Abstract
We have previously shown that the microenvironment of human embryonic stem cells (hESCs) is able to change and reprogram aggressive cancer cells to a less aggressive state. Some mechanisms implicated in the phenotypic changes observed after this exposure are mainly associated with the Nodal signaling pathway, which plays a key role in tumor cell plasticity. However, several other molecular mechanisms might be related directly and/or indirectly to these changes, including microRNA (miRNA) regulation and DNA methylation.
Aim:
To further explore the epigenetic mechanisms potentially underlying the phenotypic changes that occur after exposing metastatic melanoma cells to a hESC microenvironment.
Materials & Methods:
A total of 365 miRNAs were screened using the TaqMan® Low Density Arrays. We also evaluated whether DNA methylation could be one of the factors regulating the expression of the inhibitor of Nodal, Lefty, in hESCs (where it is highly expressed) vs melanoma cells (where it is not expressed).
Results:
Using these experimental approaches, we identified miRNAs that are up- and down-regulated in melanoma cells exposed to a hESC microenvironment, such as miR-302a and miR-27b, respectively. We also demonstrate that Notch4 is one of the targets of miR-302a, which is upstream of Nodal. Additionally, one of the mechanisms that might explain the absence of the inhibitor of Nodal, Lefty, in cancer cells is silencing by DNA methylation, which provides new insights into the unregulated expression of Nodal in melanoma.
Conclusion:
These findings suggest that epigenetic changes such as DNA methylation and regulation by microRNAs might play a significant role in tumor cell plasticity and the metastatic phenotype.
Keywords: DNA methylation, hESC microenvironment, Lefty, microRNAs, Nodal, Notch
The diagnosis of human melanoma is becoming more frequent, especially in Caucasians, largely as a result of increased exposure to ultraviolet radiation from the sun owing to increased outdoor activity [1]. Often, early diagnosis leads to prompt medical intervention and surgical excision resulting in high cure rates. Unfortunately, diagnosis of late-stage disease, characterized by the metastatic spread of malignant melanoma to the deeper layers of the skin, regional lymph nodes and distant organs, significantly reduces patient survival to a few months [2]. Furthermore, treatment with specific drugs alone or in combination has not shown any significant survival increase for patients with advanced stage metastatic melanoma [3]. Thus, understanding the molecular underpinnings responsible for the enhanced growth rate in the metastatic phenotype and drug resistance of advanced malignant melanoma would provide more efficient therapeutic strategies.
Recent work has demonstrated that it is possible to reprogram the aggressive phenotype of melanoma cells by regulating the expression of certain stem cell-related molecules and associated signaling pathways, which contribute to melanoma's biological plasticity [4]. Two stem cell signaling pathways that have been implicated in melanoma plasticity are Nodal and Notch [5]. Nodal, a member of the TGF-β superfamily, is involved in stem cell maintenance and differentiation [6]. Notch is an evolutionarily highly conserved transmembrane receptor involved in cellular proliferation and differentiation [7]. We have previously shown experimental evidence suggesting that molecular cross-talk exists between these two signaling pathways in melanoma; however, our understanding of this interaction remains unclear [5]. One of the promising reprogramming models employed by our laboratory has been the microenvironment of human embryonic stem cells (hESCs), which contains Lefty, an inhibitor of Nodal. Exposure of metastatic melanoma and breast carcinoma to this hESC microenvironment results in suppression of Nodal expression, proliferation, clonogenic potential and tumorigenicity, thereby reprogramming their metastatic phenotype [8]. However, we believe that additional mechanisms such as epigenetic regulation by DNA methylation and microRNAs (miRNAs) might also be implicated in these changes, and this related information would provide novel insights into the molecular regulation of tumor cell plasticity and the malignant phenotype.
Epigenomics has become a field of increasing interest in the last 5 years, including the cataloging of epigenetic modifications that occur in the different cell types of the body and in diseases such as cancer. Epigenetic regulation has been known to involve three interacting events – DNA methylation, histone modifications and nucleosomal remodeling. Alterations in gene expression through aberrant epigenetic modifications such as DNA methylation in promoter regions and the first exons of genes can lead to initiation, promotion and maintenance of tumorigenesis [9]. In addition, other mechanisms such as signaling via ion flows can act as an epigenetic modification that is able to change cell function at the post-translational levels, being relevant for complex signaling pathways that control gene transcription and, consequently, protein levels [10]. Epigenomics also includes gene regulation by miRNAs, which are a large group of noncoding RNAs (ncRNAs) that can block mRNA translation and affect mRNA stability [11]. It is estimated that the human genome has thousands of miRNA genes, and approximately 900 have been described to date [101]. Interestingly, it is speculated that this class of noncoding genes is able to regulate at least 30% of all human protein-coding genes by targeting their 3′-untranslated region (UTR) sequences [12]. There is also evidence that miRNAs can regulate the expression of large ncRNAs, indicating that these small genes have a big impact on cellular transcriptome networks [13]. An important concept in the miRNA biology of therapeutic relevance is that one miRNA can downregulate hundreds to thousands of target proteins by interacting with different target mRNAs (the ‘one-hit multiple targets’ concept) [14].
Recently, miRNA-deregulated expression has been extensively described in different types of cancers [15,16]. Some of these reports in human cancers indicate that miRNAs can function as classic tumor suppressors or oncogenes [17]. miRNAs have been associated with the initiation of tumorigenesis [18], in addition to important tumor-suppressor gene networks involving the p53 gene [19], and in the metastatic process [20,21]. Furthermore, miRNAs have been used to classify different tumor types [22]. Thus, there is growing evidence that miRNAs are implicated in multiple steps of tumorigenesis.
In this study, we focused on identifying some of the epigenetic mechanisms underlying the phenotypic changes that occur as a consequence of reprogramming aggressive melanoma tumor cells exposed to a hESC microenvironment. In order to achieve this goal, we have used TaqMan® Low Density Arrays ([TLDAs]; Applied Biosystems, CA, USA) to screen 365 miRNAs by real-time (RT) PCR before and after melanoma cell reprogramming. Using this approach, we identified miRNAs that are up- and down-regulated, such as miR-302a and miR-27b, respectively. We also demonstrate that Notch4 may be one of the target genes controlled by miR-302a contributing to the molecular cross-talk with the Nodal pathway. We also discovered that DNA methylation could be one of the factors regulating the expression of Nodal's inhibitor, Lefty, thus providing new insights into the silencing of this gene in cancer cells resulting in the unregulated expression of the Nodal plasticity gene.
Materials & methods
Cell culture
Human cutaneous melanoma cell lines (C8161, C81-61) have been well characterized with respect to their gene expression, invasiveness, metastatic potential and clinical significance, and were maintained as previously described [23,24]. HEK293T cells were purchased from the American Type Culture Collection ([ATCC]; VA, USA) (CRL11268) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). The normal human neonatal epidermal melanocytes ([HEMn-LP]; Cascade Biologics, OR, USA) were maintained in Medium 254 (Invitrogen, CA, USA) with human melanocyte growth supplements (HMGSs). The amniotic fluid-derived stem cell line (GM00957A – AF MSC) and adult bone marrow-derived mesenchymal stem cells ([BM MSC]; Stem Cell Technologies, BC, Canada) were maintained under the recommended conditions. The HTR-8/SVneo is a well-characterized immortalized human extravillous cytotrophoblast cell line and was maintained as previously described [8]. The H9 human embryonic stem cell line ([hESCs]; WiCell, WI, USA) was maintained as previously described [25]. For the conditioned matrix experiments, hESCs were maintained in stem cell medium preconditioned by irradiated mouse embryonic fibroblasts or StemPro® hESC SFM medium (Invitrogen Life Technologies, CA, USA) as previously described [25].
3D conditioned matrix experiments
Conditioned matrices were prepared using H9 hESCs seeded onto growth factor-reduced Matrigel™ (14 mg/ml; BD Biosciences, CA, USA) for 3–4 days as previously described [25]. In all cases, cells were 70–90% confluent during the conditioning of the matrix. Subsequently, the hESCs were removed from the Matrigel with a treatment of 20 mM NH4OH, followed by rapid H2O washes and a final phosphate-buffered saline (PBS) rinse, leaving behind a denuded, conditioned 3D matrix (hESC CMTX). C8161 melanoma cells (2.5 × 105 cells/six-well dish) were then either seeded onto this conditioned matrix or unconditioned Matrigel for 3–4 days. At the end of the incubation period the cells were harvested and total genomic DNA (gDNA), RNA and protein were isolated.
Genomic DNA extraction & methylation analyses
Genomic DNA was prepared from all cell lines grown on either plastic or 3D matrices using the Puregene® DNA isolation kit (Gentra Systems, MN, USA) following the manufacturer's protocol for 1–3 × 106 cells/sample or by digestion with proteinase K (Qiagen, CA, USA) followed by phenol/chloroform extraction. gDNA was subjected to sodium bisulfite treatment using the CpGenome™ DNA Modification Kit (Zymo Research, CA, USA). Bisulfite-treated DNA was amplified by a nested-PCR protocol as previously described for other genomic regions [26] using specific primers for Lefty B as shown in Table 1. Amplified products were purified using the Gel Purification Kit (Qiagen) and were ligated to a vector using the TOPO® TA Cloning® Kit (Invitrogen). A total of 24 positive clones were sequenced for each sample using the vector's forward and reverse primers. DNA sequencing reactions were performed using the DNA dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and an ABI3730xl sequencer (Applied Biosystems) according to the manufacturer's instructions. The DNA sequence generated was subjected to bioinformatic analyses to summarize the CpG methylation status of these clones. Sequence reads were first trimmed of low-quality sequence by the Phred program (University of Washington, WA, USA) [27] and vector sequences removed using the RepeatMasker program (Institute for Systems Biology, WA, USA) [102]. The sequences were correctly oriented with respect to the wildtype sequence (NCBI Human Genome Build 36.1) with the use of the bl2seq program [28], and then aligned by using the Multalin program [29]. Finally, perl scripts were written to identify the methylation status of all CpG sites in the sequences and then to output a summary of all the methylation sites in text and graphical formats. All clones displayed in the figure showed more than 98% bisulfite conversion efficiency.
Table 1.
Primers used for amplifications after DNA bisulfite conversion in a nested-PCR reaction for the CpG island of Lefty B.
| Primer | Sequence |
|---|---|
| LEFTY 1 (B) (60 CpGs) | |
| LEFTY B EF | 5′-TAG TTT TTA AGG TTT AGG GTG TG-3′ |
| LEFTY B ER | 5′-TAC TAA CCC TAC TCT TAT CCC-3′ |
| LEFTY B IF | 5′-AG TTT TAG TTG GGG TTT TTT AAG-3′ |
| LEFTY B IR | 5′-TTA AAA ACC AAC ACA CAC CTA C-3′ |
EF/ER: External primers; IF/IR: Internal primers.
RNA isolation, RT-PCR & Western blot analyses
Total RNA and miRNA were isolated using the miRvana™ kit (Ambion/Applied Biosystems) and Trizol® (Invitrogen). RT-PCR was performed on a 7500 Real Time PCR System (Applied Biosystems) using TaqMan gene expression primer/probe sets specific for Nodal (Hs00250630_s1) and Notch4 (Hs00270200_m1) (Applied Biosystems) as previously described [24]. The expression of each target gene was normalized to an endogenous control gene (RPLP0 large ribosomal protein; 4333761F, Applied Biosystems). Whole-cell lysates were obtained for Western blot analysis as previously described [8]. Densitometric analysis was performed to determine the relative expression of Western blot bands. The relative index for expression was determined after normalizing for equal loading of total protein as determined by actin expression.
Quantification of miRNAs by TaqMan low-density arrays
TaqMan MicroRNA Arrays V1.0 were used to quantify the expression levels of 365 mature miRNAs and controls according to the manufacture's instructions. In all miRNA profiling, the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was used to convert RNA into cDNA according to the manufacturer's instructions. The reactions were incubated for 30 min at 16°C, 30 min at 42°C and 5 min at 85°C. Reactions were run in an 7900HT Fast RT-PCR system (Applied Biosystems) in a 384-well plate at 94.5°C for 10 min, followed by 40 cycles at 97°C for 30 s and 60°C for 1 min.
Bioinformatic analyses
The expression of 365 miRNAs was analyzed by the StatMiner® software V 3.0 (Integromics®, PA, USA) to identify miRNAs that displayed a statistically significant (p < 0.05) expression level difference between the groups of interest. δ Ct values from each miRNA were also normalized against controls from the TLDAs (chosen by the StatMiner software). The MeV4.1 Software [103] was used to generate heat maps of unsupervised and supervised hierarchical clusterings by Pearson correlation using average linkage clustering. Target identification for miR-302a was evaluated by TargetScan, PicTar, miRanda and RNAhybrid.
miRNA confirmation & miR-302a overexpression
Differences in miRNA expression were confirmed using TaqMan assays (Applied Biosystems) for miR-27b and miR-302a according to the manufacturer's instructions. C8161 cells were transfected with miR-302a pre-miR miRNA precursor or pre-miR negative control 1 using the manufacture's recommendations for 6-well plates (Ambion/Applied Biosystems). After 48 h, cells were harvested for RNA and protein isolation. TaqMan assays for miR-302a were used to confirm the overexpression of this specific miRNA in transfected cells.
Luciferase assay for miR-302a binding site in Notch4
The Notch4 3′-UTR (containing the putative miR-302a binding site) was cloned into the 3′-UTR of the luciferase gene (pMIR-REPORT Luciferase vector, Applied Biosystems). HEK293T cells were co-transfected with Notch4 UTR-luciferase plasmid and pre-miR-302a using Arrest-In™ transfection reagent (Open Biosystems/ThermoFisher, AL, USA), as per manufacturer's suggested protocol. At 48 h, cells were lysed with Glo Lysis Buffer (Promega, WI, USA) and an equal volume of luciferase assay reagent was added. Luciferase activity was measured after 5 min in a FLUOstar OPTIMA luminometer (BMG Labtech, NC, USA). Each parameter was assayed in triplicate and the experiment performed three times. Statistical significance was determined using the Mann-Whitney U test.
Results
miRNA expression profiles after exposure of aggressive melanoma cells to a hESC microenvironment
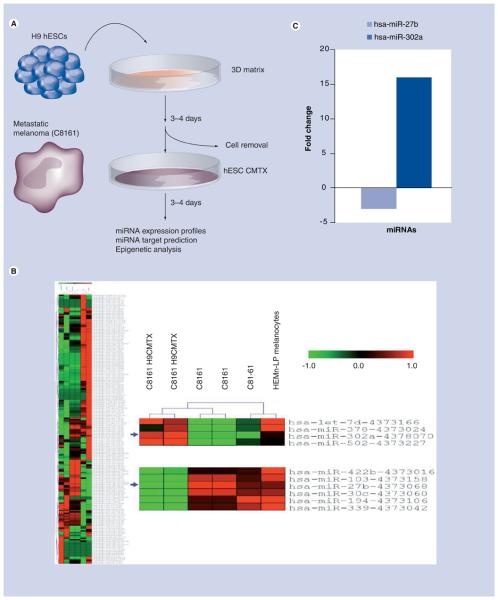
Our group has previously reported that exposing aggressive melanoma cells to a hESC micro-environment leads to a dramatic decrease in Nodal gene expression, thus reprogramming these cells to a less aggressive phenotype (experimental model is shown in Figure 1a). Some of the mechanisms implicated in the reprogramming of these tumor cells were recently described in detail [30]; however, several other unknown mechanisms may be directly and/or indirectly related to these changes, especially epigenetic modifications such as DNA methylation and/or histone modifications and gene regulation by miRNAs. In this study, we have generated miRNA expression profiles of aggressive melanoma cells (C8161) before and after exposure to a hESC microenvironment. We have also included a poorly aggressive melanoma cell line (C81–61) and normal melanocytes (HEMn-LP) as controls. After generating expression profiles for 365 miRNAs and using hierarchical clustering to generate heat maps, we were able to show that the less aggressive melanoma cells (C81–61) and normal melanocytes (HEMn-LP) cluster together, and, most noteworthy, that C8161 cells exposed to a hESC microenvironment cluster in a different branch compared with C8161 control cells (Figure 1B). Using this strategy, we were able to identify specific miRNAs that were upregulated after exposure of melanoma cells to the hESC microenvironment and some that were downregulated, such as miR-302a and miR-27b, respectively (Figure 1C). We performed additional analyses of miR-302a since this miRNA has been associated with embryogenesis [31] and developmental circuitries [32], and is highly expressed in embryonic stem cells [33]. To achieve this goal, we introduced a pre-miR-302a oligonucleotide in C8161 cells to analyze putative targets that are important in the Nodal signaling pathway.
Figure 1. miRNA expression profiles of aggressive melanoma cells (C8161) before and after exposure to a hESC microenvironment (see right).
(A) Human embryonic stem cell microenvironment experimental flow chart. The hESC-H9 embryonic stem cells (hESCs) were seeded as compact clusters onto a 3D matrix of growth factor-reduced Matrigel (BD Biosciences, CA, USA) in the presence of mouse embryonic fibroblast (mEF) conditioned stem cell medium or StemPro® hESC-SFM (Invitrogen, CA, USA) for 3–4 days. The stem cells were then removed from the 3D matrix with a NH4 OH treatment followed by thorough washes with double-distilled H2O, phosphate-buffered saline and complete medium, leaving behind a denuded, conditioned 3D matrix (hESC CMTX or H9CMTX). The metastatic cutaneous melanoma cells (C8161, 2.5 × 105 cells/well of six-well dish) were then seeded onto the denuded hESC CMTX for 3–4 days. At the end of this period, changes in phenotype, biological properties, microRNA expression profiles and epigenetic modifications were analyzed on the melanoma cells exposed to the hESC-conditioned matrix microenvironment as compared with results from melanoma cells plated onto Matrigel that was not conditioned. (B) Unsupervised hierarchical clustering using Pearson correlation for the aggressive melanoma cell line C8161, C8161 exposed to a hESC microenvironment (C8161 H9CMTX), a poorly aggressive isotype cancer cell line (C81–61) and normal melanocytes (HEMn-LP). On the left is the heat map for approximately 365 microRNAs on all cell lines, and on the right is a magnification of some differentially expressed microRNAs indicated with blue arrows. Log scale for differences in expression is also shown. (C) Individual confirmation for some examples of differentially expressed microRNAs (miR-27b and miR-302a) before and after exposure to a hESC microenvironment. Even though the Ct values for miR-302a in the TLDA arrays were low, we were able to see differences when comparing control cells with cells exposed to a hESC microenvironment using individual assays. The histograms show fold change differences between C8161 cells lines and C8161 cells exposed to hESC microenvironment. miRNA: MicroRNA.
miRNA-302a overexpression & Notch4 regulation
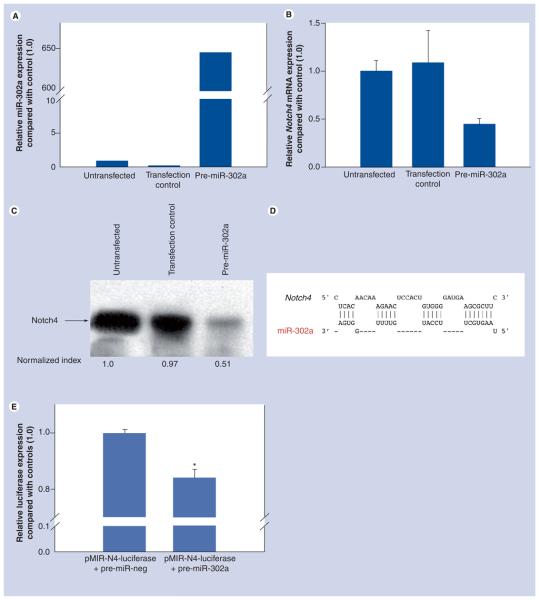
C8161 melanoma cells were transiently transfected to overexpress miR-302a and, as shown in Figure 2A, these tumor cells expressed miR-302a at very high levels when compared with untransfected controls. Our study indicated that melanoma cells transfected with pre-miR-302a demonstrate a downregulation of Notch4 by approximately 50%, at both the RNA and protein levels (Figure 2B & Figure 2C, respectively). We also discovered an alignment between miR-302a and Notch4, using the RNAhybrid software (Figure 2D), supporting the hypothesis that this downregulation might be important for Nodal signaling and molecular cross-talk between these two stem cell pathways. We have also performed a luciferase assay to confirm the regulation of Notch4 by miR-302a. The co-transfection of miR-302a and Notch4 3′-UTR (containing the region identified as a binding site for miR-302a by RNAhybrid) was able to significantly reduce luciferase expression (Figure 2E).
Figure 2. Overexpression of miR-302a and Notch signaling pathway regulation.
Human aggressive melanoma cells (C8161) transfected with pre-miR-302a (A) showed lower levels of both Notch4 mRNA (B) and protein (C) compared with untransfected and pre-miR negative controls. (D) Binding site prediction of miR-302a to the 3′-untranslated region (UTR) of the Notch4 gene using the RNAhybrid software (minimum free energy: −22.7 Kcal/mol). (E) The Notch4 3′UTR (containing the putative miR-302a binding site) was cloned at the 3′-end of a luciferase reporter gene. The reporter vector and pre-miR-302a or a pre-miR-negative control were co-transfected into HEK 293T cells and a significant downregulation of luciferase activity was observed. Data shown were normalized to control (pMIR-N4-luciferase+pre-miR-neg).
*p < 0.05
Regulation of Lefty by DNA methylation in hESCs, fetal & adult MSCs & melanoma tumor cells
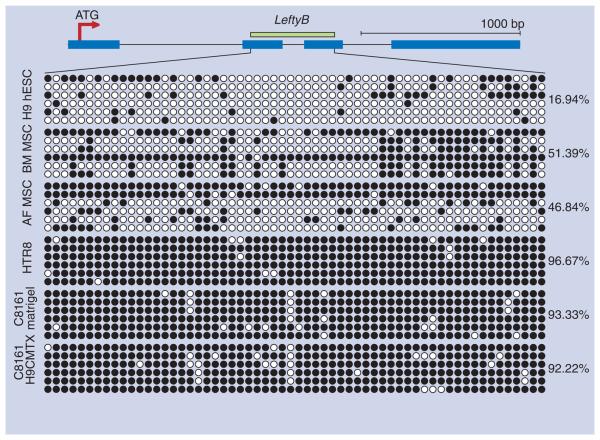
The next question we sought to address was the mechanism(s) by which Lefty A and B, natural inhibitors of Nodal signaling, might be regulated in various stem cell types, including: hESCs, adult bone marrow and fetal amniotic fluid mesenchymal stem cells (BM MSC and AF MSC, respectively), immortalized trophoblasts (HTR8) and melanoma cells before and after exposure to a hESC microenvironment. The two Lefty homologues are coordinately arranged on chromosome 1q42 and have a 97% nucleotide identity within their coding regions (with 96% amino acid identity at the protein level). Since both genes are very small and have internal CpG islands mapped to the second and third exons, we investigated whether DNA methylation in this region could play a role in their expression. We utilized sodium bisulfite sequencing to specifically amplify the CpG island within Lefty B. Our data presented in Figure 3 show that Lefty B is highly methylated in C8161 melanoma cells, indicating the unlikelihood that these cells could express Lefty, which has been previously confirmed by RT-PCR and Western blot analyses [8]. DNA methylation evaluation of H9 hESCs demonstrated that this region in Lefty B is highly unmethylated in these normal embryonic stem cells (Figure 3). This new finding supports our previous observations showing the localization of hESC-derived Lefty in the stem cell microenvironment and its ability to downregulate Nodal expression in metastatic tumor cells [8]. Additional DNA methylation analyses of Lefty's CpG island shows methylation in other human stem cells derived from amniotic fluid and adult bone marrow, as well as in human trophoblasts (Figure 3). Collectively, these data demonstrate for the first time that DNA methylation may represent a mechanism by which Lefty expression is regulated in hESC, adult stem cells, trophoblasts and melanoma cells.
Figure 3. CpG island DNA methylation analyses of Lefty B by sodium bisulfite sequencing.
Scaled map of Lefty B in human chromosome 1 is shown on the top. Exons are represented by blue boxes and introns by lines. Green box depicts the CpG island. The red arrow represents the first ATG of translation. Methylation analysis by sodium bisulfite sequencing was generated for the CpG island of Lefty B. A total of 60 CpGs were analyzed for DNA methylation. Each circle represents a CpG dinucleotide and spacing between circles is relative. Six clones or alleles are represented for each sequenced region. Each row represents a sequenced clone or allele. Cell lines analyzed by this methodology are indicated on the left and include human embryonic stem cell (H9 hESC), bone marrow-derived mesenchymal stem cells (BM MSC), amniotic fluid-derived stem cell line (AF MSC), immortalized human extravillous cytotrophoblast cell line (HTR8), C8161 on Matrigel™ and C8161 exposed to the hESC H9 microenvironment (C8161 H9CMTX). Methylation percentages are indicated on the right. White circles: Unmethylated; Black circles: Methylated.
Discussion
We have previously demonstrated that the microenvironment of hESCs suppresses the tumorigenic phenotype of aggressive cancer cells, including metastatic melanoma and breast carcinoma, by downregulating the embryonic morphogen Nodal, which is essential for embryonic and tumor cell plasticity [8]. Furthermore, we discovered that metastatic tumor cells do not express the inhibitor of Nodal, Lefty, allowing them to express this embryonic morphogen in an unregulated manner. Specifically, our studies revealed that the tumor cells can respond to the Lefty produced by hESCs (sequestered within their embryonic microenvironment), resulting in a dramatic downregulation of tumor cell-derived Nodal, concomitant with a reduction in clonogenicity and tumorigenesis. Interestingly, this ability to suppress the tumorigenic phenotype is directly related to tumor cell exposure to Lefty, which is exclusive to hESCs, because it is not detected in appreciable levels in other stem cell types or trophoblasts. Thus, the purpose of the current study was to further explore the epigenetic mechanisms potentially underlying the reprogramming of metastatic tumor cells, particularly aggressive melanoma exposed to a hESC microenvironment, to elucidate the tumor-suppressive effects of this embryonic milieu.
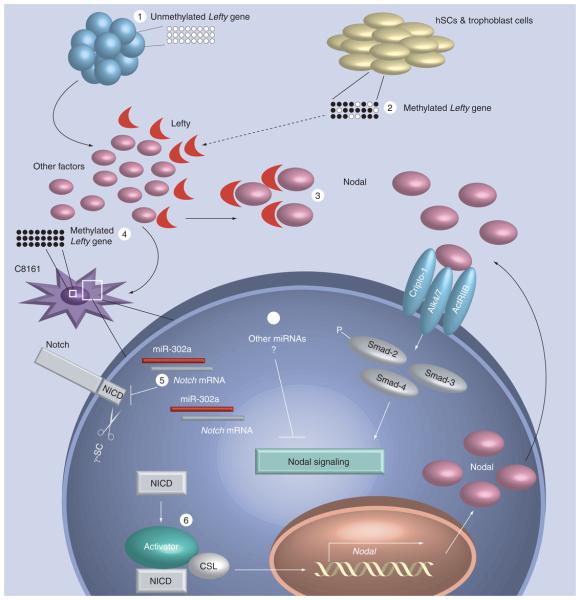
To summarize our findings, we present a hypothetical model describing select key events underlying the epigenetic reprogramming of metastatic melanoma cells (Figure 4). Human melanoma cells cultured on a hESC microenvironment containing hESC-derived Lefty (where the gene is unmethylated) demonstrate remarkably elevated levels of miR-302a and downregulated Notch4 and Nodal expression, as these cells acquire a less aggressive phenotype. The methylated status of the Lefty gene in these tumor cells may explain their inability to express this natural inhibitor of Nodal. The unmethylated Lefty gene permits high-level expression of Lefty by hESCs. By comparison, the high methylation status of the Lefty gene in human adult stem cells (hSCs) and in human trophoblasts may be responsible for the poorly detectable levels of Lefty in these cells, and support the finding that their respective microenvironments have no ability to reprogram the aggressive phenotype of tumor cells nor affect Nodal expression [8]. hESC-derived Lefty is capable of inhibiting Nodal signaling by preventing Nodal from forming an active receptor complex with Cripto-1-Alk4/7-ActRIIB, necessary for proper Smad2/3/4-dependent downstream signaling. The Lefty gene is highly methylated in C8161 cells, which supports their inability to express Lefty resulting in the unregulated expression of the Nodal plasticity gene.
Figure 4. Model of melanoma cells exposed to a hESC microenvironment including miR-302a and epigenetic changes as new players.
(1) Unmethylated Lefty gene permits high-level expression of Lefty by human embryonic stem cells (hESCs). (2) High methylation status of the Lefty gene in nonembryonic human adult stem cells (hSCs) and in human trophoblast cells may be responsible for the poorly detectable levels (dashed arrow) of Lefty in these cells. (3) hESC-derived Lefty is capable of inhibiting Nodal signaling by preventing Nodal from forming an active receptor complex with Cripto-1-Alk4/7-ActRIIB, necessary for proper Smad2/3/4-dependent downstream signaling. (4) Since the Lefty gene is also highly methylated in human aggressive melanoma cells (C8161), it is unlikely that the hESC microenvironment can induce Lefty expression in these cells. (5) miR-302a expressed in human aggressive melanoma cells after exposure to the hESC microenvironment can target and reduce Notch4 expression. (6) Binding of the γ-secretase (γ-SC) cleavage product, NICD, with an activator complex and the CBF-1/Su (H)/LAG-2 (CSL) transcription factor leads to induction of target genes, including Nodal. Since this Notch signaling can regulate Nodal expression during development, the interruption of this pathway could negatively affect Nodal expression in human melanoma cells. Epigenomic regulation of Nodal signaling by hESC-derived factors via miR302a or other miRNAs (7) would represent an alternate mechanism by which the hESC microenvironment could reprogram the aggressive phenotype of human aggressive melanoma. hSC: Human adult stem cell; NICD: Notch intracellular domain.
Elevated expression of miR-302a in melanoma cells exposed to the hESC microenvironment can target and reduce Notch4 expression. Specifically, we have demonstrated that knockdown of Notch4 expression by small interfering RNA (siRNA) is capable of reducing the expression of Nodal in C8161 cells [5]. Since Notch signaling, via binding of the Notch intracellular domain (NICD) with an activator complex and the CBF-1/Su (H)/LAG-2 (CSL) transcription factor, is known to regulate Nodal expression during the establishment of left–right asymmetry in development of vertebrates [33,34], the interruption of this pathway could negatively affect Nodal expression in human melanoma cells. Epigenomic regulation of Nodal signaling by hESC-derived factors via miR302a or other miRNAs would represent an alternate mechanism by which the hESC microenvironment could reprogram the aggressive phenotype of human aggressive melanoma.
Using miRNA TLDA arrays we evaluated the expression profiles of miRNAs capable of regulating Nodal and/or other genes in its pathway. This approach allowed us to identify specific miRNAs that were up- and down-regulated in the melanoma cells exposed to a hESC microenvironment, including miR-27b, which was downregulated, and miR-302a, which was over-expressed. Interestingly, some reports have shown that miR-27b is associated with angiogenesis [35], and is also an upregulated miRNA in early melanoma progression [36], which we plan to pursue in a subsequent study. miR-302a has been associated with embryonic pathways [37], and as a consequence we decided to evaluate the effects that overexpression of miR-302a would have on relevant genes associated with the Nodal signaling pathway in melanoma cells, which display many embryonic characteristics [4,8]. In this study, we have demonstrated that miR-302a is able to bind the putative target site in the Notch4 3′-UTR, demonstrating a specific regulation of Notch4 mRNA and protein expression. Previous work has shown that the expression of miR-302a is dependent on Oct4/Sox2 in hESCs, and miR-302a is expressed at the same developmental stages and in the same tissues as Oct4 during embryonic formation [31]. A particularly noteworthy finding is that Oct4 is upregulated in stem cell populations within aggressive melanoma cell lines [38]. miR-302a was also predicted to target many cell-cycle regulators, and its expression in primary and transformed cell lines promotes an increase in S-phase and a decrease in G(1)-phase cells, which are characteristics of an ESC-like cell-cycle profile [31]. In addition to the elevated expression of miR-302a in the reprogrammed aggressive melanoma cells, we have also observed a high expression level in hESCs [Costa FF, Children's Memorial Research Center, IL, USA. Unpublished data].
In order to evaluate other mechanisms that might regulate Nodal signaling, we performed DNA methyation analysis of the CpG island (mapped to the second exon) of Lefty B, a natural inhibitor of Nodal. We identified very high levels of DNA methylation in cancer cells, indicating their inability to express Lefty, as opposed to hESCs where the Lefty gene was unmethylated and expressed at high levels. These new findings provide novel insights into the aberrant Nodal embryonic signaling pathway that is functionally activated in aggressive tumor cells where the Lefty gene has been silenced. Importantly, we do not rule out the possibility that other epigenetic mechanisms such as histone modifications may play a role in regulating the Lefty gene in these cells.
In conclusion, our data indicate that epigenetic regulation of gene expression as shown by DNA methylation may explain why hESCs (and their microenvironment) can express high levels of Lefty capable of effectively downregulating Nodal expression and the plastic phenotype of aggressive human melanoma cells. Our data also suggest that gene expression via miRNAs (such as miR-302a) may play an important role in affecting Nodal signaling pathways in cancer cells. We are currently analyzing other miRNA candidates that are differentially expressed before and after exposure to a hESC microenvironment in melanoma and other cancers to gain new insights into the tumor suppressive effects of this unique embryonic milieu.
Executive summary.
■ Nodal signaling is an embryonic pathway that is re-expressed in aggressive tumors and is critical to maintaining plasticity and tumorigenicity.
■ The Lefty gene is highly methylated in aggressive melanoma cells, associated with its silencing and unregulated Nodal expression.
■ In human embryonic stem cells (hESCs), the unmethylated Lefty gene permits high-level expression of Lefty, which is capable of inhibiting Nodal signaling in tumor cells exposed to it.
■ During development, there is molecular crosstalk between the Notch and Nodal stem cell signaling pathways. In melanoma, there are two CSL binding sites upstream of the Nodal promoter, and Notch 4 signaling has been shown to regulate Nodal expression.
■ microRNA (miRNA) expression profiles of melanoma cells exposed to a hESC tumor suppressive microenvironment revealed the upregulation of miR-302a, whose target gene includes Notch 4, suggesting a viable mechanism underlying the reprogramming of tumor cells exposed to an embryonic microenvironment. In addition, miR-27b (associated with angiogenesis) was found to be downregulated in these tumor cells, which is the focus of ongoing studies.
■ Future investigations will address the pivotal role(s) miRNAs play in the reprogramming of metastatic tumor cells and their use as potential targets in new therapeutic approaches.
Acknowledgements
We thank Victor Ambros and Molly Hammell for helping with the RNAhybrid software analysis. We also thank Elio F Vanin, Christina Smith and Selva Musa for technical assistance and Joana Heinzelmann for critically reading this manuscript.
This work is supported by NIH grants CA59702 and CA121205 (MJCH), the Eisenberg Scholar Research Fund (LS) and the Maeve McNicholas Memorial Foundation (FFC).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
- 1.Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancer – the role of sunlight. Adv. Exp. Med. Biol. 2008;624:89–103. doi: 10.1007/978-0-387-77574-6_8. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, Soong SJ, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J. Clin. Oncol. 2001;19(16):3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 3.Lui P, Cashin R, Machado M, et al. Treatments for metastatic melanoma: synthesis of evidence from randomized trials. Cancer Treat. Rev. 2007;33(8):665–680. doi: 10.1016/j.ctrv.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Hendrix MJC, Seftor EA, Seftor REB, et al. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer. 2007;7(4):246–255. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 5.Postovit LM, Seftor EA, Seftor REB, Hendrix MJC. Targeting Nodal in malignant melanoma cells. Expert Opin. Ther. Targets. 2007;11(4):497–505. doi: 10.1517/14728222.11.4.497. [DOI] [PubMed] [Google Scholar]
- 6.Schier AF. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 7.Fleming RJ. Structural conservation of Notch receptors and ligands. Semin. Cell Dev. Biol. 1998;9(6):599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- 8.Postovit LM, Margaryan NV, Seftor EA, et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc. Natl Acad. Sci. USA. 2008;105(11):4329–4334. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morokuma J, Blackiston D, Adams DS, et al. Modulation of potassium channel function confers a hyperproliferative invasive phenotype on embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105(43):16608–16613. doi: 10.1073/pnas.0808328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa FF. Non-coding RNAs: lost in translation? Gene. 2007;386(1–2):1–10. doi: 10.1016/j.gene.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi S, Sevignani C, Nnadi SC, et al. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm. Genome. 2008;19(7–8):526–540. doi: 10.1007/s00335-008-9119-8. [DOI] [PubMed] [Google Scholar]
- 14.Wurdinger T, Costa FF. Molecular therapy in the microRNA era. Pharmacogenomics J. 2007;7(5):297–304. doi: 10.1038/sj.tpj.6500429. [DOI] [PubMed] [Google Scholar]
- 15.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Segura MF, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl Acad. Sci. USA. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12(12):580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 19.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavazoie SF, Alarcón C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Teruya-Feldstein J, Weinberg RA. Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 7163;2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 22.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008;26(4):462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 23.Welch DR, Bisi JE, Miller BE, et al. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. Int. J. Cancer. 1991;47:227–237. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
- 24.Topczewska JM, Postovit LM, Margaryan NV, et al. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat. Med. 2006;12:925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 25.Postovit LM, Seftor EA, Seftor REB, Hendrix MJC. A 3D model to study the epigenetic effects induced by the microenvironment of human embryonic stem cells. Stem Cells. 2006;24:501–505. doi: 10.1634/stemcells.2005-0459. [DOI] [PubMed] [Google Scholar]
- 26.Costa FF, Paixão VA, Cavalher FP, et al. SATR-1 hypomethylation is a common and early event in breast cancer. Cancer Genet. Cytogenet. 2006;165(2):135–143. doi: 10.1016/j.cancergencyto.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postovit LM, Costa FF, Bischof JM, et al. The commonality of plasticity underlying multipotent tumor cells and embryonic stem cells. J. Cell Biochem. 2007;101(4):908–917. doi: 10.1002/jcb.21227. [DOI] [PubMed] [Google Scholar]
- 31.Card DA, Hebbar PB, Li L, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008;28(20):6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin SL, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14(10):2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs LT, Iwai N, Nonaka S, et al. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17(10):1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 2007;101(1):59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 36.Glud M, Klausen M, Gniadecki R, et al. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J. Invest. Dermatol. 2009;129(5):1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 37.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell. 2009;16(4):517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Strizzi L, Postovit LM, Margaryan NV, et al. Nodal as a biomarker for melanoma progression and a new therapeutic target for clinical intervention. Expert Rev. Dermatol. 2009;4(1):67–78. doi: 10.1586/17469872.4.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
■ Websites
- 101.Mirnamap: a website that offers tools to cross-reference between miRNA profiles and expression profiles of its targets. http://mirnamap.mbc.nctu.edu.tw/
- 102.Repeat Masker: a program that screens DNA sequences for interspaced repeats and low complexity DNA. www.repeatmasker.org.
- 103.TM4: a software designed for gene-expression analyses. www.tm4.org/mev.html.