nagakubo D, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 27.Vasseur S, et al. DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc Natl Acad Sci USA. 2009;106:1111–1116. doi: 10.1073/pnas.0812745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D, Makkinje A, Kyriakis JM. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J Biol Chem. 2005;280:2924–2933. doi: 10.1074/jbc.M408907200. [DOI] [PubMed] [Google Scholar]
- 30.Anastasi S, et al. Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene. 2005;24:4540–4548. doi: 10.1038/sj.onc.1208658. [DOI] [PubMed] [Google Scholar]
- 31.Anastasi S, et al. Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene. 2003;22:4221–4234. doi: 10.1038/sj.onc.1206516. [DOI] [PubMed] [Google Scholar]
- 32.Nishikawa R, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoscheck CM, Carpenter G. Down regulation of epidermal growth factor receptors: Direct demonstration of receptor degradation in human fibroblasts. J Cell Biol. 1984;98:1048–1053. doi: 10.1083/jcb.98.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells A, et al. Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science. 1990;247:962–964. doi: 10.1126/science.2305263. [DOI] [PubMed] [Google Scholar]
- 35.Grandal MV, et al. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis. 2007;28:1408–1417. doi: 10.1093/carcin/bgm058. [DOI] [PubMed] [Google Scholar]
- 36.Han W, Zhang T, Yu H, Foulke JG, Tang CK. Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer Biol Ther. 2006;5:1361–1368. doi: 10.4161/cbt.5.10.3226. [DOI] [PubMed] [Google Scholar]
- 37.Stoorvogel W, Strous GJ, Geuze HJ, Oorschot V, Schwartz AL. Late endosomes derive from early endosomes by maturation. Cell. 1991;65:417–427. doi: 10.1016/0092-8674(91)90459-c. [DOI] [PubMed] [Google Scholar]
- 38.Masui H, Wells A, Lazar CS, Rosenfeld MG, Gill GN. Enhanced tumorigenesis of NR6 cells which express non-down-regulating epidermal growth factor receptors. Cancer Res. 1991;51:6170–6175. [PubMed] [Google Scholar]
- 39.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: An emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 40.Luwor RB, et al. The tumor-specific de2-7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene. 2004;23:6095–6104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- 41.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 42.Friedman LM, et al. Synergistic down-regulation of receptor tyrosine kinases by combinations of mAbs: Implications for cancer immunotherapy. Proc Natl Acad Sci USA. 2005;102:1915–1920. doi: 10.1073/pnas.0409610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.