Abstract
The AcrB trimeric multidrug efflux transporter of Escherichia coli pumps out a very wide spectrum of compounds. Although minocycline and doxorubicin have been cocrystallized within the large binding pocket in the periplasmic domain of the binding protomer, nothing is known about the binding of many other ligands to this protein. We used computer docking to evaluate the interaction of about 30 compounds with the binding protomer and found that many of them are predicted to bind to a narrow groove at one end of the pocket whereas some others prefer to bind to a wide cave at the other end. Competition assays using nitrocefin efflux and covalent labeling of Phe615Cys mutant AcrB with fluorescein-5-maleimide showed that presumed groove-binders competed against each other, but cave-binders did not compete against groove-binders, although the number of compounds tested was limited. These results give us at least a hypothesis to be tested by more biochemical and genetic experiments in the future.
Keywords: computer modeling, drug efflux, multidrug resistance, RND transporter
The gene acrA of Escherichia coli was identified as the gene whose mutation makes the host strain hypersusceptible to acridine dyes (1). It was thought for a long time that the gene affects the global barrier property of the outer membrane (for example, see refs. 2, 3), until a 1993 cloning and sequencing effort found that the neighboring gene acrB codes for a homolog of CzcA, a known toxic metal cation efflux transporter (4), and the measurement of cellular accumulation of acriflavine provided evidence that AcrB is a multidrug efflux transporter (5). We now know that AcrB is a member of RND (resistance-nodulation-division) family of transporters (6) and forms a complex with AcrA, the periplasmic “adaptor” protein (7), and TolC (8), a member of outer membrane channel family involved in the export of small molecules and some proteins (9). This tripartite structure allows the direct efflux of drugs into the external medium, rather than into the periplasm, and makes AcrAB-TolC and its homologs a very efficient machinery for increasing drug resistance levels in Gram-negative bacteria (10, 11). The transporter AcrB, which presumably captures the drug molecules mainly from the periplasm (12), shows an extremely wide substrate specificity (Fig. 1). Based on the comparison of susceptibility of acrAB+ and ΔacrAB strains [measured by minimal inhibitory concentration (MIC)], AcrAB-TolC can pump out not only basic dyes (such as acriflavine and ethidium) but also most antibiotics (such as macrolides, fluoroquinolones, β-lactams, tetracyclines, chloramphenicol, rifampin, novobiocin, fusidic acid, but not aminoglycosides), detergents (such as bile salts, Triton X-100, SDS) (13). Even simple solvents such as hexane, heptane, octane, and nonane (14) or cyclohexane (15) are pumped out by AcrAB-TolC (Fig. 1). It is important to find out how such a diverse range of substrates are recognized by AcrB and to design antimicrobial agents that would not be pumped out quickly and better inhibitors of these pumps, for which there are already a few examples (16, 17).
Fig. 1.
Some known substrates of the AcrB efflux pump.
An obvious approach to this question of specificity would be the cocrystallization of AcrB with the drugs. AcrB is a large protein containing more than 1,000 residues, and was found to exist as a trimer through the pioneering crystallographic study of Murakami et al. (18). Each protomer consists of a transmembrane domain containing 12 transmembrane helices and an equally large periplasmic domain that in turn is composed of the porter domain (which is thought to carry out the export of drugs) and the TolC-docking domain (Fig. 2A). Earlier attempts with the wild-type (19) or a mutant AcrB (20) led to models where the drug molecules were found within the central cavity in the transmembrane domain or at the entrance of the large periplasmic cleft. However, there was no way to distinguish the binding to the correct binding site on the export pathway from the nonfunctional binding of these mostly lipophilic substrates to random hydrophobic pockets on the surface of AcrB molecule. In fact, site-directed mutagenesis of residues near the drug molecules within the central cavity produced no detectable change in AcrB activity (20). In 2006–2007, however, three laboratories published asymmetric trimer models of AcrB (21–23), in which the periplasmic domain of each protomer assumes a unique conformation, called Access, Binding, and Extrusion according to the terminology of the Murakami group. Most importantly, Murakami and associates (21) showed that substrates (minocycline or doxorubicin) are bound to the binding pocket that branches off the large tunnel that putatively functions for drug export (Fig. 2 B and C). Unlike the drug-binding sites previously reported in the symmetric AcrB model (19, 20), this particular site appears much more convincing for several reasons. (i) Unlike the previously reported sites that were wide open, this site is surrounded on most sides, except an opening into the tunnel (Fig. 2C). (ii) Only one protomer among the three, the Binding protomer, binds the drug molecule in this pocket, and this led to the proposal of the functionally rotating mechanism of transport (21–23), which was confirmed by biochemical studies (24–26). (iii) The open pocket of this binding site collapses in the Extrusion protomer, in which the path to the central funnel opens up. (iv) Many of the residues lining up this pocket, when mutated into alanine, decrease the pumping activity (27).
Fig. 2.
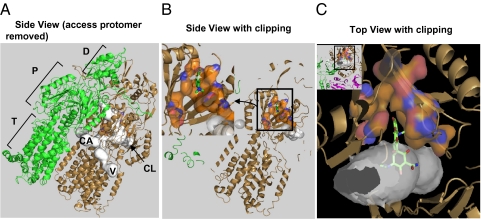
An overview of the substrate-binding pocket in the Binding protomer of AcrB. (A) The side view of the asymmetric trimer of AcrB (21), with the Access protomer removed, so that the putative substrate tunnels (detected by using the program Caver (41) and shown in white) in the Binding protomer (in the bronze color) can be seen. The three possible points of entry for substrates are shown: CL (periplasmic cleft), V (vestibule between protomers, close to the membrane surface), and CA (a large internal cavity in the transmembrane domain). The Extrusion protomer is shown in green. The porter (P) and the TolC-docking subdomains (D) of the periplasmic domain, as well as the transmembrane domain (T) are also identified. (B) The view similar to A, but the proximal portion clipped away, to reveal the binding pocket composed of F136, V139, F178, I277, A279, E280, P285, Y327, F610, V612, F615, F617, I626, and F628, shown as surface with carbons in orange. The minocycline molecule cocrystallized in PDB file 2DRD (21) is shown as green sticks. (Inset) The enlargement of the binding pocket, in a view similar to Figs. 3 and 4 and Figs. S1 and S2. (C Inset) A top view, in which the proximal part is clipped away. The Access protomer is shown in mauve. The binding pocket, shown enlarged in C, is a narrow extension of the substrate tunnel, as shown. A part of minocycline is in the pocket, whereas the hydrophilic portion protrudes into the large tunnel.
Despite these results, however, we know only how minocycline and doxorubicin bind to the site, and we have no information on the binding of dozens or even hundreds of other substrates. In this study, therefore, we tried a computer docking algorithm to predict how other substrates may interact with the binding pocket in the Binding protomer. This exercise showed us that there are two large areas that can bind ligands within this very large binding pocket and that many ligands prefer one area over the other. However, computer docking has many serious limitations. We therefore sought to evaluate the reliability of the docking predictions by using two experimental approaches recently developed in our laboratory (25, 28). The initial results, although limited in scope, indeed appear to suggest that the prediction by the computer docking may not be far from reality, at least in those cases examined experimentally.
Results
Computer-Predicted Ligand Binding to the Binding Protomer of 2DRD.
Studies of this type became possible for scientists outside the fraternity of computational biologists, like the present authors, with the recent availability of Autodock Vina (29), which is reported to accelerate the simulation process by a factor of 60 in comparison with the standard Autodock programs. We used the Autodock Vina program with the complex of Binding and Extrusion protomers of the minocycline-bound AcrB (21) (PDB file 2DRD) as the rigid “receptor” and examined the binding of about 30 known AcrB substrates, whose conformation was set as flexible. (The Extrusion protomer was included because it forms the back side of the binding pocket). The grid for docking was set as a 30 × 30 × 30 Å cube, centered at the position of C8 of the bound minocycline, which was removed for the docking exercise.
To examine the reliability of this approach, we first examined minocycline. It was shown to bind to the apo-protein approximately in a location and conformation found in the crystal structure, although this was the complex predicted to be the third most stable one (Fig. 3), suggesting either the protein model (at the resolution of 3.1 Å) is somewhat inexact or that the empirical energy scoring function of the program does not always work perfectly (29).
Fig. 3.
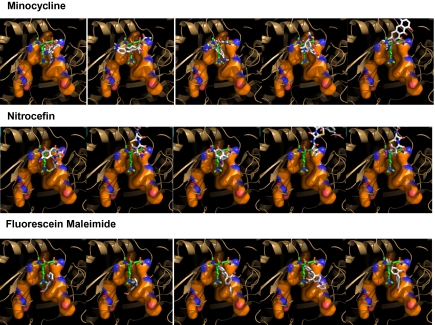
Top five binding modes of minocycline (Top), nitrocefin (Middle), and fluorescein maleimide (Bottom) for the Binding protomer of AcrB (2DRD) predicted by Autodock Vina. The view in this figure, as well as in Fig. 4 and Figs. S1 and S2, is the side view of the binding pocket (shown as a surface with carbons in orange), similar to Fig. 2B Inset. It is from the outside of the Access protomer, which was removed by clipping together with much of the Extrusion and Binding protomers. The computer-predicted poses of the ligands are shown in stick models in CPK colors, and the minocycline in the crystal structure (2DRD) is shown for reference in a stick model with carbons in green. The poses are arranged in the order of calculated binding energy, beginning with the one predicted to be most stable at extreme left.
We note that the minocycline molecule, both in the crystal structure (21) and in the docking poses, binds to the upper portion (farther away from the membrane surface) of the binding pocket (Fig. 3). Because this part of the pocket forms a narrow groove in this model (PDB 2DRD), we will call this binding mode “groove binding.” We shall see later that other ligands are predicted to bind to the lower part of the pocket, where the pocket becomes wider in this model. We will call this binding mode, “cave binding” for this reason.
In one test of the docking program, Autodock Vina was used to see if minocycline could bind to the corresponding position in the Extrusion protomer. Some binding poses were found, but the predicted binding energy was smaller (between −7.4 and −7.0 kcal here in contrast to −8.1 and −7.7 kcal with the Binding protomer) and none involved the groove, which is totally collapsed (Fig. S1; the calculated binding energies are shown in Table S1).
To test further the reliability of the docking exercise, we tried docking of minocycline with other published asymmetric structures of AcrB. With the unliganded crystal structure 2DHH published by Murakami and associates (21), the results were similar to 2DRD, although the minocycline pose similar to that in the crystal structure was achieved only in the structure calculated to be the fourth best according to the calculated binding energy (Fig. S1 and Table S1). With the unliganded, DARPin-bound structure 2J8S, which has the best resolution among the published AcrB structures, the minocycline pose similar to that found in the 2DRD structure again was achieved in the structure calculated to be the third best one (Fig. S1). These results seem to reinforce the idea that Autodock Vina predictions are indeed rather reliable. However, with the unliganded AcrB structure 2GIF, minocycline molecule was predicted to bind always to the part of the binding pocket far away from the groove (Fig. S1). This is apparently caused by the large flipping of F628 residue to close the back side of the pocket, making the lower part more efficient in binding ligands. (We note that the binding pocket itself here has a shape quite different from other models). Limitations such as this happen mainly because the binding occurs in a relatively flexible portion of the protein. Although the program allows us to rotate specified bonds in amino acid side-chains, there is no way to include the movement of the backbone. Thus the details of the predicted binding interactions of various drugs may give us only broad hints. Nevertheless, examination of the possible binding modes of many substrates can be instructive, in a way somewhat similar to the usefulness of comparison of many homologous sequences.
We therefore examined the predicted docking of many known substrates of AcrB, using the Binding and Extrusion protomers of 2DRD structure as receptor. Many substrates seem to bind, at least predominantly, with their hydrophobic domain bound to the upper groove of the binding site and with their hydrophilic groups often exposed in the very wide substrate tunnel. This group includes minocycline, nitrocefin, and fluorescein maleimide (Fig. 3), as well as tetracycline, chlortetracycline, doxycycline, novobiocin, erythromycin, nafcillin, rifampin, doxorubicin, levofloxacin, cholate, taurocholate, and rhodamine 6G (Fig. 4; for the top five poses predicted for each compound, see Fig. S2A, and for the calculated binding energies for the poses, see Table S2). This is very similar to the binding of minocycline (for structure, see Fig. 1), which is bound to the groove mainly by using its hydrophobic middle of the molecule and has its major polar domain (containing the acidic OH, a dimethylamino group, and an amide group) on top and extended into the wide-open tunnel and its minor polar group (a dimethylamino group) at the bottom close to the entrance of the cave area (Fig. 2 B and C). Thus the top and bottom of minocycline fit into more hydrophilic, larger cavities, which may even be filled with water. We also note that some of the poses predicted, for example the top poses for cholate and taurocholate, are totally outside the binding pocket. Presumably this is caused by the extensive grid we used for docking. Because the collapse of the pocket in the Extrusion protomer will not squeeze these ligands out, we do not consider them as functionally significant binding modes, and they have been disregarded in this analysis.
Fig. 4.
Some groove binders and cave binders predicted by Autodock Vina for the Binding protomer of AcrB. The numbers in parentheses indicate whether the pose shown is for the most favored one (1) or the second most favored one (2). The latter pose was chosen because the most favored pose showed binding outside the binding pocket (Fig. S2A).
Another group of compounds, including 1-(1-naphthylmethyl)piperazine (NMP), chloramphenicol, dodecylsulfate, ethidium, carbenicillin, cyclohexane, and cefamandole, appeared to bind predominantly to the lower (closer to the membrane) part of the binding pocket (Fig. 4 and Fig. S2B). As indicated earlier, we call this subdomain of the binding pocket “cave,” to distinguish this mode of binding from the minocycline-like binding to the groove. The members of this group, in contrast to most of the groove-binders, seem to have simpler structures, each containing one major lipophilic domain and no or only one charge.
Some compounds seem to use both subdomains. These include Phe-Arg-β-naphthylamide, Nile Red, Triton X-100, cephalothin, cephaloridine, oxacillin, fluorescein, linezolid, and ciprofloxacin (Fig. S2C). Finally, all binding poses for cefazolin, for which we could not find any evidence for AcrB-mediated efflux in our quantitative efflux assay (28), were outside the binding pocket (Fig. S2C), confirming that prediction of binding at these locations has no functional significance.
Competition with Nitrocefin Efflux.
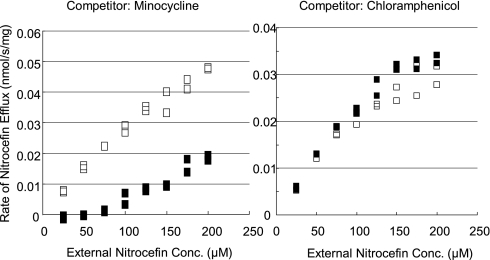
Computer prediction of ligand docking is just a prediction, subject to serious limitations as described above. We therefore sought to obtain biochemical evidence that the division of ligands into groove-binders and cave-binders has real functional consequences. We therefore examined the possible competition between the substrates by measuring the efflux of nitrocefin (a groove-binder; Fig. 3) quantitatively (28) in the presence and absence of potential competitors. Because the assay depends on the β-lactamase-catalyzed hydrolysis of nitrocefin, we could not use β-lactams as competitors. We were also unable to use large, lipophilic compounds such as macrolides, novobiocin, or rifampin, as they would be pumped out very efficiently by the functional AcrB and their periplasmic concentrations would become too low to compete against nitrocefin efflux. The assay also excluded the use of detergents and bile acids that would affect the outer membrane permeability. Thus we could use so far only a few compounds. Nevertheless, minocycline (50 μM)(a groove-binder; Fig. 3) strongly inhibited the efflux of nitrocefin (a groove-binder), whereas no inhibition at all was observed by chloramphenicol (a cave-binder; see Fig. 4) (Fig. 5). At 200 μM, both chlortetracycline and doxycycline (groove-binders; Fig. S2A) showed strong inhibition, whereas chloramphenicol did not (Fig. S3).
Fig. 5.
Competition by minocycline and chloramphenicol for AcrB-catalyzed efflux of nitrocefin. Cells were incubated with varying external concentrations of nitrocefin in the presence of a potential competitor (0 or 50 μM), and the efflux rate was calculated as described in (28). Open symbols, no competitor; filled in symbols, with competitor.
Competitive Binding of Fluorescein-5-Maleimide and Various Drugs.
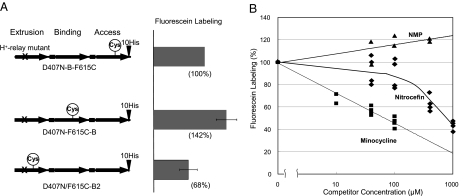
To study the binding of ligands to the AcrB binding pocket experimentally, we used the approach we introduced recently, that is the use of artificial AcrB trimers that are covalently linked together (25). In this giant molecule, we can change the conformation of one of the three protomers, for example into the Extrusion protomer by introducing mutations in the proton-relay residues, which we found to force that protomer to take the Extrusion conformation (30, 31). We can then expect that the neighboring protomers will be forced to take either the Access or Binding protomer conformations because the periplasmic domains of the three protomers in the AcrB trimer interact strongly with each other and the conformation of each protomer is complementary to those of the neighbors (21–23). To test this assumption, we changed one of the residues lining the binding pocket (Phe615) into cysteine and labeled this residue with fluorescein 5-maleimide. [Labeling of cysteine residues with maleimide reagents was pioneered by Lomovskaya's group with an RND family multidrug efflux transporter (32), and indeed resulted in inhibition of efflux of some, but not all drugs.] Because this labeling reagent is large and somewhat lipophilic, we expected that it would behave like a substrate of AcrB, and would enter the expanded binding pocket of the binding protomer most easily (Fig. 3). As shown in Fig. 6A, this prediction was borne out: the Binding protomer was labeled 42% better than the Access protomer in which the channel leading to the pocket is not fully open and more than twice better than the Extrusion protomer, in which the pocket is presumably collapsed.
Fig. 6.
Competition for fluorescein maleimide labeling of Phe615Cys mutant AcrB Binding protomer by some substrates of the pump. (A Left) composition of each construct tested. (Right) The extent of labeling was tested with 40 μM fluorescein maleimide and without competitors on all protomer units of the covalently linked AcrB trimer, in which the first protomer was forced to take the Extrusion conformation by the introduction of Asp407Asn mutation. (B) Potential competitors were added at different concentrations, and the extent of fluorescein labeling of the mutant AcrB was assessed as described in the Experimental Procedures, by using 20 μM fluorescein-5-maleimide.
Having convinced ourselves of the validity of this approach, we next labeled, again with fluorescein maleimide, the binding protomer in the D407N-F615C-B construct, in the presence of various drugs whose possible binding behaviors we examined in the computer docking study. One advantage of this approach is that the AcrB protein is inactive due to the mutation in the proton relay system (25) so that the potential competitor drugs are expected to remain in the periplasm, in contrast to the situation in the nitrocefin competition assays described above. However, because the positive outcome required the penetration, into the periplasm, of fluorescein maleimide, ligands that could affect the outer membrane barrier, such as detergents and bile salts, could not be used.
In the docking study, fluorescein maleimide binds to the “groove” with strong preference (Fig. 3). Thus we would expect that groove-binding drugs will efficiently compete with the binding of fluorescein maleimide, and thus the covalent labeling of the cysteine residue. In fact, as shown in Fig. 6, minocycline was the most effective competitor among the compounds tested. Nitrocefin, which also preferentially binds to the groove (Fig. 3), also showed a rather strong inhibition but at low concentrations the inhibition was weak. This is probably due to the hydrolysis of nitrocefin by the periplasmic AmpC β-lactamase (28), which produced, during the incubation, a red-colored hydrolysis product that binds presumably much less tightly to the binding pocket as it is less lipophilic (33). In contrast, a cave binder, NMP (Fig. 4), showed no evidence of inhibition. Although novobiocin, which appeared to prefer binding to the groove, also showed little sign of inhibition, this is likely to be due to the low permeability across the outer membrane of this large antibiotic and to its significant efflux by non-AcrAB pumps, such as MdtABC (34, 35). On the other hand, there were drugs that were classified as weak but significant competitors because, although they showed marginal (about 10%) inhibition at 100 μM, they showed clearly increasing degrees of inhibition at higher concentrations. In preliminary experiments tetracycline, a groove-binder (Fig. 4), belonged to this class, as the degree of inhibition reached around 30% at 400 μM. Similarly, erythromycin, another groove binder (Fig. 4), apparently belonged to this class, as the extent of inhibition reached 40% at 1 mM.
Discussion
One outstanding feature of some RND efflux pumps, especially the AcrB trimer of E. coli, is an extremely wide substrate specificity (12)(Fig. 1). The binding site for minocycline and doxorubicin was identified as a phenylalanine-rich pocket in the Binding protomer (21). It is likely, then, that the binding of ligands to this pocket would explain, to a large extent, the surprisingly wide substrate specificity of this transporter. However, we know the binding modes of only two ligands, and it is totally unknown how other ligands would bind to this pocket. We thus undertook the computer prediction of the possible modes of binding of other known substrates.
We first tried to confirm that the docking program, Autodock Vina (29), predicted the binding positions of minocycline correctly. Among the modes calculated to be energetically most stable with the apoprotein of minocycline-liganded AcrB structure (2DRD) (21), three were close to the position of minocycline in the crystal structure (Fig. 3). Attempted docking using crystal structures 2DHH and 2J8S produced at least one pose close to that in 2DRD. However, no such pose was found with the structure 2GIF (Fig. S1). We therefore must be conservative in interpreting the docking predictions.
We felt, nevertheless, that examination of a large number of ligands may produce useful pieces of information. Indeed, many ligands were found within the narrow groove in the part of the binding pocket farther away from the membrane surface, whereas others were always predicted to be associated with a wider cave area closer to the membrane surface. As was pointed out by Neyfakh in 2002 (36), by citing examples ranging from multidrug efflux regulators to odorant sensing proteins, the very large binding pockets of these proteins can accommodate many different ligands by using different subdomains within the pocket. In some cases, such sites are known even to allow the simultaneous binding of two drugs (37).
We used two approaches to test if these docking predictions could be reinforced by biochemical data. One was the quantitative efflux assay of nitrocefin (28), for which other drugs were used as potential competitors. A major advantage of this approach is that it uses the natural function of the efflux pump without its artificial modification. A weak point in this approach, however, was that competitors that diffuse across the outer membrane slowly could not be used, as their effective efflux would make their periplasmic concentrations vanishingly low. The other approach was the competition with the covalent labeling of the Phe615Cys residue, within the binding pocket, in a covalently linked AcrB trimer (25), whose one protomer was forced to take the Extrusion conformation thanks to the mutation introduced in the proton translocation pathway. Since the transporter is inactivated by this mutation, we could use low permeability competitors, although the assay is based on an artificial construct. Although only a limited number of ligands were tested for each of these approaches, the results were largely consistent with the division of ligands to groove-binders and cave-binders through the docking prediction (Results).
Another approach may be the site-directed mutagenesis of the residues within the binding pocket. Bohnert and coworkers (27) changed several Phe residues into Ala. Mutation of Phe610, which forms the back side of the groove, had the most wide-ranging effect on MICs of most substrates, a satisfying result. Phe178Ala mutation, on the wall of the groove, also lowered the MICs of groove-binders such as novobiocin and erythromycin, but mutations of the residues on the walls of the cave (Phe136, Phe615) had little effect on MICs of the cave-binders such as NMP and ethidium, perhaps because the cave area is so wide. Phe615Ala mutation unexpectedly affected only the MICs of macrolides, and a later study (38) showed the effect of 615–617 area on macrolide specificity. Although erythromycin was predicted to bind predominantly to the groove area, parts of the large molecule may spill over to the cave area (Fig. S2A), and thus this result is not necessarily contradictory to the docking predictions.
Because the well-known AcrB inhibitors are themselves substrates of this transporter, their effectiveness might be affected by the binding modes of antimicrobial agents. When the inhibitor of the RND efflux pumps, Phe-Arg-β-naphthylamide, was discovered by using the inhibition of levofloxacin efflux by MexAB-OprM as the indicator (16), surprisingly it was much less effective in inhibiting the efflux of ethidium and carbenicillin. These results are not for AcrB and Phe-Arg-β-naphthylamide was here classified as a mixed binder (Results). However, if we consider that this compound predominantly binds in the groove (Fig. S2C), the published results (16) are consistent with our data as levofloxacin is a groove-binder, whereas both ethidium and carbenicillin are typical cave-binders (Fig. 4 and Fig. S2B).
The effect of NMP (a typical cave-binder) in decreasing the MIC values of AcrB-overproducing E. coli varies widely depending on the agent used (17). In the presence of NMP, the AcrB-mediated efflux of chloramphenicol (a cave-binder) and of linezolid (a mixed binder) seems to be completely inhibited as the MICs are decreased to the levels shown by an AcrB-deficient strain. In contrast, NMP does not decrease the MIC to such a level for clarithromycin (likely a groove-binder on the basis of data on erythromycin). In contrast, the same study found that Phe-Arg-β-naphthylamide, which predominantly binds in the groove (see above), apparently inhibits the efflux of groove-binders such as clarithromycin or rifampin extremely strongly, yet not so strongly the efflux of linezolid or chloramphenicol.
We believe, on the basis of these observations, that computer prediction of substrate docking is a useful approach for our understanding of the substrate selectivity in the RND-pump-mediated multidrug efflux, although much more direct molecular genetic and biochemical experiments are needed to reinforce such predictions.
Experimental Procedures
Computer Prediction of Ligand Binding.
This was carried out by using Autodock Vina (29), with the exhaustivity parameter set at 16 (twice the default value) to cover the possible space as exhaustively as possible. The ligand files were prepared by using the Autodock Tools (http://autodock.scripps.edu) by accepting all rotatable bonds as such. The protein (receptor) files were derived from the binding protomer and the extrusion protomer of the AcrB protein, PDB files 2J8S, 2GIF, 2DHH, and the ligand-free version of 2DRD. With 2DHH and 2DRD, polar hydrogens could not be added to a few residues, mostly on the external surface of the protein, and the side-chain conformations of these residues had to be modified to the most stable one, using Swiss PDB viewer (http://spdbv.vital-it.ch). Furthermore, because N and O atoms in the amide structures of Asn and Gln are practically indistinguishable in crystal structures, each protein file was improved by the flipping of the amide structures at the Molprobity website (http://molprobity.biochem.duke.edu) (39). During this process, flipping was accepted when the program indicated “clear evidence for flipping,” and for other cases flipping was limited only to the situations where the flipped structure was more stable, by more than 2 kcal, than the unflipped structure.
Nitrocefin Efflux Assay.
This was carried out essentially as described earlier by using an AcrB overproducer HN1157 (28), except that possible competitors were added at 50 or 200 μM.
Competition with Fluorescein-Maleimide Labeling of the AcrB Binding Pocket.
To examine the labeling of the binding pocket in the Binding protomer with fluorescein-5-maleimide (Invitrogen), we first changed one of the putative proton-relay residues, Asp407, into a neutral residue Asn (30, 40), in the most N-terminal protomer of the linked AcrB trimer construct (25), so that this and other protomer units would take one of the desired conformations (Results). Site-directed mutagenesis here as well as elsewhere was carried out as described earlier (25). Derivatives of pS(D407N-B2) plasmid containing additionally the F615C mutation in one of the protomers were made similarly.
The covalent labeling of Phe615Cys was examined in the presence of potential competitors as follows. The strain BL21YBR (25) harboring one of these plasmids was grown in 20 mL LB at 30 °C overnight with 10 μM IPTG (without shaking) up to OD660 of about 0.5. Cells were centrifuged down, washed and resuspended at OD660 of around 20 in 10 mM Hepes-KOH, pH 7.5. The potential competitor was added, and 5 min later fluorescein maleimide was added to 20 or 40 μM, and the labeling was terminated after 30 min at room temperature by the addition of DTT to 10 mM. Cells were centrifuged, washed twice with 10 mM Hepes-KOH, broken by sonication in the presence of Complete, EDTA-free Protease Inhibitor Mixture (Roche), and the linked AcrB trimers with the C-terminal His-tag were isolated with Co2+-TALON resin essentially as described (24). The protein was analyzed by SDS/PAGE using 7.5% acrylamide after the removal of imidazole by gel filtration with Biogel P-6DG. The fluorescent bands were detected by phosphorimager (Typhoon 9400; Molecular Dynamics) with 488-nm and 520-nm filters for excitation and emission, respectively. The gel was stained with Coomassie Blue R-250, and the extent of staining was determined with a 1D scanner. The extent of fluorescent labeling was calculated by dividing the fluorescence intensity of the trimer band with the color intensity of Coomassie-stained band.
Supplementary Material
Acknowledgments
This study was supported in part by a grant (AI-09644) from the Public Health Service. C.C. was supported by the Robert and Colleen Haas Scholars Program. We thank A. N. Glazer for his encouragement.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001460107/DCSupplemental.
References
- 1.Nakamura H. Gene-controlled resistance to acriflavine and other basic dyes in Escherichia coli. J Bacteriol. 1965;90:8–14. doi: 10.1128/jb.90.1.8-14.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman WG, Jr, Leive L. Two mutations which affect the barrier function of the Escherichia coli K-12 outer membrane. J Bacteriol. 1979;139:899–910. doi: 10.1128/jb.139.3.899-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leive L, Telesetsky S, Coleman WG, Jr, Carr D. Tetracyclines of various hydrophobicities as a probe for permeability of Escherichia coli outer membranes. Antimicrob Agents Chemother. 1984;25:539–544. doi: 10.1128/aac.25.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nies DH, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma D, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 7.Dinh T, Paulsen IT, Saier MH., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen IT, Park JH, Choi PS, Saier MH., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 10.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci USA. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 14.Tsukagoshi N, Aono R. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol. 2000;182:4803–4810. doi: 10.1128/jb.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomovskaya O, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohnert JA, Kern WV. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother. 2005;49:849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 19.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 20.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: A crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 22.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 23.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J Bacteriol. 2007;189:8677–8684. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeger MA, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 27.Bohnert JA, et al. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2009;106:5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su CC, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol. 2006;188:7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takatsuka Y, Nikaido H. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J Bacteriol. 2006;188:7284–7289. doi: 10.1128/JB.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao W, et al. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: The large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol Microbiol. 2002;46:889–901. doi: 10.1046/j.1365-2958.2002.03223.x. [DOI] [PubMed] [Google Scholar]
- 33.O'Callaghan CH, Morris A, Kirby SM, Shingler AH. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol. 2002;184:4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol. 2002;184:4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neyfakh AA. Mystery of multidrug transporters: The answer can be simple. Mol Microbiol. 2002;44:1123–1130. doi: 10.1046/j.1365-2958.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher MA, Miller MC, Brennan RG. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J. 2004;23:2923–2930. doi: 10.1038/sj.emboj.7600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wehmeier C, Schuster S, Fähnrich E, Kern WV, Bohnert JA. Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob Agents Chemother. 2009;53:329–330. doi: 10.1128/AAC.00921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovell SC, et al. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 40.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Damborský J, Petrek M, Banás P, Otyepka M. Identification of tunnels in proteins, nucleic acids, inorganic materials and molecular ensembles. Biotechnol J. 2007;2:62–67. doi: 10.1002/biot.200600208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.