Abstract
Certain types of human papillomaviruses (HPVs) are etiologically linked to cervical cancer. Their transforming capacity is encoded by a polycistronic premRNA, where alternative splicing leads to the translation of functional distinct proteins such as E6, E6*, and E7. Here we show that splicing of HPV16 E6/E7 ORF cassette is regulated by the epidermal growth factor (EGF) pathway. The presence of EGF was coupled to preferential E6 expression, whereas depletion of EGF, or treatment with EGF receptor (EGFR) neutralizing antibodies or the EGFR inhibitor tyrphostin AG1478, resulted in E6 exon exclusion in favor of E6*. As a consequence, increased p53 levels and enhanced translation of E7 with a subsequent reduction of the retinoblastoma protein pRb could be discerned. E6 exon exclusion upon EGF depletion was independent from promoter usage, mRNA stability, or selective mRNA transport. Time-course experiments and incubation with cycloheximide demonstrated that E6 alternative splicing is a direct and reversible effect of EGF signal transduction, not depending on de novo protein synthesis. Within this process, Erk1/2-kinase activation was the critical event for E6 exon inclusion, mediated by the upstream MAP kinase MEK1/2. Moreover, siRNA knockdown experiments revealed an involvement of splicing factors hnRNPA1 and hnRNPA2 in E6 exon exclusion, whereas the splicing factors Brm and Sam68 were found to promote E6 exon inclusion. Because there is a natural gradient of EGF and EGF receptor expression in the stratified epithelium, it is reasonable to assume that EGF modulates E6/E7 splicing during the viral life cycle and transformation.
Keywords: cervical cancer, transformation, gene expression, signal transduction, cell differentiation
Particular types of human papillomaviruses (HPVs) such as HPV16, 18, 31, and 33, respectively, are the etiological agents for the development of anogenital tumors. Their transforming potential is encoded by the viral oncoproteins E6 and E7, where among other functions, E6 labilizes p53 and prevents apoptosis, and E7 promotes cell cycle progression by degrading the retinoblastoma protein pRb (1). HPV gene transcription is regulated by two main promoters, the early p97 and the late p670 promoter (2). Activation of either promoter is regulated by differentiation, resulting in the synthesis of polycistronic mRNAs, which are further regulated by differential splicing (3, 4). For polycistronic mRNAs, it is known that only the first ORF is translated efficiently when intercistronic distances are short (5).
Alternative splicing can be regulated dependent on the developmental stage or by extracellular stimuli, where an increasing number of pathways and splicing factors have been identified (6). For splicing reaction, the spliceosome recognizes exon-intron boundaries of the 5′-donor and 3′-acceptor splice site. Furthermore, specific sequence motifs within exons can positively or negatively influence the recognition of nearby splice sites (7). The activity of so-called exonic splicing enhancers or exonic splicing silencers can be modulated by splicing factors like SR proteins or hnRNPs, which in turn allow precise gene expression in a spatiotemporal manner (6). Signaling pathways attributed to growth factor receptor activation are known to influence splicing patterns of different genes, such as CD44 v5 and fibronectin, respectively. A common feature of these examples is that activation of the growth factor pathway leads to enhanced exon inclusion by activation or suppression of different splicing factors (6).
Referring to HPV E6/E7 splicing, selection of the first 5′ alternative splice site gives rise to a new ORF termed E6* (Fig. S1A), which has so far only been described for high-risk but not for low-risk HPVs (8). For HPV16, E6* is produced via exclusion of a 183-bp fragment of the E6 ORF, which leads to a frameshift and a premature termination codon. Only full-length E6 was shown to exhibit transforming capacity, whereas experimentally modified viruses containing only E6* are not able to replicate efficiently in organotypic cultures (9). Conversely, the E6* ORF is often found to be the most abundant transcript in progressed CIN lesions, cervical carcinoma biopsies, and in explanted carcinoma cell lines (10). The 8-kDa E6* protein was shown to modulate E6 functions. Studies show that HPV18 E6* can counteract E6 protein functions by rescuing p53 levels in vivo (11). Furthermore, though interaction of HPV16 E6 with procaspase-8 accelerates its degradation, binding of E6* results in procaspase-8 stabilization and therefore may sensitize the cells to tumor necrosis factor-alpha (TNFα)-mediated apoptosis (12).
Although these biochemical differences of the different E6 isoforms have been known for some time, the in vivo regulation of HPV E6/E6* splicing is still unexplored. Controversial publications exist on the function of splicing and its effect on the efficiency of E7 translation. In vitro experiments favor the assumption that E7 can be only translated on E6* mRNA in a distance-dependent manner by reinitiation of translation (13, 14). Moreover, modulation of splicing could so far only be linked to the autoregulatory functions of viral E6 and E2 themselves under in vitro conditions (15). However, no information exists on the regulation of alternative splicing in vivo and its biological function within the epithelia and during virus-induced carcinogenesis.
Here we show that HPV16 E6 alternative splicing is dependent on the presence of EGF, and identify the MEK1/2-Erk1/2 MAP kinases as key regulatory components, either promoting or inhibiting exon inclusion. Using siRNA knockdowns, we further uncovered the splicing factors involved in this process. Our findings may shed light on the role of HPV E6 splicing during differentiation and the viral life cycle, and could also provide insights in the regulation of viral oncogene expression during malignant transformation.
Results
EGF Presence Induces HPV16 E6 Exon Inclusion.
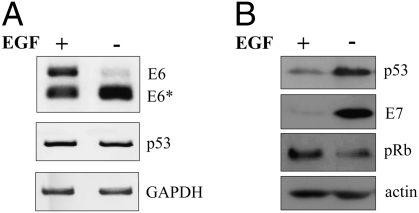
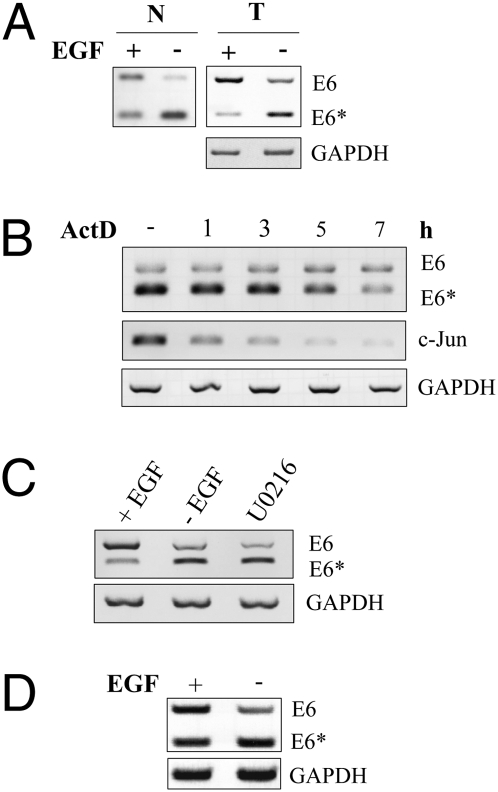
To monitor HPV16 oncogene alternative splicing, we first determined the cytoplasmic E6*/E6 ratios with primers covering the complete first alternative exon (Fig. S1A). Using HPV16 immortalized keratinocytes expressing HPV16 E6 and E7 as a bicistronic mRNA (16), depletion of EGF resulted in a strong decline of E6 full-length mRNA in favor of increased E6* mRNA (Fig. 1A). The change in E6*/E6 ratio could also be detected using both nuclear and total RNA (Fig. 2A), ruling out the possibility that alterations in nuclear transport of E6* mRNA account for this effect. E6 exon exclusion was accompanied by elevated p53 expression, which can be taken as an indicator for a decrease of the E6 protein and an increase of E7 expression (Fig. 1B). Consequently, higher E7 protein levels led to a reduction of the retinoblastoma protein (pRb; Fig. 1B). Treatment of cells with actinomycin D ensured that the observed increased exon exclusion was not due to a shorter mRNA half-life of E6 as compared with E6* mRNA. Both could still be detected after 5–7 h of treatment, whereas E6 mRNA turned out to be even more stable than the E6* isoform (Fig. 2B). The efficacy of the drug was demonstrated using the short-lived c-jun mRNA as a control (17), whereas GAPDH as housekeeping mRNA was not found to be affected during this timeframe (18). EGF depletion also led to enhanced exon exclusion in different immortalized cell lines expressing HPV16 E6/E7 either from its own or from a heterologous promoter (Fig. 2C) or in cells immortalized with the complete HPV16 genome (Fig. 2D). This suggests that EGF modulation of splicing is a general feature, independent of the cell line or the promoter directing E6/E7 expression.
Fig. 1.
EGF depletion leads to HPV16 E6 exon exclusion. “1321” cells were cultured in presence or absence of EGF for 24 h. (A) RT-PCR showing E6, E6*, p53, and GAPDH cytoplasmic mRNA levels. (B) Western blot analysis of total protein levels of p53, E7, pRb, and actin.
Fig. 2.
E6 splicing pattern is not dependent on mRNA transport, RNA stability, or promoter regulation. (A) “1321” cells were depleted for EGF, and nuclear RNA (N) or total RNA (T) was harvested for RT-PCR analysis. (B) “1321” cells were treated with actinomycin D (5 μg/mL) for indicated periods of time. Cytoplasmic RNA was harvested and E6*/E6 levels were determined with RT-PCR. c-Jun was analyzed as a control. (C) “1637” cells were depleted from EGF or treated with MEK1 inhibitor U0126 for 24 h. Cytoplasmic RNA was extracted for RT-PCR. (D) RT-PCR showing E6, E6*, and GAPDH cytoplasmic mRNA levels in FK16A cells in presence and absence of EGF.
HPV16 E6 Alternative Splicing Is Reversible and Directly Mediated by EGF Signaling.
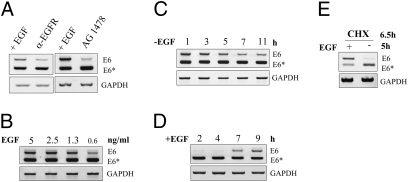
To verify that EGF receptor (EGFR) activation is responsible for the observed changes in exon inclusion, we specifically inhibited its activation with either neutralizing anti-EGFR antibodies (α-EGFR) or with the inhibitor tyrphostin AG 1478. This drug can specifically bind to and inhibit EGFR autophosphorylation (19), thereby blocking its activation and subsequent downstream signaling events. Treatment with both agents led to a similar increase in E6 exon exclusion (Fig. 3A) when compared with untreated controls. Modulation of viral alternative splicing is also dependent on the amount of EGF within the culture medium. As demonstrated in Fig. 3B, reduction of E6 in favor to E6* can be clearly seen when EGF levels drop below 1 ng/mL.
Fig. 3.
HPV16 E6 alternative splicing is directly mediated by EGFR signaling. (A) “1321” cells were treated with neutralizing anti-EGF receptor antibody or the specific EGF receptor inhibitor AG1478 for 24 h. (B) “1321” cells were grown in presence of indicated amounts of EGF for 24 h. (C and D) Time course of E6/E6* splicing after EGF depletion (C) or readdition (D). (E) “1321” cells were pretreated with cycloheximide (CHX) for 1.5 h. Subsequently, EGF was depleted for additional 5 h in the presence of CHX. In each case, cytoplasmic RNA was analyzed by RT-PCR.
To estimate the time course of EGF signaling on E6 alternative splicing, EGF depletion kinetics were performed. Here, discernible changes in E6*/E6 ratio could be detected within 5–7 h after EGF depletion (Fig. 3C), which is consistent with the half-life of the corresponding preexisting viral RNA (Fig. 2B). Conversely, readdition of EGF after 24 h of starvation led to a visible increase of E6 mRNA after 7 h of treatment (Fig. 3D), showing that EGF-mediated alternative splicing is a dynamic and reversible process. Moreover, E6 alternative splicing did not require de novo protein synthesis, because the shift from E6 to E6* after EGF omission also occurred in the presence of cycloheximide (Fig. 3E). This indicates that EGF-mediated viral splicing is achieved by preexisting splicing factors rather than by newly synthesized proteins.
EGF-Directed Alternative Splicing of HPV16 E6 Is Mediated by MAP Kinase Signaling.
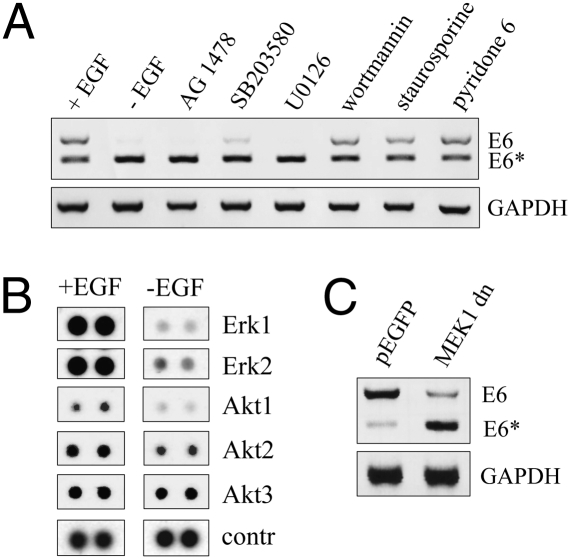
Because it is known that there is crosstalk between signal transduction pathways and alternative splicing, cells were treated with various inhibitors, targeting different downstream pathways known to be modulated by EGF (20). As shown in Fig. 4A, neither wortmannin nor staurosporine or pyridone 6, which block the phosphatidylinositol (PI) 3-kinase-dependent cascade, protein kinase C (PKC) signaling, or Janus kinase (JAK)/signal transducer and activator of transcription (STAT; Jak-STAT) signal transduction, respectively, had any impact on the E6*/E6 ratio. In contrast, inhibition of p38 signaling with SB203580, which specifically targets p38α and p38β (21), resulted in enhanced exon exclusion. The effect was even more pronounced after treatment with U0126, an inhibitor of mitogen-activated protein kinase kinase (MEK1/2) signaling (22), where the efficacy was almost comparable to that of AG 1478 (Fig. 4A). This finding could also be observed in keratinocytes expressing HPV16 E6/E7 under the control of the HPV native promoter (Fig. 2C), again excluding the possibility that differential promoter usage determined splicing site selection (23).
Fig. 4.
EGF-regulated E6 alternative splicing is mediated by MAP kinase signaling. (A) “1321” keratinocytes were depleted from EGF or treated with various inhibitors of the EGFR signaling pathways as indicated for 24 h. Cytoplasmic RNA was harvested and E6 splicing pattern analyzed by RT-PCR. (B) Phospho-MAPK Array (R&D Systems) showing the phosphorylation status of Erk1/2 and Akt1/2/3, respectively, after 24 h in presence of absence of EGF. Whole-cell extracts were prepared and processed as described. (C) 293 cells were cotransfected with 2 μg pLXSN-E6 and either of 5 μg pEGFP or with a MEK1 dominant negative mutant, respectively. Twenty-four hours posttransfection, cells were harvested and cytoplasmic RNA was extracted for RT-PCR analysis.
To further analyze the kinase signaling activation pattern in the presence and absence of EGF, a human phospho-MAPK antibody array was applied. This represents a rapid and sensitive assay to simultaneously detect the relative phosphorylation levels of various MAPKs and other serine/threonine kinases. Based on this method, the most prominent effect after EGF depletion was a strong reduction of extracellular signal-regulated kinases (Erk1/2) phosphorylation (Fig. 4B). Other intracellular kinases like Akt 1 showed only a negligible decrease of phosphorylation upon EGF-depletion, whereas Akt 2 and 3 were not involved.
Having identified Erk1/2 as the major MAP kinase affected by EGF, we next tried to modulate splicing by cotransfection of a dominant negative mutant of the mitogen-activated protein kinase kinase 1 (MEK1dn), which is the upstream kinase of Erk1/2. Using human embryonic kidney (293) cells, a heterologous cell line, which can be more easily transfected than immortalized human keratinocytes, a similar E6/E6* ratio was detectable as shown for HPV-positive cells (compare Figs. 1 and 4C). Moreover, consistent with the inhibitor studies shown in Fig. 4A, cotransfection of E6 with MEK1dn also revealed a reduction of E6 exon inclusion (Fig. 4C), when compared with EGFP transfected control cells. These data indicate that EGF-mediated alternative E6 splicing is regulated via the MEK1-Erk1/2 pathway.
To further investigate the effect on the E6*/E6 splicing ratio and to ask whether other growth-promoting factors can substitute or block the effect of EGF-depletion, the cells were treated with 12-O-tetradecanoyl-phorbol-13-acetate (TPA), insulin-like-growth factor (IGF1), and TNF-α in the presence and absence of EGF. As depicted in Fig. S2A, neither IGF1 nor TNF-α treatment had an influence on EGF-modulated splicing of E6, as neither of these factors could induce Erk1/2 phosphorylation in the absence of EGF (Fig. S2B). In contrast, TPA strongly induced Erk1/2 phosphorylation in the absence of EGF (Fig. S2B), mimicking the effect of EGF presence on E6 splicing (Fig. S2A). To investigate the influence of different growth factors on the overall cellular fate, cell cycle analyses were performed (for methods, see SI Text). Here, no major changes in cell cycle stages could be observed (Fig. S2C).
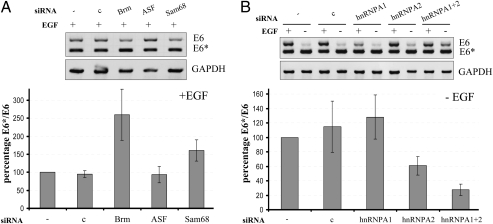
hnRNPA1 and hnRNPA2 Promote HPV16 E6 Exon Exclusion, Whereas Brm and Sam68 Mediate Exon Inclusion.
There is growing evidence that splicing and splicing factors are also dysregulated in many forms of human cancers (24). To determine which of them is involved E6 splicing, we focused our attention on hnRNPA1 and A2, ASF/SF2, Sam68, and Brm (6), which are all expressed in HPV16 immortalized keratinocytes (Fig. S3) and were shown to be modulated by growth factor pathways (6). Using siRNA knockdown experiments (Fig. S3A), Brm and Sam68 could be identified as splicing factors promoting exon inclusion in the presence of EGF (Fig. 5A), whereas siRNAs against ASF/SF2 had no effect under these experimental conditions. Conversely, individual and combinatorial knockdown by siRNA delivery revealed that hnRNPA1 and hnRNPA2 are promoting HPV16 E6 exon exclusion. In EGF-depleted cells, knockdown of hnRNPA1 or hnRNPA2 alone had no effect on E6 exon retention. Knockdown of hnRNPA1 in the presence of EGF even decreased E6 exon inclusion as compared with control cells, which were transfected with scramble siRNA. Nevertheless, in cells with decreased levels of both hnRNPA1 and hnRNPA2, inclusion of E6 alternative exon by EGF depletion could partially be retained (Fig. 5B). Conversely, EGF- depletion has no obvious effect on the quantity of the splicing factors (Fig. S3B), arguing for a posttranslational modification upon EGF withdrawal.
Fig. 5.
siRNA knockdowns of splicing factors. “1321” keratinocytes were transfected with the indicated siRNAs as described. Cytoplasmic RNA was harvested and analyzed by RT-PCR. (A) Splicing factors found to promote exon inclusion were analyzed in the presence of EGF. (B) hnRNP knockdown in the presence and absence of EGF. Band intensities were quantified from three independent experiments using ImageJ and E6*/E6 ratios were calculated. Ordinate: percentage of the E6*/E6 ratio, where values for untransfected samples were set as 100%. Abscissa: (−), untransfected; (c), scramble siRNA transfection.
Discussion
Alternative splicing within the E6/E7 transcription cassette of high-risk HPV types was discovered several years ago (8), but little is known about splicing regulation under in vivo conditions. Referring to a potential function of alternative splicing, however, examination of clinical samples has shown that the prevalence of the E6* mRNA isoform apparently increases with lesion severity, being finally the most abundant viral transcript found in cervical cancer cells and cell lines derived there from (10, 25).
Using monolayer cultures of HPV16 immortalized keratinocytes, we showed that E6/E6* alternative splicing is coupled to EGF signal transduction. In the presence of EGF, distinct levels of E6 full-length mRNA in conjunction with the E6* splicing isoform could be discerned, whereas EGF depletion resulted in a shift toward E6* mRNA (Fig. 1A). Due to the lack of functional anti-HPV16 E6 antibodies, we were not able to examine E6 or E6* protein levels directly. Nonetheless, decreased E6 levels can be deduced from enhanced p53 stabilization (Fig. 1B), which is consistent with the finding that E6* can counteract full-length E6-mediated p53 degradation (11).
It has been postulated that translation of the E7 ORF is rather inefficient in E6 full-length polycistronic mRNAs, as only two base pairs separate the stop codon of the E6 ORF from the downstream E7 start codon. With exclusion of E6 alternative exon, however, enhanced translation can occur, because ribosome assembly at the E7 AUG might be facilitated (14). In fact, as further consequence of E6 exon exclusion, higher levels of E7 protein could be observed (Fig. 1B), supporting data already predicted from in vitro studies (14). Elevated E7 protein levels also affected the amount of the retinoblastoma protein pRb (Fig. 1B), known to be targeted by E7 for proteasomal degradation (1).
We could further rule out a selective mRNA export from the nucleus on the splicing pattern, because an increased ratio of E6*/E6 was detectable independently of whether nuclear, total, or cytoplasmic RNA was analyzed (Figs. 1A and 2A). Unspliced E6 mRNA even showed a longer half-life than E6* mRNA after actinomycin D treatment (Fig. 2B), arguing against the possibility that different mRNA stability may account for this effect. Because certain promoter/enhancer elements or recruitment of transcription factors may influence alternative splicing by coupling transcription and pre-mRNA processing (26), we tested different immortalized keratinocyte cell lines, where the HPV16 E6/E7 transcription was driven by the β-actin promoter, the viral specific upstream regulatory region (16), or a long-terminal repeat (LTR) of the Moloney sarcoma virus. In each case, however, the same shift of E6 toward E6* was detected (Figs. 1A, 2C, and 4D), clearly supporting the notion that E6 splicing is entirely modulated by EGF.
Inhibition of receptor activation with neutralizing antibodies and with the EGFR inhibitor AG1478 (19) mimicked EGF depletion with respect of increased E6 exon exclusion (Fig. 3A). Therefore, EGFR activation is necessary and sufficient to maintain high levels of E6 full-length mRNA. To further decipher the signaling events launched at the EGFR upon ligand addition (20), inhibitor treatment showed that MAP kinase activation contributed substantially to the maintenance of E6 exon inclusion. Treatment with MEK1/2 inhibitor U0126 or SB203580 (a p38α/β inhibitor) or overexpressing a dominant negative MEK1 mutant, resulted in enhanced E6 exon exclusion (Fig. 4 A and C). Protein array data directly demonstrated that the extracellular signal-regulated kinase 1 and 2 (Erk1/2) are the major downstream kinases, which were strongly dephosphorylated upon EGF depletion (Fig. 4B). Consistent with this finding was that TPA, which also induced Erk1/2 phosphorylation, could overcome the effects of EGF depletion on E6 exon exclusion (Fig. S2 A and B).
As an initial attempt to identify the splicing factors that are involved in this process, siRNA knockdown experiments were performed. Considering the heterogeneous nuclear ribonucleoproteins (hnRNP) isoforms A1 and A2 as negative splicing regulators, already known to be increased during epithelial cell differentiation and involved in HPV16 gene late expression (27), pronounced exon exclusion was only detectable when both splicing factors were knocked down, whereas reduction of only hnRNPA2 showed a moderate effect on exon retention (Fig. 5). Similar observations were also made for BRCA1 exon18 mutant alternative splicing (28). The positive splicing regulator ASF/SF2 is an SR-protein, often described as antagonist of hnRNPA1 (29). Although it can promote inclusion of fibronectin alternative exons upon activation by PI3-kinase signaling (30) and is involved in HPV16 late gene splicing (31), ASF/SF2 had no influence on HPV16 E6/E6* alternative splicing in our experimental system (Fig. 5A). In contrast, knockdown of Sam68, a member of the STAR (signal transduction and activators of RNA) family of splicing factors (32), and Brm, a protein of the SWI/SNF chromatin remodeling complex (33), revealed the role of both factors in E6 alternative exon retention (Fig. 5B). Notably, Brm is already known to be recruited to the HPV promoter where it acts as a transcription factor (34).
What could be the relevance of EGF-mediated E6/E6* alternative splicing regarding the viral life cycle and malignant progression? Expression of the EGFR can be detected throughout the epidermis, with elevated levels in the basal and parabasal layers (35) (Fig. S1B). Hence, the connection of EGFR activation and HPV early gene alternative splicing might be an important regulatory mechanism for viral gene expression and the differentiation-dependent viral life cycle. In fact, analyzing E6 mRNA splicing during cellular differentiation, enhanced E6 exon exclusion could be discerned (Fig. S4A). Here, the stages of differentiation can be monitored by expression of distinct cytokeratins (36) (for methods, see SI Text). As shown in Fig. S4B, panel A, only single cells express cytokeratin K10 when cultivated in K-SFM/EGF medium, but their number increased after EGF depletion Fig. S4B, panel B. This was also observed by incubating the cells in DMEM/10% FCS (37) (Fig. S4B, panel C). Shifting cells into differentiation can even be more recognized by a strong reduction of cytokeratin K18-positive cells (Fig. S4B, panels E and F). This particular cytokeratin is a common cytoskeletal component in nondifferentiated keratinocytes grown under in vitro conditions (Fig. S4B, panel D), but becomes rapidly suppressed during differentiation from monolayer toward focally stratified areas (38).
Based on our observations, it is reasonable to assume that during the first events of viral infection of the basal keratinocytes, HPV might require high levels of full-length E6 to prevent apoptosis. With increasing differentiation and decreasing EGF/EGFR levels (Fig. S1B), it might be beneficial for the virus to use E7 expression to overcome reduced proliferation of the cells (1). Moreover, during natural infection, there is an additional expression of the E5 protein, which is also able to enhance EGFR signaling through up-regulation of both EGFR and ERK1/2 signaling (39). Therefore we propose that depending on the localization and the status of the cell within the stratified epithelium, EGFR-dependent regulation of E6/E6* alternative splicing can be considered a fine-tuning process of oncogene expression to fulfill the intracellular requirements necessary for viral maturation.
In the context of HPV-mediated immortalization and subsequent transformation, it is also tempting to speculate that enhanced E6 exon inclusion and in turn full-length E6 expression is first required for p53 labilization, thereby favoring chromosomal destabilization as initial early event in carcinogenesis (1). In a second selection step, cells with preferential E6 exon exclusion and high E7 expression gain a selective growth advantage during progression (40), providing a reliable explanation for the prevalence of E6* mRNA found in most cervix carcinoma cell lines and CIN lesions (10, 25). Analyzing the E6*/E6 ratio in two well-established HPV16 positive cervical carcinoma cells, both expressing mainly E6*, only SiHa cells showed a moderate isoform shift toward full-length E6 after EGF addition, whereas CaSki cells were not responding (Fig. S5A). Accordingly, analysis of SiHa and CaSki in comparison with immortalized cells also revealed an apparent correlation between E6*/E6 splicing ratio, the expression levels of p53 and E7 (Fig. S5B). This indicates that these cells were selected for robust E6 exon exclusion during multistep progression to cervical cancer, ensuring higher levels of E7 protein for enhanced proliferation, independently of the cellular environment. It will be of interest to investigate whether long-term selection of immortalized cells in the presence of the tyrphostin inhibitor AG1478 results in an expansion of subclones with differences in splicing pattern, levels of E7, and in vivo growth properties in nude mice.
Materials and Methods
Cell Culture.
“1321” (β-actin promoter driven HPV16 E6/E7), “1637” (HPV16 URR driven HPV16 E6/E7), and FK16A (complete HPV16 genome) immortalized keratinocytes (16, 41) were cultured in Keratinocyte-Serum Free Media (K-SFM; GIBCO) containing 5 ng/mL EGF and 50 ng/mL bovine pituitary extract. For passaging, trypsinized cells were collected in DMEM (DMEM; Sigma) supplemented with 10% calf serum (Linaris) and subsequently washed twice with PBS before seeding in a 1:3 dilution in K-SFM. SiHa and CaSki cervical carcinoma cell lines as well as HEK 293 cells were maintained in DMEM, supplemented with 10% FCS.
EGF Depletion and Cell Treatment.
For each experiment, 8 × 105 cells were seeded in 6-cm plates (Greiner) for 24 h. For EGF depletion, plates were washed twice with PBS (Linaris) and K-SFM without EGF was added. For treatment, inhibitors and cytokines were added directly to the K-SFM growth media at the following final concentrations: 30 μM tyrphostin AG1478 (Axxora), 50 nM staurosporine (Calbiochem), 500 nM JAKI inhibitor pyridone 6 (Calbiochem), 10 μg/mL neutralizing anti-EGF-receptor antibody (Upstate), 10 nM wortmannin (Calbiochem), 10 μM SB203580 (Calbiochem), 20 μM U0126 (Calbiochem), 5 μg/mL actinomycin D (Sigma), 10 μg/mL cycloheximide (Sigma), 100 ng/mL IGF1 (Millipore), 250 U/mL TNF-α (MACS Miltenyi Biotec), and 100 μM TPA (Sigma). Cells were incubated for 24 h before harvesting.
RNA Isolation and Semiquantitative RT-PCR.
Cytoplasmic RNA was isolated with the RNeasy Kit (Qiagen) according to the manufacturer's instructions after cell fractionation. Total and nuclear RNA was isolated with TRIZOL (Invitrogen). RNA was quantified using the NanoDrop1000 system (Thermo Scientific). For reverse transcription, 1 μg RNA was diluted in H2O along with 1 μL of Oligo dT Primer (500 μg/μL; Promega) to obtain a total volume of 12.5 μL. Samples were incubated at 70 °C for 10 min and chilled on ice for 3 min. Then, 0.5 μL of RNase OUT (Invitrogen), 2 μL 0.1 M DTT (Invitrogen), and 4 μL 5× First Strand Buffer (Invitrogen) were added and samples were then incubated at 25 °C for 10 min. Finally, 0.5 μL SuperScriptII Reverse Transriptase (Invitrogen) was added to obtain a final volume of 20 μL. For semiquantitative RT-PCR, cDNA was diluted 1:40 in H2O and 2 μL were amplified in 20 μL with GoTaq Green (Promega) Mastermix using 0.5 μL of a 20 μM primer mix. For primer sequences, product length, annealing temperature, and cycle number, see Table S1. Quantification of band intensities was done with ImageJ. For objective analysis, splicing rate was calculated as E6*/E6 ratio and the control sample was set to 100%.
Protein Detection and MAP Kinase Array.
RIPA protein extracts or total cellular extracts (for MAPK analyses) were prepared as described previously (42). Equal amounts of protein extracts were separated in 12% SDS-PAGE and electrotransferred to Immobilon-P membranes (polyvinylidene difluoride; Millipore) using 1 mA/cm2 for 70 min. Membranes were blocked in 5% skin milk /TBS-Tween for 1 h and subsequently incubated with different antibodies overnight at 4 °C. The following antibodies have been used: anti-p53 (DO-1; Santa Cruz), anti-HPV16 E7 (NM-2; a generous gift from M. Müller; DKFZ), anti-pRb (Beckton Dickinson), anti-Erk-1/2 (NEB), anti-phospho-Erk-1/2 (NEB), anti-p38 MAPK (NEB), anti-phospho-p38 MAPK (NEB), anti-hnRNP A1 (Abcam, 9H10), anti-hnRNP A2B1 (DP3B3; Abcam) anti-ASF (Abcam), anti-Sam68 (Santa Cruz), anti-Brahma (Abcam), and anti-actin (ICN). For detection of the respective bands, HRP-conjugated secondary antibodies anti-mouse (Dianova), anti-mouse IgG1 (Santa Cruz), and anti-rabbit (Dianova) were incubated for 1 h and visualized using the ECL detection system (PerkinElmer Life Sciences) or the SuperSignal West Femto Substrate (Thermo Scientific). For stripping, membranes were washed for 5 min in TBS-Tween, 0.2 M NaOH, 5 min H20, and 5 min TBS-Tween. MAP kinase array (Proteome Profiler Array; R&D Systems) was performed according to manufacturer's instructions.
Transient Transfection Studies.
E6 coding sequence was amplified from SiHa cells and cloned into pLXSN retroviral vector. MEK1-dominant negative expression vector was a generous gift from Roger Davis (University of Massachusetts Medical School, Worcester, MA). A total of 8 × 105 293 cells were seeded in 6-cm plates. After 24 h, cells were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. Cells were harvested 24 h posttransfection.
siRNA Knockdown.
Using HiPerfect (Qiagen) transfection reagent and following manufacturer's instructions, 7 × 105 cells were plated in 6-cm well plates and transfected with 100 nM siRNA (Dharmacon). siRNA sequences were described previously for Brm (33), ASF, hnRNPA1, hnRNPA2 (28), Sam68 (43), and siScramble (44). Twenty-four hours posttransfection, fresh media was added and, where indicated, EGF was depleted for an additional 24 h. Seventy-two hours posttransfection, cells were harvested.
Supplementary Material
Acknowledgments
The authors thank Dr. Richard Schlegel (Georgetown University, Washington, DC) for providing the immortalized human keratinocytes, Dr. Martin Müller (German Cancer Research Center, Heidelberg) for providing the HPV16 E7 antibody, and Dr. Roger J. Davis (University of Massachusetts Medical School) for providing the dominant negative mutant of MEK1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002620107/DCSupplemental.
References
- 1.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 2.Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia R, et al. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 2009;83:167–180. doi: 10.1128/JVI.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S. HPV-16 RNA processing. Front Biosci. 2008;13:5880–5891. doi: 10.2741/3123. [DOI] [PubMed] [Google Scholar]
- 5.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaustein M, Pelisch F, Srebrow A. Signals, pathways and splicing regulation. Int J Biochem Cell Biol. 2007;39:2031–2048. doi: 10.1016/j.biocel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Matlin AJ, Moore MJ. Spliceosome assembly and composition. Adv Exp Med Biol. 2007;623:14–35. doi: 10.1007/978-0-387-77374-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Schneider-Gädicke A, Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 1986;5:2285–2292. doi: 10.1002/j.1460-2075.1986.tb04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009;23:181–194. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhm S, Wilczynski SP, Pfister H, Iftner T. The predominant mRNA class in HPV16-infected genital neoplasias does not encode the E6 or the E7 protein. Int J Cancer. 1993;55:791–798. doi: 10.1002/ijc.2910550517. [DOI] [PubMed] [Google Scholar]
- 11.Pim D, Banks L. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene. 1999;18:7403–7408. doi: 10.1038/sj.onc.1203134. [DOI] [PubMed] [Google Scholar]
- 12.Filippova M, et al. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J Virol. 2007;81:4116–4129. doi: 10.1128/JVI.01924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacey SN, et al. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J Virol. 2000;74:7284–7297. doi: 10.1128/jvi.74.16.7284-7297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang S, Tao M, McCoy JP, Jr, Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodaghi S, Jia R, Zheng ZM. Human papillomavirus type 16 E2 and E6 are RNA-binding proteins and inhibit in vitro splicing of pre-mRNAs with suboptimal splice sites. Virology. 2009;386:32–43. doi: 10.1016/j.virol.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Münger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blattner C, et al. UV-Induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol. 2000;20:3616–3625. doi: 10.1128/mcb.20.10.3616-3625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dani C, et al. Characterization of the transcription products of glyceraldehyde 3-phosphate-dehydrogenase gene in HeLa cells. Eur J Biochem. 1984;145:299–304. doi: 10.1111/j.1432-1033.1984.tb08552.x. [DOI] [PubMed] [Google Scholar]
- 19.Levitzki A, Gazit A. Tyrosine kinase inhibition: An approach to drug development. Science. 1995;267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 20.Laurent-Puig P, Lievre A, Blons H. Mutations and response to epidermal growth factor receptor inhibitors. Clin Cancer Res. 2009;15:1133–1139. doi: 10.1158/1078-0432.CCR-08-0905. [DOI] [PubMed] [Google Scholar]
- 21.Lee JC, et al. Inhibition of p38 MAP kinase as a therapeutic strategy. Immunopharmacology. 2000;47:185–201. doi: 10.1016/s0162-3109(00)00206-x. [DOI] [PubMed] [Google Scholar]
- 22.Duncia JV, et al. MEK inhibitors: The chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8:2839–2844. doi: 10.1016/s0960-894x(98)00522-8. [DOI] [PubMed] [Google Scholar]
- 23.Amelio AL, Caputi M, Conkright MD. Bipartite functions of the CREB co-activators selectively direct alternative splicing or transcriptional activation. EMBO J. 2009;28:2733–2747. doi: 10.1038/emboj.2009.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häfner N, et al. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. 2008;27:1610–1617. doi: 10.1038/sj.onc.1210791. [DOI] [PubMed] [Google Scholar]
- 26.Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- 27.Cheunim T, Zhang J, Milligan SG, McPhillips MG, Graham SV. The alternative splicing factor hnRNP A1 is up-regulated during virus-infected epithelial cell differentiation and binds the human papillomavirus type 16 late regulatory element. Virus Res. 2008;131:189–198. doi: 10.1016/j.virusres.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goina E, Skoko N, Pagani F. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol Cell Biol. 2008;28:3850–3860. doi: 10.1128/MCB.02253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cáceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 30.Blaustein M, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 31.Mole S. RNA splicing factors regulated by HPV16 during cervical tumour progression. J Pathol. 2009;219:383–391. doi: 10.1002/path.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matter N, Herrlich P, König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 33.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 34.Kumar RA, Naidu SR, Wang X, Imbalzano AN, Androphy EJ. Interaction of papillomavirus E2 protein with the Brm chromatin remodeling complex leads to enhanced transcriptional activation. J Virol. 2007;81:2213–2220. doi: 10.1128/JVI.01746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanney LB, Magid M, Stoscheck CM, King LE., Jr Comparison of epidermal growth factor binding and receptor distribution in normal human epidermis and epidermal appendages. J Invest Dermatol. 1984;83:385–393. doi: 10.1111/1523-1747.ep12264708. [DOI] [PubMed] [Google Scholar]
- 36.Moll R, Divo M, Langbein L. The human keratins: Biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman L, Schlegel R. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J Virol. 1996;70:3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryle CM, et al. Density-dependent modulation of synthesis of keratins 1 and 10 in the human keratinocyte line HACAT and in ras-transfected tumorigenic clones. Differentiation. 1989;40:42–54. doi: 10.1111/j.1432-0436.1989.tb00812.x. [DOI] [PubMed] [Google Scholar]
- 39.Crusius K, Rodriguez I, Alonso A. The human papillomavirus type 16 E5 protein modulates ERK1/2 and p38 MAP kinase activation by an EGFR-independent process in stressed human keratinocytes. Virus Genes. 2000;20:65–69. doi: 10.1023/a:1008112207824. [DOI] [PubMed] [Google Scholar]
- 40.Jabbar SF, Abrams L, Glick A, Lambert PF. Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res. 2009;69:4407–4414. doi: 10.1158/0008-5472.CAN-09-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steenbergen RD, et al. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: Activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene. 1996;13:1249–1257. [PubMed] [Google Scholar]
- 42.van Riggelen J, et al. Loss of net as repressor leads to constitutive increased c-fos transcription in cervical cancer cells. J Biol Chem. 2005;280:3286–3294. doi: 10.1074/jbc.M409915200. [DOI] [PubMed] [Google Scholar]
- 43.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider G, et al. IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 2006;25:3801–3812. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.