Abstract
Inactivation of mismatch repair (MMR) is the cause of the common cancer predisposition disorder Lynch syndrome (LS), also known as hereditary nonpolyposis colorectal cancer (HNPCC), as well as 10–40% of sporadic colorectal, endometrial, ovarian, gastric, and urothelial cancers. Elevated mutation rates (mutator phenotype), including simple repeat instability [microsatellite instability (MSI)] are a signature of MMR defects. MicroRNAs (miRs) have been implicated in the control of critical cellular pathways involved in development and cancer. Here we show that overexpression of miR-155 significantly down-regulates the core MMR proteins, hMSH2, hMSH6, and hMLH1, inducing a mutator phenotype and MSI. An inverse correlation between the expression of miR-155 and the expression of MLH1 or MSH2 proteins was found in human colorectal cancer. Finally, a number of MSI tumors with unknown cause of MMR inactivation displayed miR-155 overexpression. These data provide support for miR-155 modulation of MMR as a mechanism of cancer pathogenesis.
Keywords: colorectal cancer, DNA repair, microRNA
Mismatched nucleotides may arise from polymerase misincorporation errors, recombination between heteroallelic parental chromosomes, or chemical and physical damage to the DNA (1). MutS homologs (MSH) and MutL homologs (MLH/PMS) are highly conserved proteins that are essential for the mismatch repair (MMR) excision reaction (2). In human cells, hMSH2 and hMLH1 are the fundamental components of MMR. The hMSH2 protein forms a heterodimer with hMSH3 or hMSH6 and is required for mismatch/lesion recognition, whereas the hMLH1 protein forms a heterodimer with hMLH3 or hPMS2 and forms a ternary complex with MSH heterodimers to complete the excision repair reaction (2, 3). Human cells contain at least 10-fold more of the hMSH2-hMSH6/hMLH1-hPMS2 complex, which repairs single nucleotide and small insertion-deletion loop (IDL) mismatches, compared with the hMSH2-hMSH3/hMLH1-hMLH3 complex, which primarily repairs large IDL mismatches (4–6). In addition to MMR, the hMSH2-hMSH6/hMLH1-hPMS2 components have been uniquely shown to recognize lesions in DNA and to signal cell cycle arrest and apoptosis (7, 8).
Mutations in the hMSH2, hMSH6, hMLH1, and hPMS2 core MMR genes have been linked to Lynch syndrome (LS), also known as hereditary nonpolyposis colorectal cancer (HNPCC) (9). These observations have provided considerable support for the mutator hypothesis, because defects in the MSH and MLH/PMS genes significantly increase cellular mutation rates, which then may drive the evolution of numerous oncogene and tumor-suppressor gene mutations found in cancers (10). The most obvious signature of a mutator phenotype is instability of simple repeat sequences or microsatellite DNA [ie, microsatellite instability (MSI)] (11, 12). Virtually all LS/HNPCC tumors display MSI resulting from mutation or inherited epigenetic inactivation of the core MMR genes (13–15). Most of the 10–40% of sporadic CRC, endometrial, ovarian, gastric, and urothelial tumors that display MSI are a result of acquired hMLH1 promoter methylation (16). Approximately 95% of MSI tumors can be accounted for, at least partially, by mutation and/or epigenetic inactivation of the core MMR components (14). The remaining 5%, as well as a significant proportion of the biallelic MMR inactivation mechanism, remain poorly understood.
MicroRNAs (miRs) are noncoding RNAs that play a role in the posttranscriptional regulation of >30% of the human genes controlling critical biological processes, including development, cell differentiation, apoptosis, and proliferation (17, 18). Overexpression of miR-155 has been observed in CRC (19–21), and appears to be more frequent in MSI than MSS CRCs (22). These data are consistent with the hypothesis that miR-155 might regulate components of the MMR machinery and, consequently, rates of mutation and MSI.
Results
hMLH1, hMSH2, and hMSH6 Are Targets of miR-155.
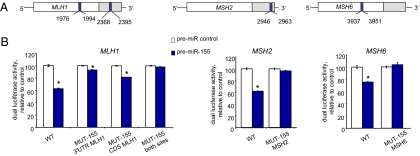
We used in silico prediction models to identify potential binding sites for miR-155 in the mRNA of the core MMR genes (23, 24). Two putative sites were found in hMLH1 using RNAhybrid (BiBiServ; NCBI NM_000249.2), one in the 3′-UTR and the other in the coding sequence (CDS) (NCBI NM_000249.2). One site was found in the 3′-UTR of hMSH2 using TargetScan (Whitehead Institute, MIT; NCBI NM_000251.1), and one site was found in the CDS of hMSH6 using RNAhybrid (BiBiServ; NCBI NM_000179.2) (Fig. 1A and Fig. S1). As a functional screen, we subcloned the coding and 3′-regions including the predicted miR-155 seed regions of MLH1, MSH2, or MSH6 downstream of the luciferase gene and recorded luciferase protein activity (25). We used MMR-proficient Colo-320 DM CRC cells for these studies, because they contain a low basal level of miR-155 (26). The Colo-320 DM cells were transfected with the luciferase reporter constructs and with a miR-155 precursor (premiR-155) or control precursor (premiR control). We observed luciferase activity reductions of 37%, 38%, and 24% in the presence of premiR-155 with constructs containing the miR-155 seed regions of MLH1, MSH2, and MSH6, respectively (P < 0.001) (Fig. 1B; see WT for each). No changes in luciferase activity were seen when the constructs contained a deletion of the miR-155 seed region (Fig. 1B; see MUT-155 for each construct). Interestingly, the effect of the individual miR-155–binding sites in the hMLH1 gene appeared to be additive (Fig. 1B; compare hMLH1 MUT-155 3′-UTR and MUT-155 CDS with WT).
Fig. 1.
hMLH1, hMSH2, and hMSH6 are direct targets of miR-155. (A) Locations of the target sites of miR-155 in the 3′ UTRs and/or the CDS of the indicated genes (see also Fig. S1). The base position is counted from the first nucleotide in the CDS. (B) Colo-320 DM cells were transfected with the phRL-SV40 construct as a control and either the luciferase construct WT or MUT-155 and with premiR-155 or premiR control. After 24 h, dual luciferase assays were performed. *P < 0.005 relative to premiR control.
Ribosomes have been recently shown to displace the miR-RISC complex from a CDS target site (27). As a result of this process, miRs cannot modulate a target protein. Both MLH1 and MSH6 contain CDS seed regions (Fig. 1 and Fig. S1). To examine the possibility of miR-RISC displacement, we cotransfected cell lines lacking MLH1 (HCT116) or MSH6 (DLD1) with a mammalian expression vector containing the corresponding full-length cDNA with or without the miR-155 precursor and examined protein expression (Fig. S2). We deleted the CDS miR-155 target site in the hMLH1 and hMSH6 cDNA sequences to examine whether expression might be affected by miR-RISC ribosome displacement. In-frame deletion of the CDS miR-155 seed sequence resulted in a truncated hMLH1 and hMSH6 protein (Fig. S2); the size reduction of the 160-kDa hMSH6 can be detected only in longer gel runs. Cotransfection of the MMR cDNAs with the miR-155 expression vector resulted in down-regulation of hMLH1 and hMSH6 proteins (Fig. S2; see hMLH1 WT and hMSH6 WT). Deletion of the CDS miR-155 seed sequence from the cDNA resulted in partial recovery of hMLH1 expression (Fig. S2; see hMLH1 MUT CDS) and complete recovery of hMSH6 expression (Fig. S2; see hMSH6 MUT). Mutation of both the hMLH1 CDS and 3′-UTR seed sequences resulted in a complete recovery of hMLH1 protein expression (Fig. S2; see hMLH1 double mutant). These results are qualitatively similar to the luciferase-based system containing the miR-155 seed sequence from hMLH1 and hMSH6 (Fig. 1B), and suggest that miR-RISC ribosome displacement is unlikely to be an issue in miR-155 modulation of MMR proteins.
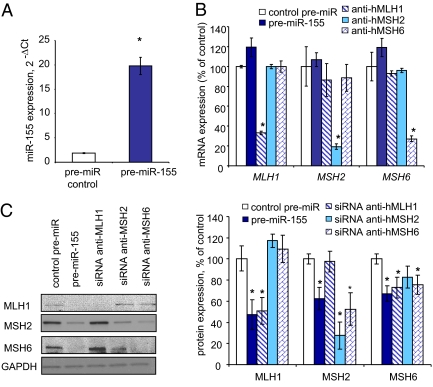
We examined the effect of miR-155 on endogenous MMR gene and protein expression in colo-320 DM cells (Fig. 2). A consistent increase in premiR-155 compared with a scrambled premiR control was determined by quantitative PCR (qPCR) for the transfected colo-320 DM cells (Fig. 2A). We found that the mRNA expression of hMLH1, hMSH2, and hMSH6 was unaffected by premiR-155 overexpression (Fig. 2B). In contrast, the hMLH1, hMSH2, and hMSH6 proteins were reduced by 53% ± 14% (P = 0.02), 37% ± 10% (P = 0.01), and 32% ± 7.4% (P = 0.004), respectively, in cells transfected with premiR-155 (Fig. 2C). To ensure that we could detect changes in mRNA expression, we performed siRNA knockdown studies in the colo-320 DM cells (Fig. 2 B and C). We found siRNA MMR gene-specific reduction of mRNA expression, as expected (Fig. 2B). The reduction in mRNA translated to a reduction in corresponding MMR proteins (Fig. 2C). Moreover, we found that the stability of the hMSH6 protein was linked to the expression of hMSH2 protein, as has been reported previously (28). These results suggest that miR-155 exerts its greatest affect on MMR proteins by posttranslational inhibition, a common characteristic of miR regulation (18, 29).
Fig. 2.
Overexpression of miR-155 decreases the expression of MLH1, MSH2, and MSH6 in CRC cells. Colo-320 DM cells were transfected with premiR-155, premiR control, or siRNA against selected genes for 48 h. (A) Transfection efficiency was confirmed by real-time PCR. miR-155 expression was normalized to the expression of RNU44. Error bars represent SEM. *P < 0.001 (n = 3). (B) miR-155 exerts a posttranscriptional effect on MMR core proteins. mRNA expression of indicated genes was normalized to that of vinculin. Error bars represent SEM.*P < 0.05 compared with control premiR (n = 3). (C) Representative blot of Western blot analysis along with the mean and SEM of three independent experiments. *P < 0.005 compared with control premiR.
To confirm the modulation of MMR proteins by miR-155, we examined MV4-11 cells that overexpressed miR-155 (Fig. S3). In these studies, MV4-11 cells were transfected with a sequence-specific locked nucleic acid (LNA) modified oligonucleotide that targets miR-155 knockdown (anti–miR-155; Exiqon), as well as a control LNA anti-miR (30). A 6.7-fold reduction in miR-155 was observed by qPCR (Fig. S1A) and confirmed by Northern blot analysis (Fig. S1B) when the LNA anti–miR-155 oligonucleotide was transfected compared with the nonspecific LNA anti-miR control. We also observed an increase in hMLH1, hMSH2, and hMSH6 proteins in MV4-11 cells transfected with LNA anti–miR-155 compared with the LNA anti-miR control transfections by Western blot analysis (Fig. S3). Together with the colo-320 DM miR-155 overexpression data, these results suggest that cellular miR-155 level has a direct effect on core MMR protein expression.
Overexpression of miR-155 Induces a Mutator Phenotype.
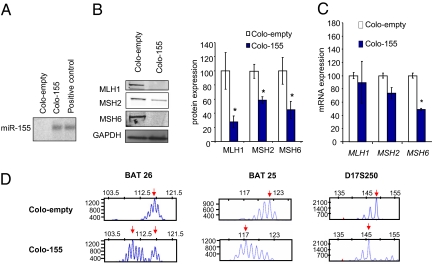
To assess the biological role of miR-155 in MMR regulation, we used a lentiviral vector system to generate stable clones of the Colo-320 DM CRC cells that overexpressed miR-155 (Fig. 3). We found that MLH1, MSH2, and MSH6 protein expression was reduced by 72%, 42%, and 69%, respectively, in Colo-155 DM cells overexpressing miR-155 (Colo-155) compared with cells expressing an empty vector (Colo-empty; Fig. 3B). We also observed a reduction in MSH6 mRNA (Fig. 3C). Because a direct effect on transcription is unlikely, these findings suggest that miR-155 may target other regulatory factors that affect transcription from the MSH6 promoter.
Fig. 3.
Stable clones with overexpression of miR-155, indicating the functional effect of miR-155 overexpression on CRC cell lines. MMR proficient Colo-320 DM cells were stably infected with a lentiviral vector overexpressing miR-155 (Colo-155) or an empty vector (Colo-empty). (A) miR-155 expression assessed by Northern blot analysis, with MV-411 cells used as positive controls. (B) Representative Western blot analysis results, along with mean ± SEM of three independent experiments. *P < 0.05 relative to controls. (C) mRNA expression of selected genes assessed by real-time PCR (normalized to vinculin). Error bars represent SEM. *P < 0.05 (n = 3). (D) Microsatellite analysis of Colo-155 and Colo-empty cells using the BAT-26 and BAT 25 (mononucleotide repeats) and D17S250 (dinucleotide repeat) markers. Size markers are given at the top of the panel.
Length changes in simple repeat sequences (ie, MSI) are a diagnostic indicator of MMR defects (11, 31). We examined MSI in the Colo-320 DM cells overexpressing miR-155 (Fig. 3D). All three of the markers examined displayed microsatellite instability. (Fig. 3D) (11, 31). These results suggest that miR-155 overexpression induces replication errors consistent with reduced or absent core MMR functions.
miR-155 Expression Inversely Correlates With hMLH1 and hMSH2 in CRC Tissues.
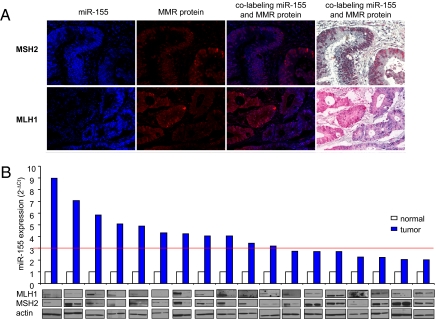
We examined the expression of miR-155, MLH1, and MSH2 in a tissue microarray containing 70 unselected cases of CRC and 5 benign intestinal lesions. A co-labeling method was used in which the LNA anti–miR-155 or nonspecific LNA anti-miR control was combined with immunohistochemical (IHC) antibodies to hMLH1 or hMSH2 (32). The miR-155 and MMR protein expression was scored positive when detected in >15% and >5% of cells, respectively (Fig. S4 A and B). More than 50% of the CRCs exhibited elevated expression of miR-155 (Fig. S4C). Reduced hMSH2 and hMLH1 expression was observed in 28% and 22% of the CRCs, respectively (Fig. S4C). Coexpression of miR-155 with MSH2 or MLH1 showed an inverse correlation, with r values of −0.74 for MSH2 (P < 0.001) and −0.6 for MLH1 (P < 0.001). miR-155 was not coexpressed with hMSH2 in 67% of the CRC tissues and was not coexpressed with hMLH1 in 50% of the CRC tissues. When the analysis was conducted with miR-155 positive CRC tissues only, a significant inverse correlation was still evident for both hMLH1 (P = 0.0003) and hMSH2 (P = 0.001). In the subgroup of tissues with miR-155 expression >50%, there was still an inverse correlation with an r value of −0.7 for MSH2 (P < 0.0002) and −0.55 for MLH1 (P < 0.005). Interestingly, in the CRC tissues that were positive for miR-155 and either hMSH2 or hMLH1, coexpression in the same cancer nest was never observed (Fig. 4A).
Fig. 4.
miR-155 expression is inversely related to MLH1 and MSH2 in CRC tissues. (A) Paraffin-embedded, formalin-fixed CRC tissues were incubated with LNA-probe anti–miR-155 or scrambled probe and IHC antibodies against MSH2 and MLH1. Representative photographs were captured with the Nuance system software (Ventana Medical System Inc.), with staining positive for both miR-155 and MSH2 or MLH1 shown. Blue and red staining identifies miR-155 and the target protein, respectively. No colocalization of miR-155 and MLH1 or MSH2 in the same cell nest is seen. (B) RNA and proteins extracted from fresh frozen human colorectal tissue. miR-155 expression was assessed by real-time PCR, and MMR protein expression was assessed by Western blot analysis. CRC with up-regulation (>2-fold) of miR-155 expression and down-regulation of MMR protein expression are shown. A strong correlation between the loss of hMLH1 and hMSH2 expression and miR-155 expression was increased by >3-fold in tumor compared with adjacent normal tissue (red line). Samples with an miR-155 cancer/normal ratio <3-fold displayed an uncertain effect on MMR expression.
We examined miR-155 and MMR protein expression in a cohort of 83 fresh frozen tumors for which cancer and normal adjacent tissues were available (Fig. 4B). We examined tumor and associated normal tissue by qPCR analysis and found that the miR-155 expression was increased by >2-fold in 18 of the specimens (22%). The expression of hMLH1 and MSH2 protein was measured by Western blot analysis in tumor tissue and associated normal tissue in these 18 specimens. A clear decrease in hMLH1 expression was observed in 12 of these specimens (67%), whereas a clear decrease in hMSH2 expression was observed in 11 (61%) (Fig. 4B). A threshold of uncertain effects on MMR expression appeared to occur when the miR-155 expression level dropped below a 3-fold increase compared with the associated normal tissue (Fig. 4B; red line). In general, miR-155 expression above this threshold appeared to down-regulate the expression of both hMLH1 and hMSH2. Together with the findings of the tissue array studies, these results strongly suggest that miR-155 overexpression in human tumors results in down-regulation of the core MMR proteins hMLH1 and hMSH2.
miR-155 Expression as a Cause of MMR Dysregulation in MSI Tumors.
MSI is acquired after the inactivation of both alleles of one of the core MMR genes (9). In LS/HNPCC carriers, the majority of mutations are found in the hMSH2 and hMLH1 genes (33). The “second hit” that leads to an MSI tumor in patients with LS/HNPCC is largely the result of loss of heterozygosity or somatic mutation of the unaffected allele (34). LS/HNPCC accounts for 2%–5% of all CRC and for one third of MSI tumors (14, 35–38). Approximately 10%–15% of sporadic CRC tumors display MSI that is largely (90%) a result of biallelic inactivation of the hMLH1 promoter (14, 36, 39). In an unselected series of 1,066 CRC patients, 135 (12.7%) were found to display MSI (14). Of these, 23 (5.9%) were determined to have germline mutation in one of the core MMR genes, and 106 (78.5%) were found to contain methylation of the hMLH1 promoter. Approximately 5% of these MSI tumors displayed loss of expression of at least one of the core MMR proteins with no clear genetic or epigenetic cause (14).
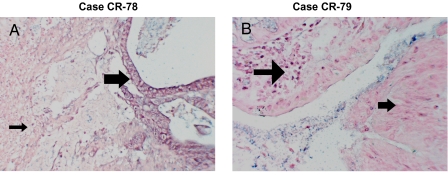
We retrospectively examined a series of 40 CRC that displayed MSI and IHC loss of expression of at least hMLH1 and hPMS2. Thirty-four of these tumors were found to have mutations in the hMLH1 gene sequence or methylation of the hMLH1 promoter. We examined six specimens that retained sufficient sample for miR-155 expression analysis by qPCR following laser capture microdissection (LCM) of tumor and adjacent normal tissue (Table S1). Because one specimen contained very little tissue, we examined miR-155 and hMLH1 expression by in situ and IHC analysis (Fig. 5). All six of the remaining MSI tumors displayed at least a 2-fold increase in miR-155 expression compared with adjacent normal tissue (Table S1). Overexpression of miR-155 in these specimens did not correlate with tumor grade or stage. Three of the samples demonstrated an miR-155 increase over the 3-fold threshold that generally results in reduced expression of hMLH1 and/or hMSH2. In addition, CR78 exhibited elevated miR-155 expression in >50% of the tumor tissue and a corresponding loss of hMLH1 expression (Fig. 5 A and B). These results are consistent with the conclusion that MSI tumors with unknown MMR defects may result from miR-155 overexpression.
Fig. 5.
miR-155 expression in hMLH1-negative tumors. miR-155 expression was assessed by in situ hybridization on paraffin-embedded tissues. (A) CR-78 cancer tissue (with unknown causes of MLH1 loss) showing strong miR-155 expression (large arrow), with stroma (small arrow) negative for miR-155 expression. (B) CR-79 tissue (MLH1 loss due to promoter methylation) showing faint expression of miR-155 only in inflammatory cells (large arrow), with no signal detected in cancer tissue (small arrow).
Discussion
The mutator phenotype that results from MMR dysfunction induces the acquisition of additional gene mutations that promote cancer progression (40). In addition to germline mutations, various pathogenic events, including promoter methylation (16) and reduced histone acetylation (41), result in reduced or absent expression of core MMR proteins, as do microenvironmental factors, such as inflammation and hypoxia (42, 43,). Our results suggest that miR-155 plays a role in this multifactorial regulation by causing down-modulation of the core MMR heterodimeric proteins MSH2-MSH6 and MLH1-PMS2. The simultaneous inhibition of these essential MMR components by miR-155 results in a mutator phenotype.
The significant effects of miR-155 on the MMR system are likely due to two phenomena. First, the stability of MMR protein is linked to its ability to form heterodimers; thus, the loss of hMSH2 and hMLH1 proteins results in destabilization of their respective heterodimeric complex proteins (28). Second, miR-155 appears to target the down-regulation of core MMR proteins. Together, these regulatory and stability alterations result in a significant increase in mutation rates. We cannot eliminate the possibility that miR-155 affects other related DNA repair proteins, enhancing the phenotypic effect of MMR defects through unrelated genomic processes.
Incomplete repression of MMR proteins by miR-155 is not unique to tumor-suppressor genes in cancer. A partial (50%) reduction in the expression of one allele of the APC (adenomatous polyposis coli) gene has been correlated with the development of CRC (44), and the reduced expression of a single allele of the TGFBR1 (TGF-β receptor I) gene also has been linked to CRC (45). Recently, miRs have been proposed to act as transactivating elements involved in allele and gene expression regulation (46, 47). Our results strongly support a role for miRs in the non-Mendelian regulation of MMR genes.
The complexity of miR regulation is enhanced by the tissue specificity and possible polymorphisms in the target sequences; for example, acquired mutations in the 3′ UTR of hMLH1 have been linked to disease relapse in patients with acute myeloid leukemia (48). We cannot exclude the possibility that miR-155 overexpression combined with acquired mutations in the 3′ UTR of MMR target genes contribute to the development of MSI in these selected patients. Moreover, well-defined mutations and/or epigenetic inactivation of MMR genes appear to be less frequent in specific subsets of MSI tumors, such as CRCs associated with inflammatory bowel disease (49, 50) and non-Hodgkin lymphoma associated with HIV infection (51). Our findings demonstrating down-regulation of core MMR genes suggest miR-155 overexpression as a potential alternative pathogenetic mechanism for these cancers. Although we found an inverse correlation between miR-155 and MMR protein expression in CRC, not all tumors with increased miR-155 expression are characterized by MSI. In human cells, several factors might account for the differing regulation of the core MMR proteins by miR-155, including a threshold expression effect or savage loops involved in the fine-tuning of miR and gene expression.
Patients with MMR-deficient tumors generally have a good prognosis but a lower survival advantage after 5-fluorouracil adjuvant chemotherapy compared with patients with MMR-proficient tumors (52). The contributing role of miR-155 in the down-regulation of the core MMR proteins suggests that miR-155 might be an important stratification factor in the prognosis and treatment of cancer patients. Although further confirmation of our findings is needed, our results suggest that miR-155 expression may be an additional analytical test for exploring the etiology of MSI tumors when the standard tests do not provide a conclusive diagnosis.
Materials and Methods
Colo-320 DM, HCT-116, and DLD1 cells were transfected using Lipofectamine 2000 (Invitrogen). Luciferase constructs were subcloned using primers in Table S2. Lentiviral vectors for miR-155 overexpression and empty vectors were generated by System Biosciences, according to the manufacturer's instructions. MSI was evaluated by genotyping analysis using diagnostic primers (7, 8). Genomic DNA from patients with MSI was sequenced as described previously (53). Statistical analysis results are expressed as mean ± SEM unless indicated otherwise. Comparisons between groups were performed using the two-tailed Student t test. Significance was accepted at a P value <0.05. Graphpad Prism version 5.0 was used for Pearson correlations.
More detailed information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
N.V. is supported by an American-Italian Cancer Foundation Postdoctoral Research Fellowship, and M.F is supported by a Kimmel Scholar Award. This work was supported by grants from the National Institutes of Health (GM080176 and CA067007 to R.F. and CA124541 and CA135030 to C.M.C.) and Associazione Italiana Ricerca sul Cancro (to S.V., N.V., and M.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002472107/DCSupplemental.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: American Society of Microbiology; 2006. 2nd Ed. [Google Scholar]
- 2.Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci USA. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 4.Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci USA. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannavo E, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 6.Räschle M, Marra G, Nyström-Lahti M, Schär P, Jiricny J. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 7.Fishel R. Signaling mismatch repair in cancer. Nat Med. 1999;5:1239–1241. doi: 10.1038/15191. [DOI] [PubMed] [Google Scholar]
- 8.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: Revising the mutator hypothesis. Cancer Res. 2001;61:7369–7374. [PubMed] [Google Scholar]
- 9.Boland CR, Fishel R. Lynch syndrome: Form, function, proteins, and basketball. Gastroenterology. 2005;129:751–755. doi: 10.1016/j.gastro.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 10.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 11.Dietmaier W, et al. Diagnostic microsatellite instability: Definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 12.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 13.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 14.Hampel H, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 15.Hitchins MP, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 16.Kane MF, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 17.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Bandrés E, et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng EK, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 21.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanza G, et al. mRNA/microRNA gene expression profile in microsatellite- unstable colorectal cancer. Mol Cancer. 2007;6:54–65. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 24.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler JM, et al. Mechanisms of inactivation of mismatch repair genes in human colorectal cancer cell lines: The predominant role of hMLH1. Proc Natl Acad Sci USA. 1999;96:10296–10301. doi: 10.1073/pnas.96.18.10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 29.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 30.Elmén J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 31.Boland CR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 32.Nuovo GJ, et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition: Update of the ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 35.Aaltonen LA, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 36.Cunningham JM, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salovaara R, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 38.Umar A, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herman JG, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perucho M. Tumors with microsatellite instability: Many mutations, targets and paradoxes. Oncogene. 2003;22:2223–2225. doi: 10.1038/sj.onc.1206580. [DOI] [PubMed] [Google Scholar]
- 41.Edwards RA, et al. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease–associated colorectal cancer. Cancer Res. 2009;69:6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo A, et al. Hypoxia-induced enrichment and mutagenesis of cells that have lost DNA mismatch repair. Cancer Res. 2001;61:7603–7607. [PubMed] [Google Scholar]
- 43.Mihaylova VT, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, et al. Small changes in expression affect predisposition to tumori-genesis. Nat Genet. 2002;30:25–26. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- 45.Valle L, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–1365. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahluwalia JK, Hariharan M, Bargaje R, Pillai B, Brahmachari V. Incomplete penetrance and variable expressivity: Is there a microRNA connection? Bioessays. 2009;31:981–992. doi: 10.1002/bies.200900066. [DOI] [PubMed] [Google Scholar]
- 47.de la Chapelle A. Genetic predisposition to human disease: Allele-specific expression and low-penetrance regulatory loci. Oncogene. 2009;28:3345–3348. doi: 10.1038/onc.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao G, Pan X, Gu L. Evidence that a mutation in the MLH1 3′-untranslated region confers a mutator phenotype and mismatch repair deficiency in patients with relapsed leukemia. J Biol Chem. 2008;283:3211–3216. doi: 10.1074/jbc.M709276200. [DOI] [PubMed] [Google Scholar]
- 49.Schulmann K, et al. Molecular phenotype of inflammatory bowel disease associated neoplasms with microsatellite instability. Gastroenterology. 2005;129:74–85. doi: 10.1053/j.gastro.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 50.Svrcek M, et al. Specific clinical and biological features characterize inflammatory bowel disease–associated colorectal cancers showing microsatellite instability. J Clin Oncol. 2007;25:4231–4238. doi: 10.1200/JCO.2007.10.9744. [DOI] [PubMed] [Google Scholar]
- 51.Borie C, et al. The mechanisms underlying MMR deficiency in immuno-deficiency-related non-Hodgkin lymphomas are different from those in other sporadic microsatellite-instable neoplasms. Int J Cancer. 2009;125:2360–2366. doi: 10.1002/ijc.24681. [DOI] [PubMed] [Google Scholar]
- 52.Ribic CM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mueller J, et al. Comprehensive molecular analysis of mismatch repair gene defects in suspected Lynch syndrome (hereditary nonpolyposis colorectal cancer) cases. Cancer Res. 2009;69:7053–7061. doi: 10.1158/0008-5472.CAN-09-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.