Abstract
Oscillations in patterns of expression of a large fraction of yeast genes are associated with the “metabolic cycle,” usually seen only in prestarved, continuous cultures of yeast. We used FISH of mRNA in individual cells to test the hypothesis that these oscillations happen in single cells drawn from unsynchronized cultures growing exponentially in chemostats. Gene-expression data from synchronized cultures were used to predict coincident appearance of mRNAs from pairs of genes in the unsynchronized cells. Quantitative analysis of the FISH results shows that individual unsynchronized cells growing slowly because of glucose limitation or phosphate limitation show the predicted oscillations. We conclude that the yeast metabolic cycle is an intrinsic property of yeast metabolism and does not depend on either synchronization or external limitation of growth by the carbon source.
Keywords: chemostat, fluorescence in situ hybridization, mRNA, single cells
Some essential reactions and even entire pathways in cellular metabolism are in fundamental chemical conflict with each other. It has long been supposed that metabolic incompatibilities of this kind underlie the spatial segregation of different metabolic functions into organelles bound by membranes, such as chloroplasts and mitochondria. However, spatial segregation is not the only way found in evolution that addresses metabolic incompatibilities. Temporal segregation and combinations of temporal and physical separation are found among the nitrogen-fixing bacteria, and plants naturally separate photosynthetic activities from respiratory activities according to the daily light cycle.
The phenomenon now known as “metabolic cycling” was first described in dense chemostat cultures of budding yeast, Saccharomyces cerevisiae, about 50 years ago (1–3). Highly regular oscillations in the levels of dissolved oxygen can be observed conveniently in chemostats that have been glucose starved and re-fed. These oscillations reflect periodic consumption of oxygen by the population and can persist indefinitely as long as the chemostat is fed. Recently, two groups (4, 5) conducted genome-wide gene-expression studies on such synchronized populations and found that a substantial fraction of all yeast genes exhibit periodic expression that is in rhythm with the oscillations in dissolved oxygen. Analysis of the expression patterns suggested that in such metabolic-cycling populations there is a temporal separation in the expression of genes involved in oxidative metabolism (e.g., peroxisomal functions) from that of genes encoding proteins associated with growth in cell mass (e.g., ribosomal proteins). The number of periodically expressed genes is much larger than that observed in synchronization of the classically defined cell-division cycle (using mating factors or cell-division-cycle mutants). The connection between the metabolic cycle and the cell-division cycle is still not entirely clear.
An inference, made by both groups, that cycling might be connected with the sensitivity of replicating DNA to oxidative damage caused by reactive oxygen species produced by some kinds of oxidative metabolism (e.g., β-oxidation of fatty acids) was fortified by subsequent observation of increased mutation rates in metabolic-cycling mutants that have lost the ability to restrict DNA replication to particular “safe” phases of the metabolic cycle (6). This explanation would conform with the idea that, in organisms with a metabolism adapted to earth's ancient reducing atmosphere, metabolic cycling evolved along with respiratory metabolism as the levels of oxygen in the atmosphere rose to their current levels. This reasoning assumes that metabolic cycling and the attendant periodic expression of so many yeast genes is an intrinsic property of yeast metabolism. However, nothing in these experiments could distinguish this situation from the possibility that the phenomenon is merely a consequence of the special conditions that produce synchrony.
A parallel investigation (7, 8) of genome-wide gene expression as a function of growth rate in unsynchronized chemostat cultures of the same yeast strain showed a remarkably similar division of genes on the basis of correlation of their expression levels with growth rate. Specifically, the genes whose expression was found to be negatively correlated with growth rate (i.e., genes expressed more strongly in slower growth conditions) largely overlapped with those associated with the oxidative phase of the metabolic cycle in synchronized populations.
The simplest way to reconcile this coincidence in gene-expression patterns is to suppose that all yeast cells, even those that are not in a synchronized population, display a metabolic cycle; that is, that metabolic cycling is an intrinsic property of yeast metabolism. Over the many years of study of metabolic cycling in one form or another, aspects of metabolic oscillations have been observed that suggested an intrinsic cycle (1–3, 9) but have not provided definitive evidence of a cell-autonomous cycle in individual unsynchronized cells.
In this paper we present the results of FISH of individual mRNAs in single cells (10), which we carried out to test the hypothesis that individual cells in unsynchronized cultures display gene-expression profiles associated with metabolic cycling.
We selected pairs of periodically expressed genes that, according to the expression data from metabolically synchronized cultures (4), should be expressed coincidentally and thus have significant numbers of labeled mRNA molecules for both genes in the same individual cells of an unsynchronized population. We also chose other pairs of periodically expressed genes in synchronized cells that should be expected to have significant numbers of mRNA molecules only in different individual cells of an unsynchronized population.
Quantitative analysis of the FISH data shows excellent concordance with the predictions, indicating that metabolic cycling occurs in the cells of unsynchronized yeast populations. Further, FISH experiments using phosphate as the limiting nutrient show that the metabolic cycling in unsynchronized cells does not depend on carbon limitation.
Results
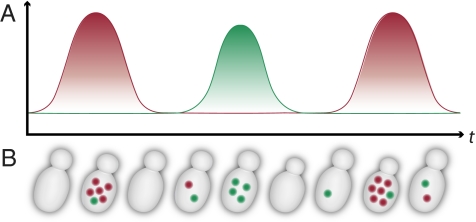
Cyclical expression (whether caused by the cell-division cycle, the metabolic cycle, or any other regular cycle) can be observed only in suitably synchronized populations if conventional measures of gene expression are used. However, single-cell methods have the potential to measure cyclical expression in unsynchronized populations, because individual cycling cells in an unsynchronized population are equivalent to cells drawn at random points in time from a synchronized population. Only if two genes are expressed coincidentally in the cycle will their mRNAs appear together in the same individual cell (Fig. 1). This principle makes it possible to predict which pairs of genes should be found coincidentally in the same individual cell from the patterns of gene expression in synchronized cultures. Thus we predict the following:
Fig. 1.
Principle of the FISH assay. (A) mRNA abundance (vertical axis) as a function of time (horizontal axis) for two anticorrelated, oscillating genes in a synchronized culture. (B) Expected mRNA abundance using FISH (colored spots) as a function of time for two anticorrelated, oscillating genes in a single cell.
First, only a fraction of the individual cells from an unsynchronized population should contain significant numbers of mRNA copies from any gene chosen on the basis of its robust cyclical expression in synchronized populations. (Most of the individual cells in an unsynchronized population should be in phases of the cycle in which such genes are expressed weakly, if at all.)
Second, individual cells from unsynchronized populations often should contain significant numbers of the mRNAs of both of a pair of genes whose expression is coincident in the metabolic cycle of synchronized cells.
Third, individual cells from unsynchronized populations should contain significant numbers of the mRNAs of one or the other, but not both, of a pair of genes whose expression peaks in different phases of the metabolic cycle in synchronized cells.
We required a quantitative method for measuring gene expression in individual yeast cells. To this end, the multicolor FISH method (11) served us very well. In this method, single mRNAs appear as diffraction-limited spots within cells, as thoroughly documented previously (12). We made some adaptations and improvements, mainly to deal with the very low levels of gene expression we encountered. In the end, some improvements in the algorithms for automated analysis of the images were required to produce the primary useful data, namely the number of mRNA molecules in each cell corresponding to each of a pair of genes.
We used chemostats as a source of unsynchronized, exponentially growing yeast cells. We chose a slow growth rate because the distinctions in gene expression between the genes associated with different phases of the metabolic cycle are expected to increase at lower growth rates (8). All cultures were grown in a defined medium at a dilution rate of 0.1 volumes per hour, corresponding to a doubling time of about 7 h. We used both glucose-limited and phosphate-limited chemostats under conditions identical to those described in ref.8. Dissolved oxygen was monitored continuously to verify that the cultures we used showed no detectable synchronous metabolic cycling (Fig. S1).
Discrete data collected from individual unsynchronized cells are fundamentally different from continuous data collected from synchronized cell populations (Methods and ref. 13). Therefore we adopted an analytical framework to compare statistically coincident expression in metabolically synchronized cultures and unsynchronized cells. We measured the Pearson correlation of each gene pair using the entire data set. To interpret these single-cell correlation coefficients better, we calculated the range of values that the correlation coefficient can assume given our discrete, single-cell data. This range is truncated relative to the full range that can be obtained using continuous data from synchronized cultures, partly because so many cells contain no signal for either gene. We also show an alternate measure of coincidence that consists of the full data with the zero-signal cells removed. The final, key step was to compare the single-cell correlations with those of Tu et al. (4) by calculating the Pearson correlation between them (correlation-of-correlations in Tables 1–3). A positive outcome in this test indicates that the correlations are ordered the same way in unsynchronized single cells and synchronized cultures.
Table 1.
Cell-cycle gene correlation measured in single asynchronous cells from glucose-limited chemostat cultures and in synchronized populations
| Gene pair | Synchronized population | Single asynchronous cells | ||
| Range | Full data | High signal | ||
| MCD1, CLB2 | −0.85 | −0.073, 0.93 | 0.034 | −0.42 |
| MCD1, POL30 | 0.86 | −0.14, 0.98 | 0.75* | 0.71* |
| CLB2, POL30 | −0.77 | −0.19, 0.92 | −0.041† | −0.2† |
| POL30, SUR7 | −0.73 | −0.17, 0.91 | −0.048† | −0.17† |
| Correlation-of-correlations | 0.99 | 0.99† | ||
Pairwise correlation between cell cycle–regulated genes in single, unsynchronized cells and synchronized populations. Synchronized population correlations were calculated using microarray data from cdc15-synchronized cultures (15). The Range column lists the minimum and maximum correlation coefficients obtainable given the mRNA count distributions from FISH data. For each gene pair, the Pearson correlation across single cells was calculated using FISH data from all cells (Full data) and from the subset of cells with nonzero FISH signal (High signal).
*, P ≤ 0.005; †, 0.005 < P ≤ 0.005–0.05.
Table 3.
Metabolic-cycle gene correlation measured in single asynchronous cells from glucose-limited chemostat cultures and in synchronized populations
| Gene pair | Synchronized population | Single asynchronous cells | ||
| Range | Full data | High signal | ||
| CTS1, SCW10 | −0.73 | −0.1, 0.94 | −0.08* | −0.24* |
| GAS1, HXK2 | 0.80 | −0.22, 0.99 | 0.42* | 0.3* |
| NOP1, SNU13 | 0.98 | −0.34, 0.95 | 0.31* | 0.16* |
| PFK26, SUR4 | −0.84 | −0.16, 0.98 | 0.18 | 0.016 |
| HXK2, YGP1 | −0.80 | −0.15, 0.97 | 0.02 | −0.13 |
| Correlation-of-correlations (glucose-limited) | 0.84 | 0.84 | ||
| Correlation-of-correlations (phosphate-limited) | 0.93 | 0.95 | ||
Correlation of metabolic cycle–regulated genes in glucose-limited chemostats. These gene pairs also were measured in phosphate-limited chemostats (first five rows of Table 2). For comparison, we show the correlation-of-correlations for these pairs under glucose limitation and under phosphate limitation. The Range column lists the minimum and maximum correlation coefficients obtainable given the mRNA count distributions from FISH data. For each gene pair, the Pearson correlation across single cells was calculated using FISH data from all cells (Full data) and from the subset of cells with nonzero FISH signal (High signal).
*, P ≤ 0.005.
We used the time-series DNA microarray-based gene-expression data from synchronized cultures [metabolic cycle (4, 5, 14) and cell division cycle (15, 16)] to choose pairs of genes for which we expected coincident expression, or the lack of it, in unsynchronized cells. In addition, we prioritized our selections based on relatively high abundance of mRNAs according to several genome-wide studies using microarrays (17–19) or high-capacity sequencing (20). Where possible, we also favored genes whose mRNAs had been reported to have relatively short half-lives (Table S1). Probe sequence design and details are given in Dataset S1.
We used FISH to assess quantitatively the mRNA content in individual cells from slowly growing unsynchronized cultures.
Analysis of Coincident Expression Based on the Classical Cell-Division Cycle.
As a proof of concept for analytical methods as well as the technology, we studied expression of four genes (MCD1, POL30, CLB2, and SUR7) whose periodic expression during the yeast cell-division cycle that operates in every growing cell is well established. MCD1 encodes a cohesin subunit whose mRNA abundance peaks during the S phase; POL30 encodes PCNA, whose mRNA abundance also peaks during S phase; CLB2 encodes a B-type cyclin whose mRNA peaks during G2 and M; and SUR7 encodes a plasma membrane protein whose mRNA abundance also peaks during G2 and M.
Our expectation in cells from an unsynchronized population was that we frequently should find cells with mRNA from neither gene, frequently should find mRNAs in the same cell for using FISH probes for MCD1 and POL30 (the two S-phase genes) and for CLB2 and SUR7 (the two G2/M genes), but only rarely, if ever, should find mRNAs in the same cell using pairs of probes such as CLB2 and POL30 or SUR7 and POL30 (i.e., one S-phase gene and one G2/M-phase gene).
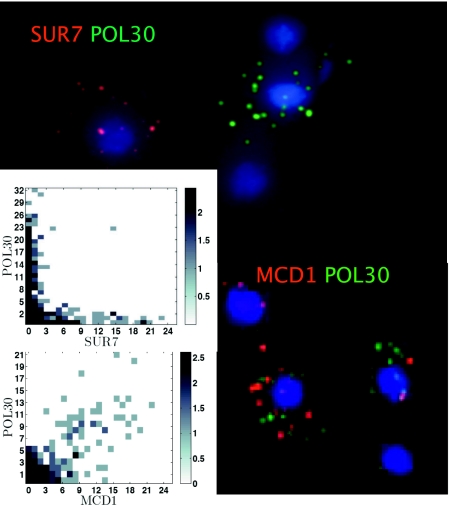
Fig. 2 shows the results with one pair (MCD1, POL30) for which coexpression was expected and another (SUR7, POL30) for which it was not. The figure summarizes data that include mRNA counts from two experiments for each pair. Hybridizations using probe pairs labeled with one or another cyanine (Cy) dye (dye-swaps) give equivalent results. (A third control gene labeled with Cy5 typically appears in >95% of cells; Fig. S2). In each case an image of a few selected cells is shown, along with a 2D histogram of the joint empirical distribution for the mRNA counts derived from >200 cells. It is clear that in both cases not all cells are labeled with probes from either gene: One sees the blue corresponding to the DAPI nuclear stain, but no red or green spots corresponding to the labeled Cy3 or Cy3.5 DNA probes. When such spots appear, the numbers are small: We rarely found more than about 40 spots and more commonly found 10 or fewer. This result is entirely consistent with expectation, because there is no doubt that the cells in this unsynchronized population growing at steady state in a chemostat are nevertheless undergoing regular cell-division cycles, with each cell at a different point in its cycle.
Fig. 2.
FISH of mRNA from genes periodically expressed during the standard cell-division cycle. Maximum projection (z dimension) composite image from a portion of a single field in the Cy3, Cy3.5, and DAPI emission channels: SUR7 Cy 3.5 (red), POL30 Cy3 (green), and MCD1 Cy 3.5. DAPI (blue) stains the nucleus. (Inset) The joint distribution is plotted as a heat-map matrix plot. To improve the visibility while accurately representing the dynamical range, the frequency counts were shifted by 1 and were log2 transformed.
For the gene pair MCD1, POL30 (both expressed in S phase in synchronized cultures), most cells that are labeled with one probe are also labeled with the other, as is clear from the images and the histogram. If we calculate a raw correlation coefficient, understanding that this calculation includes many cells that contain no labeled mRNAs, we obtain a value of 0.75 (note range, −0.14, 0.98; see discussion above) (Table 1). If we remove the zero-signal cells, we arrive at the alternative estimate of 0.71. In both cases bootstrapping P values are below 10−4. These results can be compared with the overall correlation in the synchronized population of 0.86 (15), where expression noise interferes minimally with the signal. These data accord perfectly with our second expectation—namely, that single cells from unsynchronized populations should contain mRNAs of both genes if they are coexpressed in synchronized populations.
For the gene pairs CLB2, POL30 and SUR7, POL30 (i.e., cases in which coincidences are not expected, and the overall correlation in synchronized populations is about −0.7), we find that cells with abundant labeling of one of the mRNAs show essentially no labeling of the other. Here the raw correlations are only weakly negative (about −0.05, but close to the minimum possible of about −0.2), but P values are statistically significant. Again, this result is exactly in accord with the third expectation above—namely, that single cells from unsynchronized populations should not contain mRNAs of both genes when they are expressed at very different times in synchronized populations. From these results using well-characterized cell-cycle genes, we see what can be expected when synchronized population data are compared with single-cell FISH results: The range of correlation values is restricted by the mRNA distributions featuring so many cells with no FISH signal in them.
Analysis of Coincident Expression Based on the Metabolic Cycle.
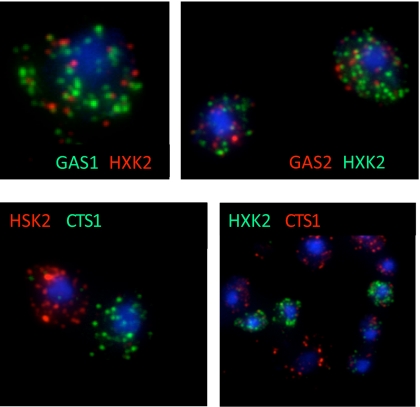
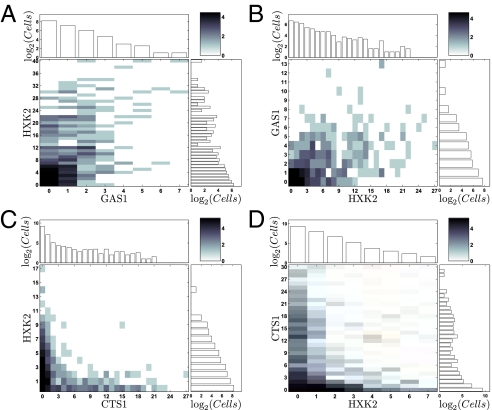
Fig. 3 A and B show selected cells from an unsynchronized culture (growing slowly under phosphate limitation) labeled with probes for genes GAS1 (encodes a beta-1,3-glucanosyltransferase) and HXK2 (encodes hexokinase 2), which show coincident expression in the metabolically synchronized cultures of Tu et al. (4). Fig. 3 C and D show selected cells in which the probes are directed to HXK2 and CTS1 (encoding endochitinase), which show expression at different points in the metabolic cycle in synchronized cells. (Note the equivalence of dye-swap results.) Histograms corresponding to these four experiments are shown in Fig. 4, and histograms for all gene pairs used in this study in are shown in Fig. S3. The data here, as in the case for the genes we chose on the basis of their periodic expression during the cell-division cycle, are clearly consistent with expectations if the unsynchronized cells are undergoing metabolic cycles. There are many cells that are totally unlabeled, many coincidences of labeling when genes in a pair are coexpressed in synchronized cultures (4), and very few coincidences of labeling when the genes in a pair are expressed at different times in the synchronized cultures.
Fig. 3.
FISH of mRNA from genes periodically expressed during the metabolic cycle. Maximum projection as in Fig. 2. (Upper Left) GAS1 Cy3 (green), HXK2 Cy3.5 (red). (Upper Right) Dye labels are reversed on these probes. (Lower Left) CTS1 Cy3, HXK2 Cy3.5. (Lower Right) Dye labels are reversed.
Fig. 4.
Joint and marginal distributions for gene pairs from the experiments shown in Fig.3. The joint distribution for each gene pair is plotted as a heat-map matrix plot (as in Fig. 2), and the corresponding marginal distributions for each mRNA are plotted above and to the right of the heat-map plot.
Table 2 shows a summary of the analyzed data for 22 metabolically cycling gene pairs (including the ones shown in Figs. 3 and 4), all growing slowly in limiting phosphate. The analysis was done in the same way as the analysis for the cell-division-cycle genes in Table 1. Specifically, the correlation-of-correlations is 0.65. In addition, a Wilcoxon rank-sum test comparing the single-cell correlations between negatively and positively correlated gene pairs (14) is significant (P = 0.002). These data are all clearly consistent with expectations, requiring us to conclude that individual unsynchronized cells growing slowly at steady state in a phosphate-limited chemostat are undergoing a metabolic cycle that is qualitatively and quantitatively similar to the cycle previously observed only in synchronized cultures growing under glucose limitation.
Table 2.
Metabolic-cycle gene correlation measured in single asynchronous cells from phosphate-limited chemostat cultures and in synchronized populations
| Gene pair | Synchronized population | Single asynchronous cells | ||
| Range | Full data | High signal | ||
| CTS1, SCW10 | −0.73 | −0.14, 0.98 | −0.12* | −0.37* |
| GAS1, HXK2 | 0.8 | −0.53, 0.98 | 0.35* | 0.3* |
| NOP1, SNU13 | 0.98 | −0.63, 0.88 | 0.25* | 0.17* |
| PFK26, SUR4 | −0.84 | −0.10, 0.79 | −0.02 | −0.38 |
| HXK2, YGP1 | −0.8 | −0.098, 0.83 | 0.015 | −0.22 |
| CTP1, SNU13 | 0.96 | −0.18, 0.98 | 0.15* | −0.0027* |
| CTS1, HXK2 | −0.76 | −0.30, 0.94 | −0.15* | −0.32* |
| GAS1, SCW10 | 0.82 | −0.18, 0.98 | 0.20* | 0.073* |
| NOP58, NOP1 | 0.98 | −0.75, 0.93 | 0.52* | 0.38* |
| RKI1, NOP1 | 0.97 | −0.57, 0.89 | 0.29* | 0.14* |
| SCW10, HXK2 | 0.9 | −0.17, 0.90 | 0.39* | 0.11* |
| NOP1, CTP1 | 0.95 | −0.65, 0.90 | 0.25* | 0.18* |
| SUR4, UTR2 | 0.9 | −0.16, 0.83 | 0.10* | −0.48* |
| OM45, PFK26 | 0.88 | −0.30, 0.85 | 0.12† | −0.016† |
| CTP1, RKI1 | 0.89 | −0.26, 0.85 | 0.057 | −0.43 |
| GAS1, YGP1 | −0.82 | −0.10, 0.65 | −0.04 | −0.081 |
| UTR2, OM45 | −0.75 | −0.22, 0.89 | 0.042 | −0.18 |
| PFK26, UTR2 | −0.75 | −0.32, 0.80 | 0.05 | −0.4 |
| OM45, SUR4 | −0.83 | −0.38, 0.96 | 0.09 | −0.089 |
| SCW10, OM45 | −0.75 | −0.26, 0.91 | 0.17 | 0.015 |
| SCW10, YGP1 | −0.8 | −0.13, 0.98 | 0.11 | −0.062 |
| HXK2, OM45 | −0.77 | −0.38, 0.98 | 0.25 | 0.08 |
| Correlation-of-correlations | 0.65* | 0.46† | ||
Correlation of metabolic cycle–regulated genes in phosphate-limited chemostats. Synchronized population correlations were calculated using microarray data from metabolically synchronized, glucose-limited chemostat cultures (4). The Range column lists the minimum and maximum correlation coefficients obtainable given the mRNA count distributions from FISH data. For each gene pair, the Pearson correlation across single cells was calculated using FISH data from all cells (Full data) and from the subset of cells with nonzero FISH signal (High signal).
*, P ≤ 0.005; †, 0.005 < P ≤ 0.005–0.05.
The Metabolic Cycle in Unsynchronized Cells Is Independent of the Nature of the Growth-Limiting Nutrient.
We used phosphate limitation in the investigations described above in part because we thought it less likely that we would adventitiously induce metabolic-cycle synchrony in the cultures, which, as may be recalled, required starvation and refeeding of glucose. Because gene expression based on growth rate is independent of the identity of the limiting nutrient (8), we believe that the limiting nutrient should not matter if our hypothesis of an autonomous metabolic cycle is correct.
We assessed the expression patterns of five pairs of genes in which the cells were drawn from unsynchronized populations growing under glucose limitation at the same rate that we used under phosphate limitation (compare Table 3 and the first five rows of Table 2). Also shown in Table 3 is a comparison of the correlation-of-correlations for glucose (0.84) and phosphate (0.93). It is clear that unsynchronized cells display the metabolic cycle under both growth regimes.
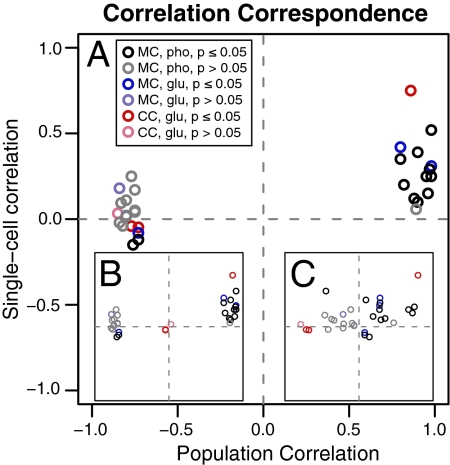
Finally, Fig. 5 summarizes graphically the relationship between the FISH correlations observed in single unsynchronized cells and the DNA microarray correlations observed in the synchronized cultures of Tu et al. (4) and Kudlicki et al. (14).
Fig. 5.
Correspondence of single-cell and synchronized population correlation coefficients (Full Data). (A) Metabolic (MC) single-cell correlations (black, phosphate; blue, glucose) compared with metabolic population correlations (4) and cell division (CC; red) compared with cell-division populations (15). (B) MC and CC compared with metabolic population correlations. (C) MC and CC compared with cell-division population correlations. Points with P > 0.05 are shown in muted colors.
Fig. 5A shows the relationship of the single-cell correlations to the population correlations, comparing cell-cycle genes with the synchronized cell-cycle and metabolic-cycle genes with the synchronized metabolic cycle. Fig. 5B compares all the correlations with the synchronized metabolic cycle: Here it is notable that the cell-division-cycle genes are significantly less correlated. Fig. 5C compares all the correlations with the synchronized cell-division cycle, and here it is notable that the metabolic genes are not correlated. These data fortify the conclusion that there is not a one-to-one correspondence between these cycles.
Discussion
The experiments described in the previous sections provide strong evidence that the periodic patterns of gene expression observed in the metabolic cycle (4, 5) are an intrinsic feature of yeast growth and are not dependent on the means used to achieve metabolic synchronization. Although direct support for the generality of this conclusion by these data across growth media and rates is limited (two limiting nutrients at a single relatively slow growth rate were studied with FISH) and also is limited even across genes (we studied 22 metabolically cycling gene pairs, all of which behaved as expected), our expectation now should be that the metabolic cycle likely underlies metabolism in a great variety, if not quite all, growth conditions in which yeast cells grow exponentially. The main reason for our confidence in the generality of cycling is the connection between the metabolic cycle and growth rate.
We found that we could observe the gene-expression oscillations directly equally well in cells growing in limited phosphate (and excess glucose) as in limited glucose. This result shows that metabolic cycling is not dependent on carbon-source limitation, even though virtually all previous observations of cycling in synchronized cultures have been conducted under carbon limitation [usually glucose, but also ethanol (21) or trehalose (22)].
Brauer et al. (8) suggested that the presence of the metabolic cycle quite generally in unsynchronized yeast cells might account for the remarkable relationship they discovered between the expression of as much as 25% of all genes and the growth rate. To correlate growth rate to metabolic cycling, one needs only to posit that the phase(s) of the metabolic cycle are partitioned in a growth-rate–dependent manner, such that the relative length of the “oxidative” phase is greater at slower growth rates. Accepting this relationship, we can see evidence for the metabolic cycle in all the conditions studied by Brauer et al. (8) (i.e., six limiting nutrients over a 7-fold range of growth rate, all with glucose as the carbon source), over the entire range of the diauxic shift in batch cultures (discussed in refs. 7, 8, and 23), during heat shock and a great variety of other stress responses in chemostats and batch cultures (24), by implication in all the data from ref. 25, and in cytoplasmic petite strains whose metabolism is confined to fermentation of glucose (24). Finally, apparently spontaneous synchronization of continuous cultures has been observed often, usually with glucose as a carbon source but also using ethanol (21) or trehalose (22).
Because under most conditions there seems to be an autonomous metabolic cycle in individual unsynchronized yeast cells, the cycles of starvation and growth, rather than cycling per se, must serve to induce synchrony in populations. Likewise, communication among cells or between cells and the environment need be invoked only for synchronization and not for cycling per se. On the other hand, the remarkable precision of the synchrony and its relationship to the instantaneous rate of growth must involve intracellular events and communication, the basis of which remains obscure. What we have learned is only that the apparently disparate phenomena of metabolic cycling and growth-rate correlation of gene expression must have a common mechanistic basis. Studies that involve perturbations in synchronized cultures (e.g., refs. 4, 22, 26) must distinguish whether the effects under study are affecting synchronization or are affecting the underlying cycling itself.
The suggestions of others (4–6, 27) regarding the evolutionary origins of the metabolic cycle all predicted that this cycle would be, as we have shown, an intrinsic feature of yeast cellular metabolism under virtually any circumstance. These same groups have provided some evidence that the cell-division cycle is gated by the metabolic cycle to avoid certain metabolic activities during DNA replication as well as evidence that mutation rates are increased when replication is allowed to occur despite these metabolic activities. It nevertheless should be recalled that we, as well as these authors, have observed that there is not a one-to-one correlation between the metabolic and cell-division cycles.
In general the idea that oxidative metabolism might be limited during DNA replication suggests derivation of energy from the glycolysis of stored carbohydrate, namely glycogen and/or, in the case of fungi, trehalose. Boer et al. (28) found that trehalose is one of only two intracellular metabolites (of more than 150 surveyed) that accumulate preferentially at slow growth rates. Given the connection between growth rate and the metabolic cycle (8), we suggest that trehalose accumulates cyclically to provide internal fermentable substrate during the phases of the metabolic cycle that do not involve reactive oxygen metabolism. The trehalose level might even be part of the system that gates entry into S phase of the cell-division cycle (29). The observation that growth on trehalose as the sole carbon source produces spontaneous metabolic cycling in batch cultures (22) may be relevant in this regard, as are the older observations that trehalose is metabolized preferentially during S phase of the cell-division cycle (30, 31).
The methods we have assembled to study metabolic cycling in single cells have made it possible to design relatively simple assays for metabolic cycling under virtually any conditions. The apparently ancient origin (32) of metabolic cycling strongly suggests that these methods can be used to discover metabolic cycling in other eukaryotes, including animal cells and tissues. Such studies may involve additional analytical and statistical theory and development, some of which are currently in preparation for publication separately.
Methods
Growth Conditions.
Diploid, S288c heritage cells, DBY12007, MATa/MATα, GAL2/GAL2, HAP1/HAP1 were used for all experiments. Cells were grown to steady state at a growth rate of 0.1 volumes per hour in phosphate-limited (20 mg/L) or glucose-limited (8 mg/L) chemostat medium (8) at 30 °C established in 500-mL vessels (Sixfors; Infors AG) containing 300 mL of culture. Pregrowth in batch phase was initiated from a 1/100 dilution of cells grown in the same medium. Continuous culture was initiated during exponential growth to avoid a starvation regimen that can induce synchronous metabolic cycling of the culture.
Fluorescence in Situ Hybridization.
FISH was performed as described by Zenklusen et al. (11) with slight modifications for chemostat-grown cells as described in SI Text and www.princeton.edu/genomics/botstein/protocols/.
Image Acquisition.
All images were taken using an IX81 inverted fluorescence microscope (Olympus) with a motorized stage (Prior), PlanApo TIRFM 100× oil objective with a numerical aperture of 1.45, X-Cite Exacte light source (EXFO), IX2-SHA motorized shutter, and ORCA II ER Mono CCD camera (Hamamatsu). Images were acquired using Slidebook 5.0 digital image acquisition software (Intelligent Imaging Innovations). Twenty-five images were taken with a 0.2-μm step size using filter sets SP100v2 (DAPI), SP102v2 (Cy3), SP103v2 (Cy3.5), and SP104v2 (Cy5) from Chroma Technology.
Image Processing.
Cell boundaries were determined using watershed segmentation of the DAPI image (33, 34). Spot locations were identified using top-hat transformation and contrast analysis (35). Spot intensities were measured using a Gaussian mask approach (36). The algorithms are described in SI Text.
Correlation Quantification.
We used the Pearson correlation coefficient to quantify expression correlation in single cells. We computed the correlation using both the full data set (“Full data”) and the subset of cells with nonzero FISH signal (“High-signal”). Correlation P values were estimated by bootstrap resampling with replacement. Pearson correlations calculated using discrete data from single cells are not necessarily numerically identical to those calculated using continuous data from synchronized populations (13). Therefore, to compare our vector of correlations with the vector of correlations published by Kudlicki et al. (14), we computed the Pearson correlation between the two correlation vectors and estimated the P values by bootstrap resampling without replacement.
Supplementary Material
Acknowledgments
We thank Benjamin Vernot for the python script for probe finding, Sandeep Raj for technical assistance, and Kara Dolinski for help with preparation of the manuscript. Research was funded by National Institutes of Health (NIH) Grant GM046406, NIH Grant GM57071 to R.H.S, and Grant GM071508 from the National Institute of General Medical Sciences Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002422107/DCSupplemental.
References
- 1.Chance B, Estabrook RW, Ghosh A. Damped sinusoidal oscillations of cytoplasmic reduced pyridine nucleotide in yeast cells. Proc Natl Acad Sci USA. 1964;51:1244–1251. doi: 10.1073/pnas.51.6.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck C, von Meyenburg HK. Enzyme pattern and aerobic growth of Saccharomyces cerevisiae under various degrees of glucose limitation. J Bacteriol. 1968;96:479–486. doi: 10.1128/jb.96.2.479-486.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh AK, Chance B, Pye EK. Metabolic coupling and synchronization of NADH oscillations in yeast cell populations. Arch Biochem Biophys. 1971;145:319–331. doi: 10.1016/0003-9861(71)90042-7. [DOI] [PubMed] [Google Scholar]
- 4.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 5.Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 7.Airoldi EM, et al. Predicting cellular growth from gene expression signatures. PLOS Comput Biol. 2009;5:e1000257. doi: 10.1371/journal.pcbi.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray DB, Klevecz RR, Lloyd D. Generation and maintenance of synchrony in Saccharomyces cerevisiae continuous culture. Exp Cell Res. 2003;287:10–15. doi: 10.1016/s0014-4827(03)00068-5. [DOI] [PubMed] [Google Scholar]
- 10.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 11.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 13.Ståhlberg A, Bengtsson M. Single-cell gene expression profiling using reverse transcription quantitative real-time PCR. Methods. 2010 doi: 10.1016/j.ymeth.2010.01.002. 10.1016/j.ymeth.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Kudlicki A, Rowicka M, Otwinowski Z. SCEPTRANS: An online tool for analyzing periodic transcription in yeast. Bioinformatics. 2007;23:1559–1561. doi: 10.1093/bioinformatics/btm126. [DOI] [PubMed] [Google Scholar]
- 15.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O, et al. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4:223–233. doi: 10.1038/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura F, et al. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics. 2008;9:574–587. doi: 10.1186/1471-2164-9-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuai L, Das B, Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:13962–13967. doi: 10.1073/pnas.0506518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipson D, et al. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 21.Keulers M, Suzuki T, Satroutdinov AD, Kuriyama H. Autonomous metabolic oscillation in continuous culture of Saccharomyces cerevisiae grown on ethanol. FEMS Microbiol Lett. 1996;142:253–258. doi: 10.1111/j.1574-6968.1996.tb08439.x. [DOI] [PubMed] [Google Scholar]
- 22.Jules M, François J, Parrou JL. Autonomous oscillations in Saccharomyces cerevisiae during batch cultures on trehalose. FEBS J. 2005;272:1490–1500. doi: 10.1111/j.1742-4658.2005.04588.x. [DOI] [PubMed] [Google Scholar]
- 23.Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu C, Brauer MJ, Botstein D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol Biol Cell. 2009;20:891–903. doi: 10.1091/mbc.E08-08-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z, Tsurugi K. Destabilization of energy-metabolism oscillation in the absence of trehalose synthesis in the chemostat culture of yeast. Arch Biochem Biophys. 2007;464:350–358. doi: 10.1016/j.abb.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 27.Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 28.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futcher B. Metabolic cycle, cell cycle, and the finishing kick to start. Genome Biol. 2006;7:107–111. doi: 10.1186/gb-2006-7-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Küenzi MT, Fiechter A. Changes in carbohydrate composition and trehalase-activity during the budding cycle of Saccharomyces cerevisiae. Arch Mikrobiol. 1969;64:396–407. doi: 10.1007/BF00417021. [DOI] [PubMed] [Google Scholar]
- 31.Paalman JW, et al. Trehalose and glycogen accumulation is related to the duration of the G1 phase of Saccharomyces cerevisiae. FEMS Yeast Res. 2003;3:261–268. doi: 10.1111/j.1567-1364.2003.tb00168.x. [DOI] [PubMed] [Google Scholar]
- 32.de Bivort BL, Perlstein EO, Kunes S, Schreiber SL. Amino acid metabolic origin as an evolutionary influence on protein sequence in yeast. J Mol Evol. 2009;68:490–497. doi: 10.1007/s00239-009-9218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer F. Topographic distance and watershed lines. Signal Process. 1994;38:113–125. [Google Scholar]
- 34.Eddins S. Cell segmentation. Steve on Image Processing. 2006 Available at: http://blogs.mathworks.com/steve/2006/06/02/cell-segmentation/. Accessed 1/26/2009. [Google Scholar]
- 35.Grigoryan AM, et al. Morphological spot counting from stacked images for automated analysis of gene copy numbers by fluorescence in situ hybridization. J Biomed Opt. 2002;7:109–122. doi: 10.1117/1.1428292. [DOI] [PubMed] [Google Scholar]
- 36.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.