Abstract
The doublesex (dsx) gene of the parasitic wasp Nasonia vitripennis is described and characterized. Differential splicing of dsx transcripts has been shown to induce somatic sexual differentiation in Diptera and Lepidoptera, but not yet in other insect orders. Two spliceforms of Nasonia dsx mRNA are differentially expressed in males and females. In addition, in a gynandromorphic line that produces haploids (normally males) with full female phenotypes, these individuals show the female spliceform, providing the first demonstration of a direct association of dsx with somatic sex differentiation in Hymenoptera. Finally, the DM domain of Nasonia dsx clusters phylogenetically with dsx from other insects, and Nasonia dsx shows microsynteny with dsx of Apis, further supporting identification of the dsx ortholog in Nasonia.
Keywords: DM domain genes, doublesex, haplodiploidy, Hymenoptera, Nasonia, sex determination
Introduction
The role of the transcription factor doublesex (dsx) in sex determination was first described in Drosophila melanogaster (Baker & Wolfner, 1988). Differential splicing of male and female dsx mRNA results in sex specific dsx proteins, which regulate downstream somatic sexual dimorphism (Baker & Wolfner, 1988). The dsx protein is characterized by two functional domains, an N-terminal DNA binding domain (DM domain or OD1) and a C-terminal dimerization domain (dsx dimer or OD2). The DM domain is a zinc module, with conserved amino acid residues for zinc chelating and promoter binding (Zhu et al., 2000). The dsx dimer is an alpha-helical motif which, by promoting dimerization of dsx, enhances DNA recognition by the DM-domain (Bayrer et al., 2005).
A strong conservation of the DM domain occurs between D. melanogaster dsx (DMdsx) and the sex determining protein mab-3 in the nematode Caenorhabditis elegans. Function of the gene in these divergent organisms is sufficiently conserved that DMdsx can partially rescue male-specific development in mab-3 deficient worms (Raymond et al., 1998). Furthermore, the mammalian Dmrt-1 gene transcribes a DM domain protein that is differentially expressed in testis, and mutations in this gene suggest that it plays an important role in male sexual differentiation (Raymond et al., 2000). Recently, dsx orthologs have been detected in a variety of insects. Doublesex orthologs have most extensively been described in Diptera, including Anastrepha obliqua (Ruiz et al., 2005), Anopheles gambiae (Scali et al., 2005), Bactrocera tryoni (Shearman & Fromer, 1998), Musca domestica (Hediger et al., 2004), but also have been found in Lepidoptera (Bombyx mori, Ohbayashi et al., 2001), and Hymenoptera (Apis mellifera; Cho et al., 2007) as all have a characteristic strongly conserved DM domain. However, in only a few cases have spliceform differences between males and females been shown, e.g. in A. mellifera (Cho et al., 2007) and functional assays are restricted to Drosophila (e.g. Baker & Wolfner, 1988; Raymond et al., 1998) and to a lesser extent to Lepdoptera (Suzuki et al., 2005). A current hypothesis predicts higher conservation at the base of sex determining gene cascades, i.e. dsx, but divergence at the level of upstream primary signals (Wilkins, 1995).
Hymenoptera, including the honeybee A. mellifera and the jewel wasp Nasonia, have haplodiploid sex determination. Males are haploid and develop parthenogenetically from unfertilized eggs, whereas females are diploid and develop from fertilized eggs. For over 60 years it has been known that different sex determining mechanisms exist within the Hymenoptera (Whiting, 1943). In haplodiploids, a relatively common mechanism is complementary sex determination (CSD). Gender is genetically determined by a single locus with multiple alleles: individuals that are heterozygous at this locus develop into females, whereas hemizygotes and homozygotes develop into haploid and diploid males respectively (Whiting, 1943, Crozier, 1971). This mode of sex determination has now been shown for more than 60 species (Van Wilgenburg et al., 2006) and for the honeybee the complementary sex determiner (csd) gene was identified and cloned (Beye et al., 2003). However, it is clear that sex determination in some groups, such as the large parasitoid wasp group Chalcidoidea to which Nasonia belongs, cannot be explained by CSD, because upon inbreeding homozygous diploids still develop into females (Skinner & Werren, 1980).
The parasitic wasp Nasonia has been extensively studied genetically and is rapidly being recognized as a model system in evolutionary and developmental biology (Werren & Stouthamer, 2002; Shuker et al., 2003; Pultz & Leaf, 2003; Beukeboom & Desplan, 2003; Werren et al., 2004; Lynch et al., 2006; Ferree et al., 2006). It has been known for a long time that its sex determination is not governed by CSD (Whiting, 1967; Skinner & Werren, 1980), but until recently little progress had been made in elucidating its mode of sex determination (Beukeboom, 1995). Recently, several studies reported on the genetics of sex determination in Nasonia (Beukeboom & Kamping, 2006; Trent et al., 2006), including the characterization of genetic strains that produce gynandromophic phenotypes (Beukeboom et al., 2007; Kamping et al., 2007).
The recent advent of the Nasonia genome sequence (Werren et al., 2004) now provides avenues for identification of genes involved in sex determination, and to investigate their potential role in Nasonia sex determination pathways and the evolution of genetic sex determining mechanisms in general. Here we report the identification and characterization of a dsx ortholog in Nasonia. We show that Nasonia dsx clusters phylogenetically with dsx in other insects, is differentially spliced in males and females, and is associated with sex determination in haploid individuals from a gynandromoph producing strain of Nasonia. We have also investigated the presence and sequence divergence of dsx in related species of Nasonia.
Results
Computational detection of the Nasonia dsx ortholog
A gene containing a DM-domain coding sequence that showed striking homology to dsx of other insects was originally detected during efforts to clone genes involved in morphological sex specific differences between Nasonia species (Werren et al., unpublished). The putative coding region of this gene, provisionally termed DM1, was represented in a fully sequenced BAC of N. giraulti (NG, GenBank accession number AC185330). Subsequently, the N. vitripennis (NV) genome was bioinformatically screened with the DM-domain coding sequence and, in addition to finding the NV DM1, three additional DM-domain genes were detected, termed DM2, DM3 and DM4. These four DM-domain containing genes were found in the following scaffolds: DM1, scaffold 23; DM2, scaffold 53; DM3, scaffold 62; DM4, scaffold 6 (NV genome version 1.0; HGSC at Baylor College of Medicine). Besides giving the strongest blast match to any insect dsx genes, the DM1 gene also contains a second conserved coding sequence characteristic of dsx proteins: the dsx dimerization domain (dsx dimer). Only one gene containing the dsx dimer was found in each of the five insect genomes searched (see below). Further analysis of scaffold 23, involving the immediate flanking sequences of DM1 revealed that the DM1 is flanked upstream by a prospero ortholog and downstream by an elongase gene (data not shown). Microsynteny of prospero - dsx gene order is also present in the genomes of A. mellifera, Tribolium castaneum, and An. gambiae, but not in D. melanogaster.
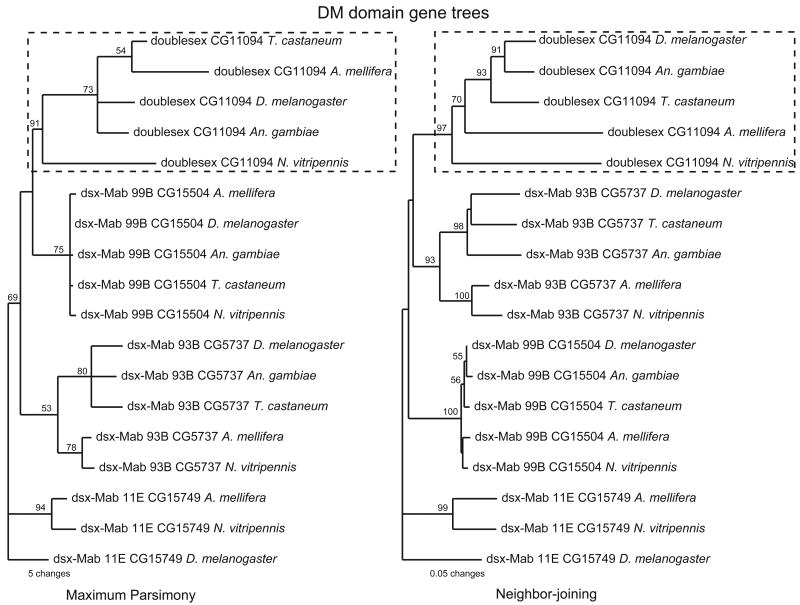
Four different DM-domain genes were also found in the genomes of D. melanogaster and A. mellifera, while only three were found in the genomes of T. castaneum and An. gambiae. Phylogenetic analysis using amino acid sequences of the DM domain showed four gene clusters, each containing a single gene from each species (Figure 1). These phylogenetic clusters are consistent with the overall gene architecture, as indicated by other identifiable structural markers, e.g. the dsx dimer in the dsx cluster and the DMRTA motif (pfam 03474) in the dsx-mab 93B cluster (data not shown). The Nasonia dsx groups with dsx of the other species with strong support. Relationships among DM domains are not fully resolved within the dsx cluster (Figure 1), due in part to a number of amino acid substitutions unique to the Nasonia lineage (Figure 2). In addition to dsx, all five species share genes homologous to dsx-mab 93B and a dsx-mab 99B genes (DM2 and DM4 respectively for Nasonia), the latter being the less divergent gene, indicating stronger selective pressure to maintain the amino acid sequence. The fourth gene dsx-Mab 11E (DM3 for Nasonia) was not found in the genome of T. castaneum and An. gambiae. Since most insect genomes are still in draft stage, at this point is difficult to be sure whether dsx-mab 11E has not been sequenced or it was really lost during T. castaneum and An. gambiae evolution. However, it appears that four DM domain genes is the ancestral condition in holometabolous insects.
Figure 1.
DM domain gene tree. The Nasonia dsx clusters with dsx of other insects. The most parsimonious tree (consensus of 20 trees with 222 steps) and a neighbor-joining tree obtained using the amino acid sequences of the DM domain region (67 total characters, 42 that are parsimony-informative) are shown with bootstrap supports above the branches. Many of the DM domain genes used in this analyses are predicted gene models.
Figure 2.
Amino acid sequence alignment of the Nasonia dsx and other insects. Only the two conserved regions are presented in the figure, including the DNA binding (DM) and the dsx dimer. The zinc chelating residues in the DM domain are in bold in the reference pfam sequence(Finn et al., 2006). Putative conserved residues that distinguish the dsx DM domain from the DM domain of other proteins are shown in dotted boxes. The female isoforms were used for the dsx dimer region alignment. The predicted amino acid sequences of the 3′ region of the male isoform are shown for Nasonia and Trichomalopsis.
Gene architecture, genomic microsynteny, and phylogenetic analysis of conserved regions of established dsx genes lead us to the conclusion that DM1 is the Nasonia ortholog of dsx.
Sex differential splicing of Nasonia dsx mRNA
Male and female forms of dsx were detected using primers designed for the predicted 3′UTR and the DM-domain exon. Results of these experiments show that from female mRNA an approximately 500 bp fragment is predominantly amplified, whereas in males the amplicon using the same primer set is approximately 110 bp longer (Figure 3).
Figure 3.

Male and female spliceforms of Nasonia dsx. RT-PCR of males, females and gynandromorph RNA. Arrows indicate male specific and female specific splice fragments. Lane 1-3: adult male; lane 4-6: adult HiCD12 haploid gynandromorph, morphologically male; lane 7-9: adult female; lane 10-11: adult HiCD12 haploid gynandromorph, morphologically female; lane 12: negative control; lane 13: 100bp Molecular marker.
The male and female spliceforms were confirmed by sequencing of the 3′ RACE products generated for NV (AsymC and HiCD12 strains) and NG. Full length transcripts were determined for the NV strain HiCD12 by sequencing of the 5′ RACE products. For the second NV strain, AsymC, and for NG several RT-PCR reactions with different primer combinations confirmed all intron-exon boundaries including the most 5′ exon (Figure 4).
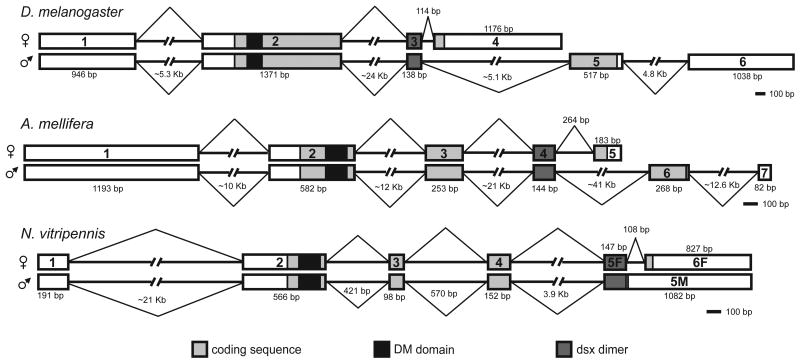
Figure 4.
Insect dsx gene models. The Nasonia dsx male and female spliceforms are compared to dsx of D. melanogaster and A. mellifera. See text for details.
The mature Nasonia male dsx mRNA is composed of five exons, with a single large exon 5M of 1082 bp (hereafter sizes are for NV). This last male exon includes the C-terminal coding sequence, starting from the dsx dimer region and does not undergo further splicing. The first four exons of Nasonia female mature dsx mRNA are identical to the male isoform. The Nasonia female dsx mRNA, however, is derived from six exons. Exons 5F and 6F are derived from the same exact genomic region as the last male exon (exon 5M) but are interrupted by an intron of 108 bp, that is not spliced out in the male mRNA, resulting in two exons of 147 bp (exon 5F) and 827 bp (exon 6F), respectively. Sex specific differences in dsx protein sequence are caused by female specific splicing of the last exon resulting in a different stop codon (see below).
To summarize, in Nasonia females an intron of 108 bp is spliced out of the primary transcript, while in males no splicing of this intron occurs (Figure 4). This process of generating the male specific dsx isoform has not been described for other insects. In other insects, including A. mellifera (Cho et al., 2007), D. melanogaster (Baker & Wolfner, 1988), and the B. mori (Ohbayashi et al., 2001), the male isoform is generated by an alternative splice that completely skips the female specific exon (see Figure 4 for a comparison).
RT-PCR conducted for different life stages (embryos, larvae, pupae, and adults), and for different adult body parts (head, thorax, and abdomen) showed that both sexes predominantly express their sex-specific form in all stages and body parts tested (Figure 3B shows adults). The female form was often detected in independent experiments as a weaker band in males, whereas the male form was rarely seen in females. The presence of the female form of dsx in males was also noted in A. mellifera (Cho et al., 2007).
Interestingly, for NV in both males and females a 564 bases longer 3′ UTR transcript was also identified (but not for NG). Apparently a second poly(a) site can be used in both males and female to generate longer dsx mRNA's. Since these longer forms are observed in spliced mRNA, i.e. they do not include intronic sequences, we believe that these products are not the result of genomic contamination, but represent a genuine dsx mRNA. However, comparison of these 3′UTR sequences to the genomic sequence of NV shows that a stretch of 25 As is present in the NV genomic sequence. Therefore, the actual alternative poly(A) site could be even more downstream, but it was not detected due to the annealing of the oligo-d(T) to the internal A-tract upon performing 3′-RACE. In addition a smaller transcript that skips exon 2 (Figure 4), i.e. a spliceform that joins directly exons 1 and 3, was consistently observed as a clear second band in RT-PCR in both sexes. This is puzzling because exon 2 codes for the DM domain and it also has the start codon (the alternative start codon maintains the frame). Obviously, these alternative transcripts will need further characterization and are not described in more detail here. It is worth noting that evidence for more than two splicing variants of dsx have been shown previously, e.g. for D. melanogaster (Baker & Wolfner, 1988) and A. mellifera (Cho et al., 2007).
dsx mRNA splice variants in gynandromorphs
Gynandromorphism (individuals with both male and female structures) has been previously described in Nasonia. A line (HiCD12) has been characterized that produces gynandromorphs from unfertilized (haploid) eggs which range from single female structures (e.g. antennal segment) in an otherwise male morphology to haploids with a complete female morphology (Beukeboom et al., 2007; Kamping et al., 2007). To evaluate the involvement of dsx in hymenopteran sex determination, we analyzed haploid individuals from the HiCD12 line that were either completely morphological male or female, for the presence of male and female specific dsx splice variants. We found that morphological male gynandromorphs always express the male specific splice variant at a considerable level, sometimes together with the female-specific variant, whereas haploid females express mostly or only the female transcript (Figure 3A). Presence of the female specific spliceform in morphological males of the gynandromorphic line may be due to presence of female-like structures in these individuals that were not apparent from their outer morphology. As stated above, however, haploid males from the non-gynandromorphic strain NV AsymC can also contain low levels of female-specific splice variants of dsx mRNA (see Figure 3B). In contrast, females of either strain show a clear predominant female-specific spliceform, confirming the female specificity of this splice variant.
The difference between a normal female and a phenotypic female from the gynandromorph line is that the former are diploid whereas the latter are derived from haploid unfertilized eggs, yet develop somatically into females. These data clearly support an association for dsx in Nasonia sex specific phenotypic differentiation, independent of ploidy.
Predicted dsx proteins in Nasonia and other insects
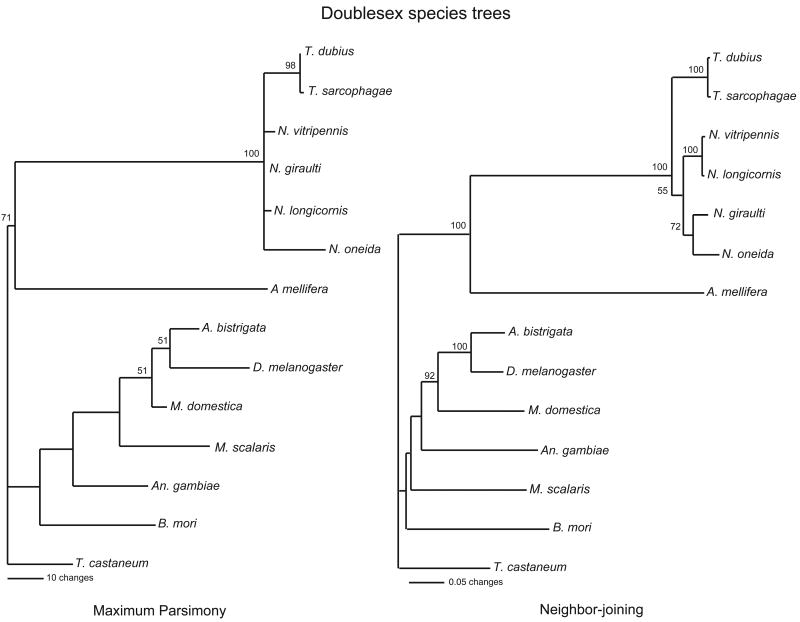
In Drosophila and other organisms, dsx has two characteristic domains: a DNA binding domain (DM or OD1) and an oligomerization domain (dsx dimer or OD2). We have sequenced the genomic region of all coding exons of dsx of two additional Nasonia species as well as of two Trichomalopsis species, a closely related pteromalid wasp. The two conserved regions of the predicted dsx proteins were aligned with known dsx sequences from eight other species: the hymenopteran A. mellifera, the dipterans, Anastrepha brisgata, D. melanogaster; An. gambiae, M. domestica, Megaselia scalaris, the lepidopteran B. mori, and the coleopteran T. castaneum (Figure 2). Parsimony and neighbor-joining dendrograms of this alignment shows that the combined protein sequence groups Nasonia dsx with A. mellifera dsx (Figure 5), as expected based on phylogenetic relationships. Apis and Nasonia did not cluster together when only the DM domain was used (Figure 1), reflecting the amino acid sequence divergence of this domain in Nasonia. The Hymenoptera combined dsx domains show longer branch lengths than found in other insects (Figure 5), probably caused by an elevated divergence rate.
Figure 5.
An insect species trees based on the conserved dsx amino acid sequences. The most parsimonious tree (consensus of 20 trees with 335 steps) and a neighbor-joining tree obtained using the combined amino acid sequences of the DM-domain region and the dsx dimer region (138 total characters, 89 that are parsimony-informative) are shown with bootstrap support above the branches.
Examination of the DM domain consensus (pfam00751, Figure 2) reveals amino acids that are conserved across the DM domains shown, relative to the consensus for DM domains. Most of these conserved amino acid residues are not dsx specific, but are common to other DM domain containing proteins and are important for zinc binding (Zhu et al., 2000). However, examination of the DM domain sequences also reveals two highly conserved amino acids are shared among all insect dsx. An insect dsx shared threonine (T) occurs four amino acids upstream of a glutamine (Q) in the second half of the DM domain (Figure 2). A third highly conserved glutamic acid (E) occurs five amino acids upstream of the conserved T. Only Trichomalopsis shows a variant at this position, with arginine (R) in place of E. Both are polar amino acids with strong hydropathy indices, whereas glutamic acid is acidic and arginine is strongly basic. The clustering of these conserved amino acids suggests that they may be important in dsx protein specificity in sexual differentiation, relative to other DM domain proteins.
Figure 2 also shows alignment of the dsx dimer domain in Nasonia, relatives and other insects, compared to the consensus pfam sequence for this region. As can be seen, there is considerable variation in the terminus of this domain, in both length and sequence. Because the dsx dimer is an alpha-helical motif (Bayrer et al., 2005), structure conservation rather than amino acid conservation is expected. Nevertheless, Nasonia and relatives retain a nine amino acid sequence in the middle of the dsx dimer region (MLYVILKDA) that is also mostly conserved in Diptera and Tribolium, but not in Apis or Bombyx. Drosophila melanogaster shows the greatest similarity to the pfam sequence, but this likely reflects the contribution of Drosophila and other Diptera to determination of this consensus.
We have also compared the sequence of NV and its sister species, NG, N. longicornis (NL), and N. oneida (NO), as well as two closely related wasps, T. sarcophagae and T. dubius. The species dsx differ in length of the C terminus for the predicted female specific protein. A 16 amino acids long extension in the female specific protein appears to be the ancestral condition, as it is shared among two Nasonia species, NV and NL, and Trichomalopsis (Figure 2). In NG, the female transcript contains a stop codon that truncates the protein 12 amino acids relative to the ancestral form. A single deletion in NO creates a frameshift in this tail, resulting in a 32 amino acids long extension in the females that has a completely different amino acid composition (Figure 2). These results indicated accelerated evolution in the female dsx in these wasps and low purifying constraints. It is in agreement with recent work by Yang et al. (2008) showing that the C-terminal’ “tail” of the dsx protein in D. melanogaster does not play an important role in dimer formation or intersex binding of the female protein. Intersex and the female form of dsx function together to repress male sexual differentiation in Drosophila (Baker & Ridge, 1980).
Discussion
We have described the Nasonia dsx, an ortholog of the Drosophila transcription factor gene doublesex. Evidence that Nasonia dsx is the Drosophila dsx ortholog are (a) Nasonia dsx has an amino-terminus DNA binding domain (DM) that clusters phylogenetically with highly conserved dsx DM domains from other insect species, as well as an oligomerization domain (dsx dimer), (b) Nasonia dsx is expressed as male and female specific spliceforms, (c) expression of Nasonia dsx in phenotypically variable gynandromorphic mutants matches their sexual phenotype differentiation, consistent with a role of Nasonia dsx in sex determination. Research on the sex-determining mechanism of D. melanogaster has shown that the transcription factor coding gene dsx is the major switch at the base of the sex determination cascade, which subsequently determines somatic sex determination (Baker & Wolfner, 1988; Raymond et al., 1998). Here we have confirmed that in Nasonia the dsx gene is conserved, both in functional domains and in sex-specific splicing, indicating a similar major switch role in Nasonia sex determination.
In D. melanogaster, males can be considered the default gender, as the male specific variant of dsx splicing requires no active upstream gene activity. This male default situation also seems to apply to the Nasonia sex determining system, because the female specific variant of dsx requires an additional splicing event. However, in Bombyx the female splice variant is the default due to suppression of the male splice variant (Suzuki et al., 2001). Therefore, without knowledge of upstream regulatory factors, the default gender in Nasonia remains unknown.
In Drosophila, the active specification of the sex determining cascade ultimately leading to sex-specific splicing of dsx has long been considered the ratio of X chromosomes to autosomes (X:A). However, recent evidence indicates that the X-chromosome dosage rather than the X:A ratio is the key factor in sex determination in Drosophila (Erickson & Quintero, 2007). This dosage effect is exemplified through the timing of blastoderm formation: haploid embryos undergo an extra nuclear division cycle that prolongs the period in which X-encoded signal element proteins are expressed, whereas triploid embryos cellularize a cycle earlier than diploids. These authors recognize that sex determination in haplodiploids may work in a similar way. Crozier (1977) proposed that a chromosomal-cytoplasmic (maternal effect) balance could be the distinguishing signal between haploid and diploid embryos. Kamping et al. (2007) found evidence for a strong maternal effect on Nasonia sex determination. If haploid embryos develop slower than diploid embryos, a differential dosage effect of the chromosomal constitution of the embryo may tilt the balance towards males.
Sex determining mechanisms vary greatly at the chromosomal level, ranging from male heterogamety (XX-XY) to female heterogamety (ZZ-ZW) to haplodiploidy without heteromorphic sex chromosomes. The underlying genes for sex determination are organized in cascades and can also vary between closely related species or even within species (Werren & Beukeboom, 1998). Nevertheless, there is now ample evidence for evolutionary conservation of dsx at the base of these cascades in holometabolous insects. During evolution, genes appear to be added to the top of the cascade (Wilkins, 1995), but the responsible selective forces are not well understood.
We do not yet know which genes regulate alternative splicing of dsx in Nasonia, although transformer (TRA), or a hymenopteran homolog, is a likely candidate. The Drosophila transformer and transformer-2 (TRA2) proteins bind to regulatory elements, a 13-nucleotide sequence repeated 6 times, to activate the female specific splicing of the dsx primary transcripts (Inoue et al., 1992). We have not been able to identify these regulatory elements in Nasonia, and they are not present in Bombyx (Ohbayashi et al., 2001) and Apis (Cho et al., 2007) either. Recently, the feminizer (fem) gene was characterized in the honeybee sex determination pathway, and fem is also present in Nasonia (Hasselmann et. al., 2008). It was shown that knockdown of the female form of fem results in diploid male bees. Sex in honeybees is determined by complementation of two alleles of the complementary sex determiner gene (csd; Beye et al., 2003), fem is the ancestral paralog of csd and is also located near the csd locus (Hasselmann et. al., 2008). Molecular details of how fem and csd regulate dsx are not yet known. Since sex determination in Nasonia does not depend on heterezygosity, a system with the csd gene as the primary regulator of dsx could be ruled out in this species, nevertheless an involvement of fem seems probable.
Many models for sex determination in Nasonia have been proposed (reviewed in Cook, 1993, Beukeboom, 1995). Sex determination in Nasonia is currently viewed as an interaction of maternal effects, chromosome dosage and genomic imprinting (Beukeboom et al., 2007). Identification of the upstream genes to dsx in Nasonia is needed for a better understanding of its sex determination. This will also provide more insights in the evolution of sex determination in insects in general and that of Hymenoptera in particular.
Experimental Procedures
Nasonia rearing, strains, and gynandromorphy induction
Nasonia and the closely related genus Trichomalopsis were reared at 25°C on Sarcophaga bullata or Calliphora vicina pupae under constant light. Strains of four Nasonia species, AsymCX or WM114 (N. vitripennis), RV2X(u) (N. giraulti), IV7X (N. longicornis), NONYBr36/11 (as yet undescribed N. oneida), and two Trichomalopsis species, T. sarcophagae and T. dubius were studied. In addition, the N. vitripennis strain HiCD12 was also investigated, which produces gynandromorphic individuals (having both male and female characteristics) from unfertilized haploid eggs. Culturing details of the HiCD12 strain were described in Kamping et al. (2007) and Beukeboom et al. (2007). In short, high temperature (31°C) was used to induce maximum gynandromorph production as follows: virgin females were kept at 25°C for 2-3 days with fly pupae for host feeding, hosts were removed and females were left at 31°C for 12 hours, after which they were given new hosts for oviposition during 12 hours at 31°C, and parasitized hosts were put at 25°C for completion of Nasonia offspring development. Emerging gynandromorphs were selected based on their morphological resemblance to either males or females.
Tissue dissection and RNA purification
Male or female RNA was isolated from Nasonia tissue from a variety of life stages including embryos, larvae, pupae, and adults, and body parts including head, thorax, abdomen, and wing and leg imaginal discs. Tissue was dissected under RNAse free conditions in 1×PBS. After dissection, tissue was placed immediately on dry ice and if necessary stored at -80°C until RNA was isolated. Total RNA was isolated using Trizol (Invitrogen, CA) or Invisorb Spin Tissue RNA Purification Kit (Invitek, Germany). Poly(A) RNA was isolated using the Dynabeads mRNA DIRECT Kit (Dynal Biotech, Norway) by the mini volumes protocol. When necessary, RNA was quantified using a Qubit fluorometer (Invitrogen, CA) and a Quant-iT RNA Assay Kit (Invitrogen, CA) or a ND-1000 Spectrophotometer (Nanodrop technologies, UK). All steps were performed according to the respective manufacturer's protocols.
Exonic mRNA composition determination by RT-PCR and 5′ and 3′ RACE
Poly(A) RNA or total RNA were reverse transcribed using SuperScript III (Invitrogen, CA) and Oligo(dT)12-18 primer (Invitrogen, CA) or using RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas, MD) and 3′ RACE adapter supplied with FirstChoice RLM-RACE kit (Ambion, TX). Both methods yielded similar results. The resulting cDNA was used for PCR with Taq polymerase (Invitrogen, CA) under the following conditions: 94°C for 2 min; 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min; and 72°C for 10 min. PCR screening for male versus female spliceforms was done with forward primer dsdsx_FF (5′-CTG CCG AGT ATA CCA ATA CC-3′) and reverse primer dsdsx_FR2 (5′-GTA ATA GAA CAT TGG CAT CAA CC-3′). For full mRNA sequences, 5′- and 3′-RACE were conducted on 150 ng of Poly(A) RNA from NV and NG using the GeneRacer Kit (Invitrogen, CA). Reverse transcription was done with a mixture of the included oligo-dT primer and a gene specific primer dsx_1R (5′-TCG AGT CCT TTA AGA TTA CGT ACA T-3′).
Standard PCR and DNA Sequencing
Genomic sequencing of all coding exons was performed for all four Nasonia species and the two Trichomalopsis species. DNA was obtained from single wasps (DNAeasy; Qiagen, CA) and long PCRs were performed with AmpliTaq (Invitrogen, CA). Primers used are available from the authors upon request. Sequencing reactions were sent to the Cornell Biotechnology Resource Center or the University of Rochester Functional Genomics Center. Sequences were edited using the program Sequencher 4.7 (Gene Codes Corp, MI).
Sequence data from this article have been deposited with the GenBank Data Libraries under accession nos. FJ440989-FJ440998
Bioinformatics and phylogenetic analyses
Five insect genomes, D. melanogaster (Build 5.1), An. gambiae (AgamP3), T. castaneum (Build 1.1), A. mellifera (Amel 4.0), and N. vitripennis (Nvit 1.0) were queried for genes containing DM domains using blast searches (Altschul et al., 1990). The two conserved domains, DM and dsx dimer, in the dsx protein were aligned using the consensus sequences in the Pfam database as references (Finn et al., 2006); pfam00751: DM DNA binding domain (DM domain) and pfam08828: doublesex dimerization domain (dsx dimer). Visual inspection of the alignment was performed with MacClade 4.08 (Maddison & Maddison, 2000).
Maximum parsimony (1000 tree-bisection-reconnection replicates) and neighbor-joining trees were inferred in Paup 4.0 (Swofford, 2002) using amino acid sequences. Since unambiguous alignment is restricted to the two conserved domains (DM domain and dsx dimer), only these regions were used in phylogenetic inferences. To construct a DM domain gene tree 67 aligned characters (42 parsimony-informative) were included, the entire DM domain plus the five flanking N-terminal amino acids and the 15 flanking C-terminal amino acids. To construct an insect phylogeny based on the dsx gene the same DM domain region was used combined with the dsx dimer region and the female specific region. The region included the dsx dimer, the flanking five N-terminal amino acids and five flanking C-terminal amino acids, generating 83 additional characters. In total 138 characters were included in the dsx insect phylogeny (88 parsimony-informative). Branch support was accessed by 500 bootstrap replicates (Felsenstein, 1988).
Acknowledgments
We are grateful to Rachael Edwards for assistance with the laboratory work. We thank anonymous reviewers for valuable comments. The work in the John H. Werren laboratory was funded by a National Institute of Health grant, 5R01 GM070026. Work in the Leo W. Beukeboom laboratory was funded by Pioneer grant, ALW 833.02.003, from the Netherlands Organization for Scientific Research.
Literature Cited
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bayrer JR, Zhang W, Weiss MA. Dimerization of doublesex is mediated by a cryptic ubiquitin-associated domain fold: implications for sex-specific gene regulation. J Biol Chem. 2005;280:32989–32996. doi: 10.1074/jbc.M507990200. [DOI] [PubMed] [Google Scholar]
- 3.Baker BS, Ridge K. Sex and the single cell: On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 1988;2:477–489. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- 5.Beukeboom LW. Sex determination in Hymenoptera: a need for genetic and molecular studies. Bioessays. 1995;17:813–817. doi: 10.1002/bies.950170911. [DOI] [PubMed] [Google Scholar]
- 6.Beukeboom L, Desplan C. Nasonia. Curr Biol. 2003;13:R860. doi: 10.1016/j.cub.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 7.Beukeboom LW, Kamping A. No patrigenes required for femaleness in the haplodiploid wasp Nasonia vitripennis. Genetics. 2006;172:981–989. doi: 10.1534/genetics.105.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beukeboom LW, Kamping A, Louter M, Pijnacker LP, Katju V, et al. Haploid females in the parasitic wasp Nasonia vitripennis. Science. 2007;315:206. doi: 10.1126/science.1133388. [DOI] [PubMed] [Google Scholar]
- 9.Beukeboom L, Kamping A, van de Zande L. Sex determination in the haplodiploid wasp Nasonia vitripennis (Hymenoptera: Chalcidoidea): A critical consideration of models and evidence. Semin Cell Dev Biol. 2007;18:371–378. doi: 10.1016/j.semcdb.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–429. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 11.Cho S, Huang ZY, Zhang J. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics. 2007;177:1733–1741. doi: 10.1534/genetics.107.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook JM. Sex determination in the Hymenoptera: a review of models and evidence. Heredity. 1993;71:421–435. [Google Scholar]
- 13.Crozier RH. Heterozygosity and sex determination in haplodiploidy. Am Nat. 1971;105:399–412. [Google Scholar]
- 14.Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dosage, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5:2821–2830. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 16.Ferree PM, McDonald K, Fasulo B, Sullivan W. The origin of centrosomes in parthenogenetic hymenopteran insects. Curr Biol. 2006;16:801–807. doi: 10.1016/j.cub.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 17.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucl Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasselmann M, Gempe T, Schiott M, Nunes-Silva CG, Otte M, Beye M. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature. 2008;454:519–522. doi: 10.1038/nature07052. [DOI] [PubMed] [Google Scholar]
- 19.Hediger M, Burghardt G, Siegenthaler C, Buser N, Hilfiker-Kleiner D, et al. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol. 2004;214:29–42. doi: 10.1007/s00427-003-0372-2. [DOI] [PubMed] [Google Scholar]
- 20.Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci USA. 1992;89:8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamping A, Katju V, Beukeboom LW, Werren JH. Inheritance of gynandromorphism in the parasitic wasp Nasonia vitripennis. Genetics. 2007;175:1321–1333. doi: 10.1534/genetics.106.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature. 2006;439:728–732. doi: 10.1038/nature04445. [DOI] [PubMed] [Google Scholar]
- 23.Maddison DR, Maddison WP. MacClade: Analysis of phylogeny and character evolution, version 4.0. Sinauer Associates; Sunderland, Massachusetts: 2000. [Google Scholar]
- 24.Ohbayashi FM, Suzuki MG, Mita K, Okano K, Shimada T. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp Biochem Physiol B Biochem. Mo Biol. 2001;1:145–158. doi: 10.1016/s1096-4959(00)00304-3. [DOI] [PubMed] [Google Scholar]
- 25.Pultz MA, Leaf DS. The jewel wasp Nasonia: querying the genome with haplo-diploid genetics. Genesis. 2003;35:185–191. doi: 10.1002/gene.10189. [DOI] [PubMed] [Google Scholar]
- 26.Raymond CS, Shamu CS, Shen MM, Selfert KJ, Hirsch B, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 27.Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- 28.Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz MF, Stefani RN, Mascarenhas RO, Perondine AL, Selivon D, et al. The gene doublesex of the fruit fly Anastrepha obliqua (Diptera, Tephritidae) Genetics. 2005;171:849–854. doi: 10.1534/genetics.105.044925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scali C, Catteruccia F, Li Q, Crisanti A. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J Exp Biol. 2005;208:3701–3709. doi: 10.1242/jeb.01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skinner SW, Werren JH. The genetics of sex determination in Nasonia vitripennis (Hymenoptera, Pteromalidae) Genetics. 1980;94:s98. [Google Scholar]
- 32.Sherman DC, Frommer M. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determining gene doublesex. Insect Mol Biol. 1998;7:355–366. doi: 10.1046/j.1365-2583.1998.740355.x. [DOI] [PubMed] [Google Scholar]
- 33.Shuker D, Lynch J, Peire Morais A. Moving from model to non-model organisms? Lessons from Nasonia wasps. Bioessays. 2003;25:1247–1248. doi: 10.1002/bies.10367. [DOI] [PubMed] [Google Scholar]
- 34.Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki MG, Ohbayashi F, Mita K, Shimada T. The mechanism of sex-specific splicing at the doublesex is different between Drosophila melanogaster and Bombyx mori. Insect Biochem Mol Biol. 2001;31:1201–1211. doi: 10.1016/s0965-1748(01)00067-4. [DOI] [PubMed] [Google Scholar]
- 36.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods), version 4.0 beta. Smithsonian Institution; Washington, DC: 2002. [Google Scholar]
- 37.Trent C, Crosby C, Eavey J. Additional evidence for the genomic imprinting model of sex determination in the haplodiploid wasp Nasonia vitripennis: isolation of biparental diploid males after X-ray mutagenesis. Heredity. 2006;96:368–376. doi: 10.1038/sj.hdy.6800810. [DOI] [PubMed] [Google Scholar]
- 38.Werren JH, Gadau J, Beukeboom L, Desplan C, Lynch J, et al. Proposal to sequence the Nasonia genome. 2004 www.genome.gov/Pages/Research/Sequencing/SeqProposals/NasoniaSeq.pdf.
- 39.Werren JH, Stouthamer R. PSR (Paternal Sex Ratio) chromosomes: The ultimate selfish genetic elements. Genetica. 2002;117:85–101. doi: 10.1023/a:1022368700752. [DOI] [PubMed] [Google Scholar]
- 40.Werren JH, Beukeboom L. Sex Determination, Sex Ratios and Genetic Conflict. Ann Rev Ecol Syst. 1998;29:233–261. [Google Scholar]
- 41.Whiting PW. Multiple Alleles in Complementary Sex Determination of Habrobracon. Genetics. 1943;28:365–382. doi: 10.1093/genetics/28.5.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiting AR. The biology of the parasitic wasp Mormoniella vitripennis. [=Nasonia brevicornis] (Walker) Q Rev Biol. 1967;42:333–406. [Google Scholar]
- 43.Wilkins AS. Moving up the hierarchy. A hypothesis on the evolution of a genetic sex determination pathway. Bioessays. 1995;17:71–77. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]
- 44.Wolf U. Reorganization of sex-determining pathway with the evolution of placentation. Human Genet. 1999;105:288–292. doi: 10.1007/s004390051103. [DOI] [PubMed] [Google Scholar]
- 45.van Wilgenburg E, Driessen G, Beukeboom LW. Single locus complementary sex determination in Hymenoptera: an “unintelligent” design? Front Zool. 2006;3:1–15. doi: 10.1186/1742-9994-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Zhang W, Bayrer JR, Weiss MA. Doublesex and the regulation of sexual dimorphism in Drosophila melanogaster: Structure, function, and mutagenesis of the female-specific domain. J Biochem Chem. 2008;283:7280–7292. doi: 10.1074/jbc.M708742200. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, et al. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]