Abstract
Aims
B-cell chronic lymphocytic leukemia (CLL) is a heterogeneous malignancy that clinically ranges from indolent to rapidly progressive. CLL, like other cancers, can be affected by epigenetic alterations.
Materials & methods
A microarray discovery-based study was initiated to determine DNA methylation in CLL cases with a range of CD38 expression (1–92%).
Results
Many loci were either methylated or unmethylated across all CD38 levels, but differential methylation was also observed for some genes. Genomic sequencing of DLEU7 confirmed extensive cytosine methylation preferentially in patient samples with low CD38 expression, whereas NRP2, SFRP2 and ADAM12 were more commonly methylated in those with high CD38 expression.
Conclusion
This study demonstrates that CLL is affected by CpG island methylation in some genes that segregate with CD38 expression levels, while most others show similar methylation patterns across all levels. The CpG island methylation in certain functional gene groups and pathway-associated genes that are known to be deregulated in CLL provides additional insights into the CLL methylome and epigenetic contribution to cellular dysfunction. It will now be useful to investigate the effectiveness of epigenetic therapeutic reversal of these alterations to develop effective treatments for the disease.
Keywords: CD38 expression, CLL, DNA methylation, epigenetics, leukemia
Formerly, B-cell chronic lymphocytic leukemia (CLL) was considered a single disease occurring mainly in the elderly, and shows a variable but generally indolent clinical course. It is now clear that CLL is a heterogeneous disease with at least two subtypes in terms of both biological makeup and prognosis (reviewed in [1]). Some patients survive for many years without any therapy, while others progress rapidly within months of diagnosis, requiring early initiation of treatment.
Several biomarkers have been identified that appear to segregate patients with CLL into low- and high-risk clinical groups. Low-risk CLL is perhaps best characterized by immunoglobulin variable heavy-chain gene (IgVH) mutation (≥2% compared with germline) and low CD38 (CD38low) and ζ-chain associated protein kinase 70 (ZAP-70) expression, while high-risk cases of CLL show a reverse pattern. However, the precise definition of CD38low and CD38high is debatable, with proposed thresholds ranging from 7 to 30% [2-5]. While it has been proposed that CD38 and ZAP-70 expression are surrogates for IgVH mutational status, more recent studies suggest that each parameter is independently prognostic, but with considerable overlap [6,7]. Expression of CD38 is tightly regulated in normal B-cell ontogeny, with low expression in resting B cells and higher expression in stimulated B cells [8]. Both CD38 and B-cell receptor (BCR) signaling are altered in, and segregate with, clinical subsets of CLL patients, but the reasons for this are unclear. Based on our previous studies suggesting variation of genome methylation in small B-cell lymphomas, including CLL [9], we hypothesized that these modifications might relate to CD38 expression and the biological behavior of these patient groups, or conversely, methylation may be a functional attribute of the disease in general.
We now present data from discovery-based DNA methylation studies of CLL patients with a range of CD38 expression and demonstrate mainly similarities, but a few differences, in the methylation status of specific genes related to CD38 expression levels. The genes affected across all CD38 levels were functionally classified into groups involving ion and solute transport, and pathways such as WNT that are known to be deregulated in CLL, thus suggesting an important epigenetic underpinning of cellular dysfunction. Those segregating with CD38 levels will require further research to define their potential role(s) in differential clinical behaviors. Nevertheless, with the ongoing and future clinical trials using epigenetic modifiers, it becomes important to understand the CLL epigenome and how demethylating agents, histone modifiers and other novel agents affect the underlying biological behavior and clinical outcomes.
Patients & methods
Samples
Blood samples were obtained from patients following diagnostic evaluation, and before any treatment, at the Ellis Fischel Cancer Center in Columbia (MO, USA), the Holden Cancer Center in Iowa City (IA, USA) and the Mayo Clinic in Rochester (MN, USA) in compliance with local Institutional Review Board requirements. DNA was isolated using the QIAmp DNA Blood Minikit (Qiagen, CA, USA). The samples (n = 38) had levels of CD38 expression on the CLL cells varying from 1 to 92% by flow cytometry [10], and all contained more than 60% (range 60–96) neoplastic cells as determined by CD19/CD5/CD23 expression (data not shown). The percentage of CD38 expression was adjusted for CD19 expression and used as a variable in the clustering analyses. Genomic DNA (Promega, WI, USA) was used as an unmethylated normal control. In addition, CD19+ nonmalignant B cells were also used as a normal control, as well as CD19+ B cells (Invitrogen, CA, USA). The source in both cases is from peripheral blood, and for the genes tested, there is no difference in methylation.
Cell culture & pharmacological treatments
Three CLL cell lines with differing levels of CD38 expression by flow cytometry (not shown) were included: WAC3CD5 (4.7%, CD38), MEC1 (69.5%, CD38) and MEC2 (96.6%, CD38). These were maintained in RPMI 1640 media as previously reported [9]. Included in this study were three CLL cell lines with differing levels of CD38 expression: WAC3CD5 (4.73%, CD38), MEC-1 (50.5%, CD38) and MEC-2 (6.6%, CD38). MEC-1 was initially obtained 3 years after diagnosis from peripheral blood lymphocytes (PBLs) of a 58-year-old Caucasian patient with CLL. A year later, a second cell line (MEC-2) was obtained from PBLs of the same patient. Analysis of IgVH showed that these cell lines have not undergone somatic hypermutation, but they differ in expression of CD23 and FMC7. The WAC3CD5 line was induced by cytokines and infected with Epstein–Barr virus.
For gene reactivation experiments, cells were cultured in the presence of a combination of a demethylating agent (5-aza-2′-deoxycytidine [5′-Aza]) and/or a histone deacetylase (HDAC) inhibitor (Trichostatin A [TSA]). Cell lines were cultured with vehicle (phosphate-buffered saline [PBS]) or 1.0 μM 5′-Aza, with medium changed every 24 h. After 4 days, cells were either harvested or further treated with TSA (1.0μM) for 12 h and then harvested. Some cells were treated with TSA alone for 12 h. Genomic DNA or total RNA was isolated using QIAgen kits and used for methylation and gene expression analysis, respectively.
Real time RT-PCR
Total RNA was extracted from the cell lines and reverse transcribed in the presence of SuperScript™ II reverse transcriptase (Invitrogen, CA, USA). The cDNA was then used for expression analysis of LHX2, NRP2, DLEU7, KCNK2, DLC-1, ADAM12, APC2 and SFRP using the Absolute™ QPCR SYBR® mix (AB gene, Rochester, NY, USA) or SuperArray RT2 Real Time™ primers and SYBR® green PCR master mix (Bioscience, MD, USA). Taqman® primers and probes were utilized to study the expression of DLC-1 and GAPDH in CLL cell lines. Real-time PCR was carried out in an iCycler iQ™ (BioRad, CA, USA). In order to examine the effects of epigenetic reversals of DNA methylation and/or inhibition of histone deacetylases, quantitative expression of these mRNAs was measured in three CLL cell lines before and after treatment with 5′-Aza, TSA, or both combined. The primer sequences and PCR conditions are summarized in Supplementary table 1.
CpG island microarrays, amplicon preparation & hybridization
Microarray slides containing approximately 12,000 CpG island (CGI) clones were obtained from the Microarray Centre, University Health Network, Toronto, Canada. The slides were processed after hybridization using the Pronto Universal Microarray Reagents (Corning Life Science, MA, USA). Procedures for amplicons preparation, labeling, hybridization and post-hybridization processing were essentially as previously reported [9]. The Toronto 12K array is comprised of DNA fragments enriched for CpG islands with a methyl-binding protein column. Of these 12,000 loci, the array was filtered for unusable spots based on criteria described below. This filter reduced the number of loci under consideration to 5853.
We also used the Methylated CpG Island Amplification with Microarrays (MCAM) protocol [11]. Briefly, 5 μg DNA was digested consecutively with SmaI and XmaI, which cut unmethylated and methylated CCGG sites, respectively. SmaI digestion results in blunt ends, and XmaI digestion in sticky ends that can be ligated to linkers. Digested DNA was purified using concentrator columns, and 500 ng was ligated to annealed RMCA12 and RMCA24 adapters then PCR amplified. Following purification of amplicons, 2 μg was labeled with Alexafluor555® or 647 using the Bioprime Plus Array CGH Indirect Genomic Labeling kit according to the manufacturers' instructions. 5 μg of each labeled DNA preparation was co-hybridized to Agilent 244K CpG island microarrays representing more than 27,000 CGIs across the human genome. Slides were then washed and scanned on a GenePix® scanner according to standard protocols. Data was extracted using the Feature Extraction Tool (Agilent Technologies, CA, USA) and imported into GeneSpring GX (Agilent Technologies, CA, USA) for downstream selection of putatively methylated probes.
Methylation confirmation
A total of 15 genes were selected for validation in the cell lines and the same patient samples from microarray studies using methylation-specific PCR (MSP) or combined bisulfite restriction analysis (COBRA) as previously reported [9]. Selection of these genes was based on their appearance in microarray experiments to be either commonly or differentially methylated across levels of CD38 expression in CLL. Successful development of primers for COBRA or MSP is sometimes difficult and an alternative method is used. In some cases, it is not feasible to develop COBRA, or conversely MSP primers, that are successful, usually depending on the number of CG dinucleotides present. Nevertheless, as confirmatory methods, these are both standard.
The DLEU7 gene was further interrogated using bisulfite genomic sequencing. Not much is known about the function of this gene, and it seems to segregate well with lower levels of CD38, thus perhaps becoming useful as a biomarker in future investigations. Sodium bisulfite converts all unmethylated cytosine, but not methylated cytosine, to uracil. Upon amplification, the uracil is converted to thymine. All cytosines present in the sequencing result are therefore methylation sites based on their resistance to deamination. Primer sequences and PCR conditions are listed in the Supplementary table 2. Relative locations of CGIs and gene promoters are shown in Supplementary Figure 1. Amplified PCR products were sub-cloned using the TOPO® TA Cloning® Kit for Sequencing (Invitrogen, CA, USA). Plasmid DNA of insert-positive clones was isolated using the Montage Plasmid Miniprep kit (Millipore Corporation, MA, USA) and sequenced using ABI 3730 sequencing systems (Applied Biosystems, CA, USA).
Statistical analysis
For each of the 15 genes validated using COBRA and MSP, 2 × 2 contingency tables were formed, with the rows indicating CD38 levels within each of the two groups (<10% and ≥10%, or >30% and ≤30%) and the columns indicating the methylation status (methylated/unmethylated). For each table (one table per gene), we carried out Fisher's exact test to determine if the CD38 level was associated with methylation status. This test was chosen as an alternative to the χ2 test of independence because, for many genes, the small expected cell counts would cause the χ2 test to perform poorly, and Fisher's does not suffer this limitation. We also computed odds ratios (ORs), log odds ratios and confidence intervals for the log odds ratio, for which a constant equal to 0.5 was added to each cell in the contingency table, because that is known to improve the bias and mean-squared error properties of the aforementioned statistics [12]. The strength of the association between CD38 level and methylation increases as the OR moves above a value of 1.
Results
Methylated gene discovery
Our previous methylation microarray experiments using the differential methylation hybridization (DMH) method resulted in hierarchical clustering that suggested at least two subgroups of CLL based on DNA methylation patterns [9]. At that time, we did not have CD38 expression data for enough of the CLL samples to assess the correlation between methylation and CD38 expression, but the study suggested another set of experiments that are now reported using larger feature microarrays.
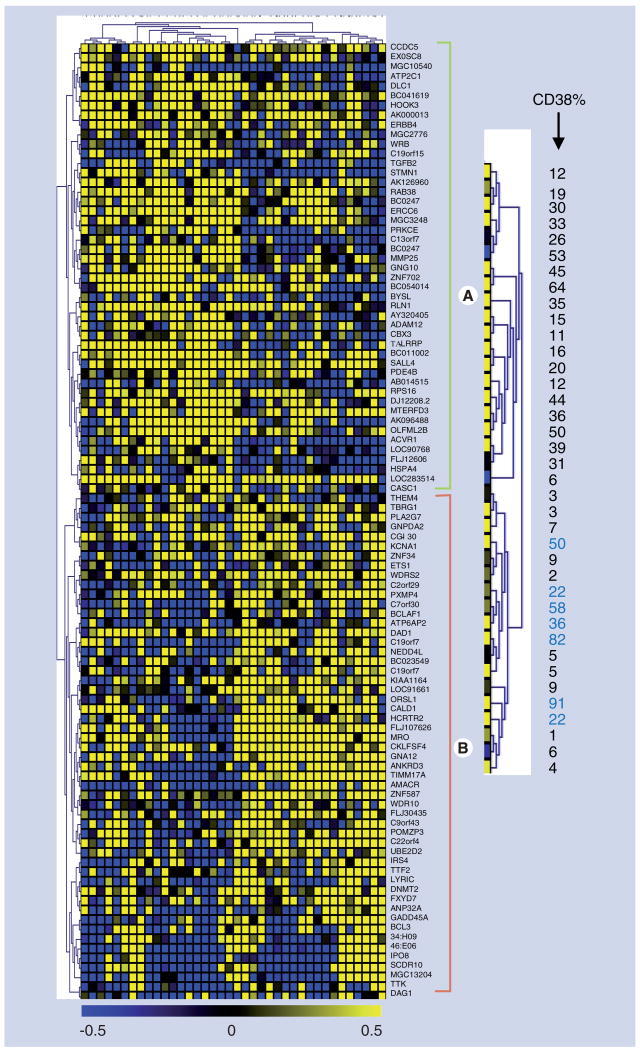
To identify loci that were methylated, we first filtered the data and considered only those loci that ruled out the following: blank probe spots, mitochondrial DNA sequences and control sequences, and included loci with a DNA sequence length of more than 100 bp, and sequences that contained methylation-sensitive enzyme restriction sites for BstUI and/or HpaII. This filter reduced the loci under consideration to 5853. The filtered loci were then subjected to statistical analysis using the Wilcoxon, Mann–Whitney and t-test. Data from each locus was then ranked in increasing order based on p-values, and the top 100 based on Mann–Whitney were used as input for clustering analysis. The statistics and procedure used to produce a hierarchical cluster is detailed in earlier work from our laboratory [9] (Figure 1 & Supplementary Figure 2). The upper dendrogram (rotated 90° and enlarged on the right) illustrates the relationship of CD38 expression values of CLL samples to each other on the basis of their DNA methylation patterns. We further examined high and low CD38-expressing cases using Agilent CpG island microarrays that interrogate more than 27,000 CGIs across the genome.
Figure 1. Hierarchical clustering analysis of DNA methylation.
This cluster is based on a 10% threshold where there are 12 samples with less than 10% of the cells expressing CD38 and 26 samples with more than 10% CD38 expression. This illustrates a measure of relatedness of DNA methylation across all loci for each sample. Each column represents a patient sample and each row represents a clone/locus on microarray chip. The florescence ratios of cy3/cy5 are measures of DNA methylation and are depicted as a color intensity (-0.5 to +0.5) in log base 2; yellow indicates loci that have a higher level of DNA methylation in chronic lymphocytic leukemia compared with normal controls, blue indicates a lower level of methylation and black indicates no change. Graded colors across the spectrum represent various levels of methylation. The dendrogram from the top of the cluster (rotated 90° and enlarged on the right) represents the CD38 expression level of each sample. Not every patient sample clustered with the expected group. The numbers shown in blue color are seven patients with CD38high that are clustered in the group of 10% or less, and one sample with 6% CD38 clustered with the CD38high group.
From the published literature, it has been suggested that clinical behavior best segregates with CD38 expression ranging from 7 to 30% [2-5]. For simplicity, we chose to initially re-examine the data from our clustering analysis with a CD38 expression threshold of 10%. The segregation at 10% resulted in a number of genes/loci with differential frequencies of methylation in CD38high (>10%) and CD38low (≤10%) CLL samples, but most appeared methylated (or not) universally across the samples. Methylated genes in group A (Figure 1) were almost exclusively (20/26 cases) associated with samples having CD38high expression. Group B genes did not segregate quite as clearly, but were slightly more likely to be methylated in CD38low CLL samples (11/18 cases), but seven CD38high samples also clustered with the CD38low CLL samples and are shown in blue (Figure 1). Since it is the oveall pattern of methylation that is used in the clustering algorithm, the specific reason for this less robust separation is not entirely clear, but warrants further investigation.
Independent confirmation of methylation
While microarrays are useful discovery tools, it remains important to independently confirm methylation in representative observations to avoid false-positive and -negative results. Therefore, we selected 16 regions/loci from 15 genes for confirmation by COBRA and MSP in three CLL cell lines with different CD38 expression levels. These genes were selected based on these microarray results, as well as previous work in our laboratory suggesting that differential methylation in CLL subsets might be present [9]. The MSP assay was used to study methylation of NRP2, DUOX2 and RLN2, and the other validations were carried out using COBRA (Figure 2) [9]. This was because not all genes were equally able to be interrogated with COBRA primers. In addition, we recently reported on the performance of ultradeep bisulfite sequencing (454 technology) of CLL for 25 gene regions [13]. Four genes, ADAM12, DLC-1, LRP1B and KCNK2, from Figure 2 were also included in that study and thus confirm the presence of DNA methylation at specific cytosine molecules of these genes in CLL from our microarray experiments.
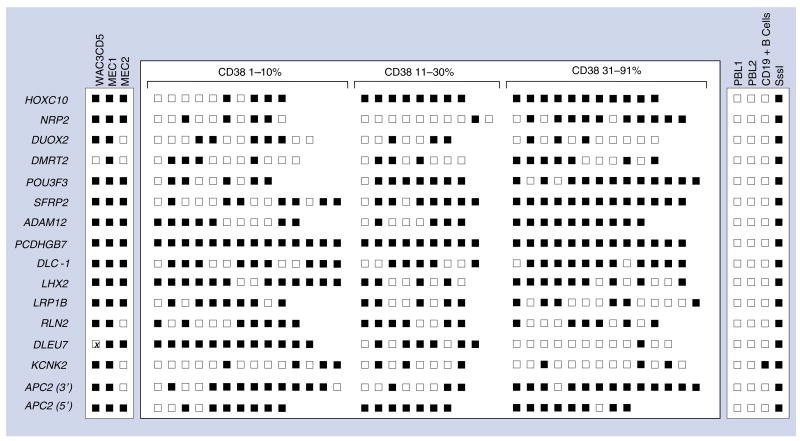
Figure 2. COBRA or MSP validation of DNA methylation.
The DNA methylation status of primary chronic lymphocytic leukemia (CLL) tumor samples was validated using either combined bisulfite restriction analysis (COBRA) or methylation-specific PCR (MSP) from the samples above, as well as primary CLL samples that were divided to three groups (1–10%, 11–30% and 31–91%) based on CD38 positivity. Results are summarized as methylated (dark square) or unmethylated (open square). Each square represents an individual patient sample and each specific gene. The right-side panel represents the results of control samples, while the left-side panel represents the DNA methylation status in CLL cell lines.
The results varied among the three cell lines (Figure 2). Methylation was present in APC2 (CGI2, CpG island located in the coding region), HOXC10, NRP2, POU3F3, SFRP2, ADAM12, PCDHGB7, DLC-1, LHX2 and LRP1B in all three cell lines, while DUOX2, APC2 (CGI3, CpG island located in the 3′ region), RLN2 and KCNK2 were only methylated in WAC3CD5 and MEC1. Fewer genes (11/16) were methylated in MEC2, which expressed the highest level of CD38 (96.6%) than in WAC3CD5 (14/15) or MEC1 (16/16), which have lower levels of CD38 (4.7 and 69.5% respectively). Of note, in multiple attempts, we were unable to amplify the DLEU7 region with COBRA or bisulfite sequencing primers in the WAC3CD5 cell line, even though they were successful in the other two cell lines and primary samples. The reason is not entirely clear, but this gene is located in a region on chromosome 13q14 where deletions occur frequently in CLL. Thus, it is possible that DNA sequence alterations might be responsible in this cell line. This precludes any statement regarding methylation in this cell line.
Results from cell lines do not always match those of primary cells, so we also examined methylation of the same 15 genes from 16 regions in primary CLL cells. As shown in Figure 2, each row represents a single gene, and columns are grouped by CD38 expression (0–10%, 11–30% and 31–92%) to represent results of methylation within three groups of patients with increasing levels of CD38 expression. The methylation results are also shown for 0–10% versus 11–30% versus 31–92% in Supplementary Tables 3 & 4. Two fragments were examined for APC2; a CGI located in the 3′ region and a CGI in a coding region. Not every row has an equal number of samples. For instance, 36 samples were successfully examined for PCDHGB7, but only 29 samples for ADAM12. All samples for which no result is reported were re-examined by COBRA at least three times, but all still failed to provide a PCR product for digestion. The reasons are not entirely clear, but one possibility is that this is a result of genomic alterations in the regions under consideration.
Based on our earlier observation suggesting that the 10% CD38 threshold might be best for class separation, contingency tables were generated for each gene based on the methylation results. The CD38low group was initially assigned to those with 1–10% expression, while the CD38high group contained those with 11–92% expression. Estimates of the OR and other statistics were computed and are reported in Supplementary Tables 3 & 4. For example, in the case of HOXC10, methylation was present in all 19 cases with more than 10% CD38 expression, and four out of ten cases with 10% CD38 expression or less. The corresponding OR for HOXC10 was over 56, which implies that the odds of methylation are more than 56-times greater when CD38 is more than 10%, as compared with odds of methylation when CD38 is 10% or less. Conversely, methylation was present in DLEU7 for all 12 cases with 10% CD38 or less, and seven out of 21 samples with more than 10% CD38, resulting in an OR of 0.02. This implies that the odds of methylation in DLEU7 are approximately 50 (1/0.02) times greater when CD38 is less than 10%, compared with methylation when CD38 is greater than 10%. Using Fisher's exact test, the p-value for the methylation differences between the two groups was p = 0.0004 for HOXC10 and 0.0002 for DLEU7. Thus, methylation of HOXC10 is found mainly in CD38high, while that of DLEU7 is present mainly in CD38low. Of the remaining genes, only SFRP2 showed a p-value less than 0.05 (p = 0.0144), with an OR of 8.2, suggesting more methylation events at higher CD38 levels (Supplementary Tables 3 & 4). Since studies have also suggested the best clinical discriminator was closer to 30% [14], we then re-analyzed our data with new definitions; cases were assigned to the CD38low group if they demonstrated 1–30% expression, while the CD38high group contained those with 31–92% expression. In this scenario, methylated DLEU7 still remained segregated with the CD38low group (p < 0.0001, OR: 0.025). The methylated genes NRP2 (p < 0.0052, OR: 8.455), SFRP2 (p < 0.0136, OR: 17.690) and ADAM12 (p < 0.0265, OR: 15.522) were all associated with the CD38high group. At these CD38 expression levels, HOXC10 and the other tested genes had p-values greater than 0.05 (Supplementary Tables 3 & 4). Therefore, DLEU7 and SFRP2 remain as candidate biomarkers for separation of CLL based on CD38 expression at both the 10% and 30% level.
Bisulfite genomic sequencing of DLEU7
Qualitative or semi-quantitative estimates of DNA methylation play an important role in epigenetic studies, but in order to define the density and specific location of cytosine methylation in DLEU7, which seemed to be preferentially methylated in the CD38low group(s), we performed bisulfite genomic sequencing from representative primary CLL samples, cell lines and appropriate controls to fully detail the methylation at a single cytosine molecular level.
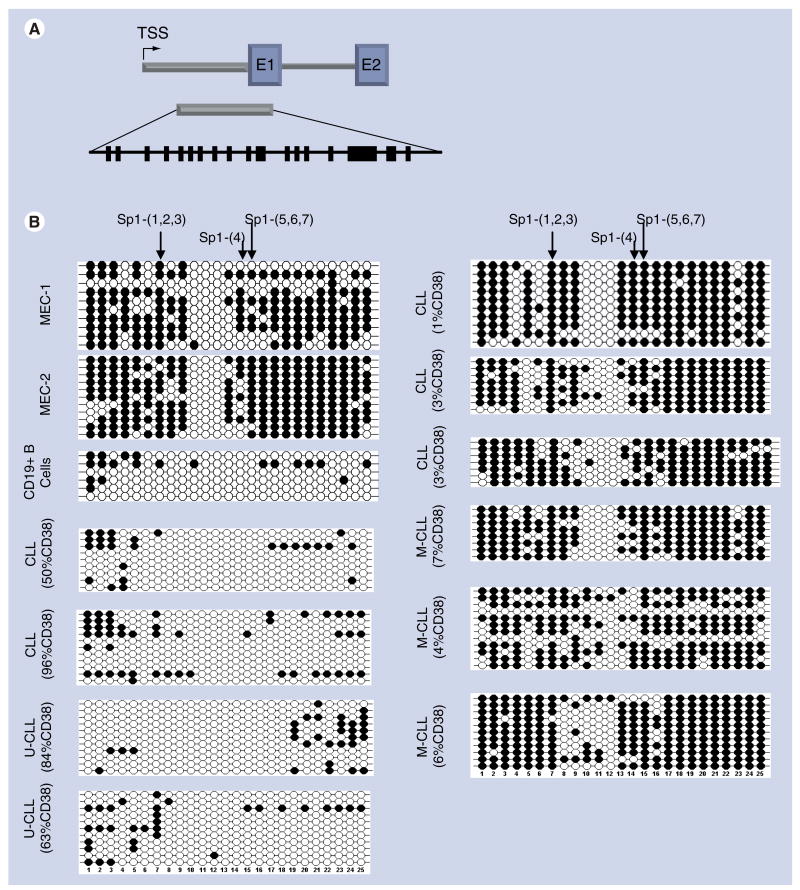
The DLEU7 gene contains a CGI that spans part of the promoter and first exon and includes binding sites for SP1 (and other) transcription factors (Figure 3). The sequencing reaction interrogated 25 CG dinucleotides with reference to potential interactions of binding sites and methylated or unmethylated cytosine. The patterns of methylation in MEC1 and MEC2 cell lines were similar, and showed a region near the middle of the CGI that was unmethylated, while the remainder was densely methylated. The CGs from 5′ cytosine positions 1–9 and 14–25 that also encompass SP1 binding sites were methylated. Multiple attempts to perform this analysis on the WAC3CD5 cell line failed owing to an inability to generate a PCR product with these primers, and therefore its methylation status was not determined. Of the primary CLL samples studied, six were CD38low (1–7% positive). IgVH mutational status was tested for three of these six and also found to have mutated IgVH (≥ 2%), a group generally associated with a favorable prognosis. These six all showed extensive methylation for CGs from 5′ cytosine positions 1–9 and 14–25 similar to that of the cell lines. A core region from CGs 10–12 remained unmethylated, but contained no SP1 transcriptional regulatory protein-binding sites. Conversely, DLEU7 was much less methylated in two representative patients with CD38high (50 and 96% CD38) and two additional CD38high (63 and 84% CD38) with unmutated IgVH genes (generally associated with a poor prognosis).
Figure 3. Bisulfite sequencing of DLEU7.
DNA methylation of 25 CG dinucleotides was examined in a 5′ region of DLEU7 that spans part of a region spanning the predicted 5′UTR and part of the first exon; the location of this region with respect to the TSS is shown. The CG dinucleotides are shown as heavy black lines. The methylation status of 25 CG dinucleotides was determined from bisulfite-treated DNA of CLL patients with different CD38 expression levels and either M-CLL or unmutated U-CLL IgVH mutational status, two CLL cell lines (MEC1 and MEC2) and CD19+ B cells from a healthy donor. Each row is the result from an individual clone across the 25 CG dinucleotides. Filled circles indicate methylated cytosine and open circles are unmethylated sites.
CLL: Chronic lymphocytic leukemia; M-CLL: Mutated CLL; TSS: Transcription start site; U-CLL: Unmutated CLL.
Pharmacologic reactivation of genes in CLL cell lines
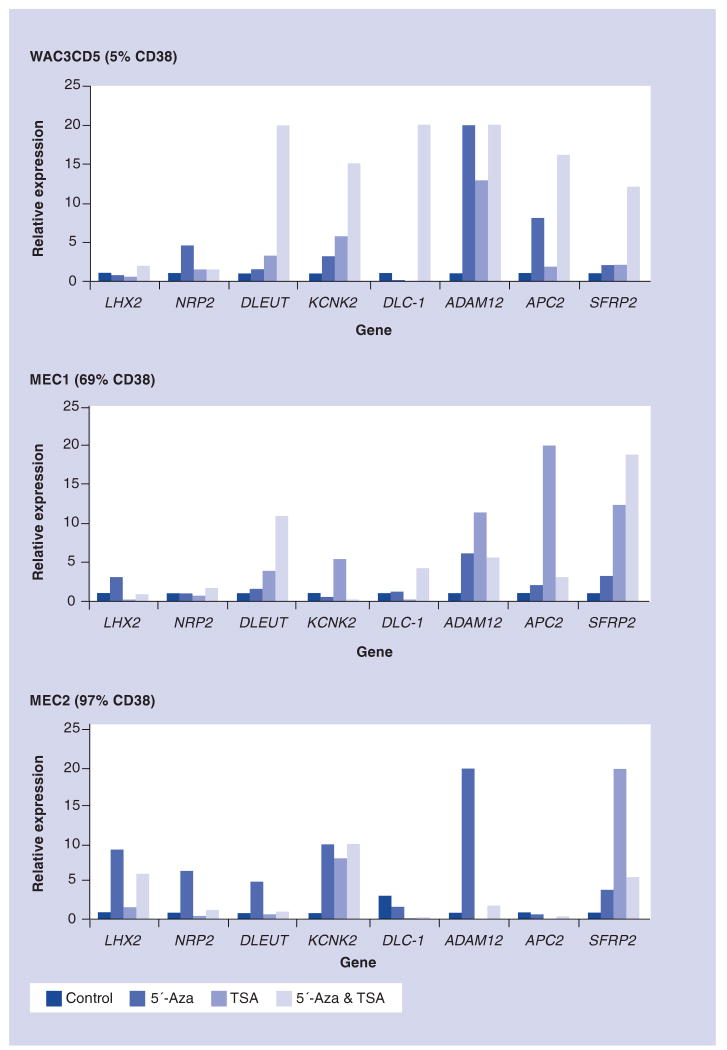
To investigate the functional impact(s) of DNA methylation and/or histone modifications on mRNA expression of selected genes (LHX2, NRP2, DLEU7, KCNK2, DLC-1, ADAM12, APC2 and SFRP2), we used CLL cell lines before and after treatment with the demethylating agent 5′-Aza, a histone deacetylase inhibitor (TSA), and both in combination. The viability after treatment was generally greater than 80%. It is quite difficult to perform such studies on primary cells, since 5′-Aza is a competitive inhibitor of DNA methyltransferases (DNMTs) and demethylation seems to require at least a few rounds of cell division; primary CLL cells are not very amenable to such in vitro conditions. Quantitative real-time reverse transcription (RT)-PCR was performed in triplicate, and the median expression level was calculated (Figure 4). As shown, the re-expression pattern varied by gene and by cell line, but in all instances, pharmacologic reversal was observed to some degree. This data suggests the involvement of additional mechanism(s) affecting chromatin compaction in addition to DNA methylation.
Figure 4. Pharmacologic reactivation in CLL cell lines.
The mRNA expression levels of eight genes were quantified using real-time RT-PCR in untreated WAC3CD5, MEC1 and MEC2 cell lines. These were also treated with the demethylating agent 5′-Aza, TSA or the combination of 5′-Aza and TSA. A housekeeping gene (HRPT1) was used as an internal control for all of the above genes, except for DLC-1 where GAPDH was used. The relative expression level of each gene was determined using 2-ΔΔCt, where Ct is the cycle threshold for positivity. 5′-Aza: 5-aza-2′-deoxycytidine; TSA: Trichostatin A.
CpG island microarray studies
Based on the need to further expand our analysis, we also used a human CGI microarray representing more than 27,000 CGIs across the genome (Agilent). Again, the majority of gene-related CGIs were either methylated or unmethylated across all CD38 levels, but we also discovered a small number of candidate differentially methylated genes. Using two samples with CD38low and two with CD38high we first filtered for only named regions where the CGI was in a promoter region, required that the fluorescence ratio be greater than 2.0 for each probe, and that at least three probes within 500 bp met these criteria. Then, we selected those genes with predicted methylation in either both CD38high samples or both CD38low samples, but not the other. This produced a list of 36 new candidate genes (beyond those already studied) in the CD38high group and five new candidates in the CD38low group (Supplementary Table 5). We also identified an additional 65 genes/loci that appeared methylated in the promoters of at least three of four primary CLL cases, regardless of CD38 expression levels (Supplementary Table 6).
Additional genes of potential importance in CLL from the new arrays were further examined to confirm methylation. The SOX family contains both transcriptional activators and repressors, and along with APC and SFRP2, are involved in WNT signaling, which is reported to be constitutively active in CLL [15]. Validation of six SOX family members using COBRA revealed methylation of SOX1 in 31 of the total 48 samples (65%), SOX3 in all 48 (100%), SOX4 in 37 (77%), SOX9 in 10 (21%), SOX11 in all 48 (100%) and SOX17 in 24 (50%) of the 48 primary CLL samples, regardless of the CD38 expression level (data not shown).
Analysis of this combined new list of over 100 candidate genes and open reading frames methylated in CLL was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resource [16]. This analysis produced a list of five highly significant (Benjamini scores of 1.6E-10 to 1.3E-4) gene ontology (GO) terms focused in four main areas: WNT pathway genes, genes involved in ion and solute transport, regulators of transcription and Homeobox genes. There were too few genes that were potentially methylated in the CD38low or CD38high class to reliably analyze these subsets independently and achieve significant Benjamini scores.
Discussion
Aberrant DNA methylation has been reported in virtually every type of human cancer examined. Previous studies have also demonstrated global [17] and gene-specific hypomethylation events in CLL [18-21], as well as gene-specific hypermethylation [9,22]. A recent study demonstrated that CD38 and ZAP-70 co-expression are functionally linked, and increased expression of these proteins marks cells with high migratory potential and more aggressive disease [23]. There is a general consensus that CD38 expression is a reliable negative prognostic indicator, and also correlates closely with other markers including ZAP70. The correlation with IgVH mutational status may not be quite as good, but both are independent prognostic factors. We did not have IgVH mutational results on all these patients, and therefore selected CD38 as a comparator and surrogate for clinical activity. Furthermore, CD38 is thought to function as a co-signaling molecule, along with the BCR, and relates to the ability to transduce transmembrane signaling via cross-linkages of BCR and CD38 [24–28]. In our prior series, which was too small for meaningful statistical analysis, there appeared to be two subgroups of CLL cases based on methylation profiles [9]. This observation motivated us to determine if methylation of some or many genes might segregate with CD38 expression and possibly contribute to the clinical behavior of each subgroup. Even though multiple genes were methylated across all levels of CD38, these are not likely to explain differences in biological behaviors between CD38low and CD38high subsets, although the alterations likely contribute to biological behavior of CLL in general.
Analysis of the combined new list of over 100 candidate genes and open reading frames methylated across all CD38 levels in CLL was performed using the DAVID bioinformatics resource [16]. This analysis produced a list of five highly significant (Benjamini scores of 1.6E-10 to 1.3E-4) GO terms focused in four main areas: WNT pathway genes, genes involved in ion and solute transport, regulators of transcription and Homeobox genes. Methylation of WNT pathway genes is a prominent feature of many solid tumors, as well as lymphoid and myeloid malignancies [29–39]. We also found a number of methylated genes involved in the WNT pathway in this analysis of CLL that further extends the list of epigenetically affected genes in this important signaling pathway. The WNT pathway is known to be aberrantly activated in CLL, and a few of these genes have previously been reported as aberrantly methylated [21,22,33,40–44]. Additional members are now added and contribute to a more detailed understanding of a disordered WNT epigenome. This signaling pathway is a target of some very recent pharmacological research showing that certain small molecule inhibitors of the pathway may be useful clinical agents [45]. Beyond APC2 and SFRP2, we also discovered promoter DNA methylation involving six SOX genes that mainly function as negative regulators of WNT signaling. Others have shown methylation of additional WNT genes including SFRP1 (100%), SFRP2 (55%), SFRP4 (30%) and SFRP5 (15%); SFRP3 has no CpG island and was not tested in their study [43]. It is therefore possible that DNA methylation may have a prominent epigenetic effect that contributes to an activated WNT pathway in CLL. We are currently exploring this possibility since it has important potential interactions with other pathways that are deregulated in CLL and may also affect certain treatment options.
Beyond the genes methylated across all levels of CD38, some genes were preferentially methylated based on CD38 expression levels; 32 in the CD38high group and five in the CD38low group (Supplementary Table 5). It is currently debatable which threshold of CD38 expression more clearly correlates with clinical behavior, with proposed thresholds ranging from 7 to 30% [2–5]. Since one part of our analysis was based on determining what, if any, differences in methylation might relate to CD38 as a surrogate of clinical behavior, we selected three different thresholds (10, 20 and 30%) to determine if there were any such correlations. In our sample set, the difference between using 10% and 7% was negligible; only two samples fell in between, and both had a value of 9% CD38. A reanalysis of our COBRA results shows that no significant changes would have been present if we used 7% or 10%.
The list of 32 genes from the CD38high group included some related to ion transport and solute transport (ATP11B, CACNB3, KCNJ3 and four members of the solute carrier family). It is known that abnormal transmembrane signaling segregates with the mutational status of IgVH, CD38 and ZAP70 in CLL, and that this involves ion fluxes. Thus, it is plausible that epigenetically deregulated ion and/or solute transport may contribute to this process. Other genes from the CD38high group included transcriptional regulators and genes involved in neural development, as well as other miscellaneous genes. Of the genes initially confirmed by COBRA, NRP2, SFRP2 and ADAM12 were mainly methylated in CD38high cases (>30% CD38). The statistical treatment of segregating methylated genes at each level is shown in Supplementary Tables 3&4. NRP2 and NRP1 comprise the neuropilin family of genes and play an important role in normal and neoplastic angiogenesis [46]. Aberrant angiogenesis is present in CLL, and increased levels of soluble VEGF and increased bone marrow vascular density are considered poor prognostic factors and signs of disease progression, and are more apparent in those with increased CD38 [47,48]. We previously reported that NRP2 was methylated in most cases of follicular lymphoma tested, and only a subset of CLL and mantle cell lymphoma cases [9].
In contrast, DLEU7, located adjacent to a critical chromosome 13q14 breakpoint region commonly deleted in CLL, was preferentially methylated in CD38low cases. The DLEU7 promoter includes binding sites for multiple transcriptional enhancing factors. This gene was previously reported to be affected by DNA methylation, but its relationship to CD38 expression was not explored [49]. It is not clear if such differential methylation in CLL affects clinical behaviors, since not much is known about the function of DLEU7, but this should be addressed in future studies.
A direct comparison of the two microarrays used in this study is not readily performed. The reasons for this are multiple. The Toronto 12K array is comprised of genomic DNA fragments enriched for CGIs using a methyl-binding protein column. Of these 12K loci, the array contains approximately 5411 loci after filtering unusable spots based on criteria described in Heisler et al. [50]. This array represents approximately 10% of predicted CGIs. We actually started this project using this array because CGI commercial arrays were not well-developed at the time. However, we later planned to expand our studies and selected the Agilent CGI array that is comprised of 244,000 well-defined oligonucleotide capture probes representing over 27,000 CGIs that are well-annotated. In using the Toronto array for discoveries, we only examined the loci of interest via direct validation by sequencing and confirmatory independent methylation analyses. Thus, even with limitations, the data produced was valid. Furthermore, the two methods of amplicon preparation are also different. For the Toronto array, we used the DMH amplicon preparation method [9,51] that employs digestion with the methylation-sensitive endonucleases, HpaII and BstUI, whereas the MCAM method for the Agilent array uses SmaI and XmaI enzymes [11]. While there is some overlap in DNA fragments produced, the cut sites recognized by these enzymes also results in different size and yield, and therefore different possible amplicons and results. Nevertheless, the independent uses of validated results of each type of array are useful even though they may identify overlapping, as well as different methylated regions. In our experiments, we generally were able to successfully validate approximately 75% of loci from the Toronto array and over 95% from the Agilent array (data not shown).
In the context of our pharmacologic reversal experiments, demethylation generally induced upregulation of the associated gene mRNA, but not always. In some cases, full re-expression required a combination of demethylation and inhibition of HDACs. These observations are not unexpected, as there are many examples of genes that seem to require both for full re-expression. These treatments can be quite toxic, and we therefore regularly monitor this by use of cell counts and viability measurements. In these short-term cultures, the cells generally show above 80% viability. In addition, cell lines can differ in their sensitivity to these treatments. The cell lines in this study have been in our laboratory for more than 10 years and have been optimized in these treatment systems. Patterns of mRNA re-expression are known to be variable in this commonly used experimental system. In some instances, gene methylation can be present, and still the re-expression can be variably affected by each, or the combination of, epigenetic modifying agents. It is still possible that for some genes, a variable amount of DNA methylation can be present, but this may not be the cause of transcriptional downregulation. Alternatively, downegulation may be heavily affected by HDACs, and therefore reactivation occurs mainly with the use of TSA ± 5′-Aza. It would be ideal to directly measure DNA demethylation, but it has also been shown that a minor level of demethylation can sometimes result in re-expression, and therefore direct measurements may not even tell the entire story [52,53]. There is clearly more to be learned about differential gene susceptibility to epigenetic modifications and their sensitivities to pharmacologic reversals. These questions are relevant in discussions of clinical epigenetic therapies.
In summary, there are clear alterations in DNA methylation of multiple genes in CLL compared with normal peripheral blood B lymphocytes. Most are methylated or unmethylated regardless of CD38 expression level, and therefore may affect the biological behavior of all CLL cases in a similar manner. A listing of newly discovered candidate gene promoters is included (Supplementary Tables 5 & 6). These need further confirmation, but provide a basis for more detailed epigenetic investigations in CLL. Some genes may be differentially methylated in cases with CD38low or CD38high expression and might be associated with more subclass-specific characteristics of the disease. Further genome-wide and gene-specific studies of DNA methylation, histone modifications and other epigenetic alterations will be needed to fully appreciate the impact of the CLL epigenome on treatment options for patients. It is clear in the case of acute myeloid leukemia that genomic regions are frequently differentially affected by treatments with the most common demethylating agents [54]. The widespread perception that 5-azacytidine, 5-aza-2′-deoxycytidine and zebularine are interchangeable as inhibitors of DNA methyltransferases should be reconsidered, and knowing which genes/regions can be effectively reactivated with these agents, as well as HDAC inhibitors, is important in the context of ongoing and future clinical trials with epigenetic modifying agents. This study and others that further enhance our understanding of cancer epigenomes will help by providing candidate biomarkers that may be useful in diagnosis or predictive of treatment outcomes, and for monitoring the success of therapies.
Conclusion
As for other cancers, CLL demonstrates DNA methylation of multiple genes that may be affected at a transcriptional level. The genes were mostly either methylated or unmethylated across all CD38 expression levels, but some methylated genes segregated with CD38low or CD38high groups that are thought to demonstrate differential biological behaviors. Certain groups of pathway-associated genes, such as those involved in WNT signaling, were overrepresented in the methylated group. The list of candidate methylated genes produced in this study should help facilitate further investigations. Characterization of epigenetic alterations in CLL is necessary before invoking certain aspects of the newer epigenetic therapeutic approaches to patient management.
Executive summary
DNA methylation in B-cell chronic lymphocytic leukemia (CLL) involves at least one hundred genes
Using two different types of microarrays, we identified over 100 candidate genes that show hypermethylation compared with normal cells.
Many of these were validated by additional independent assay methods.
Additional candidate genes have been shown to be methylated in other studies.
This gene list will provide a basis for further research by other investigators beyond our laboratory.
While most genes are either methylated or unmethylated across leukemic subtypes, a few segregate with either CD38high or CD38low levels
The level of expression of CD38 is one independent surrogate marker of biological behavior in CLL. Low-risk patients generally fall into the CD38low category and high-risk patients generally are CD38high. The absolute level of CD38 that comprises CD38low versus CD38high is debatable.
In initial microarray studies, the genes NRP2, SFRP2, and ADAM12 were found to be preferentially methylated in CD38high cases and DLEU7 in CD38low cases, while 11 other genes were methylated across all levels.
Using the Agilent CpG island microarray, compared with normal B-cell DNA, we found 65 new candidate genes methylated across all CD38 levels, 32 new candidate genes in the CD38high group and 5 in the CD38low group.
The biological significance of these differential effects awaits further investigation.
Pathway analysis of the DNA methylation patterns reveals an overrepresentation of WNT pathway genes, genes involved in ion and solute transport, regulators of transcription, and Homeobox genes
WNT pathway genes independently confirmed to be methylated were mainly those with inhibitory functions; APC, SFRP2, and 6 SOX family members.
Genes involved in ion and solute transport could affect some of the known deregulated transmembrane signaling mechanisms in CLL.
Additional methylated genes are involved in angiogenesis and stromal cell interactions, another process known to be deregulated in CLL.
Conclusion
As in virtually all other cancers, CLL displays epigenetic alterations in DNA methylation that affects multiple genes that may have a biological impact on how and how rapidly the tumor progresses and potentially on how therapeutic interventions might be improved. The candidate gene lists developed can serve as a platform for further and more extensive investigations into CLL biological and clinical behavior.
Supplementary Material
Acknowledgments
We thank Susan Souchek, Laura Jacobus, Melinda Andreski and Kathy Olson for assistance with tissue banking and Angel Surdin for expert editorial assistance.
Footnotes
Financial & competing interests disclosure: Support was provided from the National Cancer Institute grants CA 100055 and CA097880 (CWC), CA123018 (HS), CA113408 (TS) and the CRC Missouri Chair in Cancer Research (CWC). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Contributor Information
Farahnaz B Rahmatpanah, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Stephanie Carstens, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Sam I Hooshmand, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Elise C Welsh, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Ozy Sjahputera, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Kristen H Taylor, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Lynda B Bennett, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Huidong Shi, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
J Wade Davis, Department of Health Management and Informatics, University of Missouri School of Medicine, Columbia, Missouri 65212, USA, Department of Statistics, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Gerald L Arthur, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Tait D Shanafelt, Department of Internal Medicine, Mayo Clinic, Rochester, MN 55905, USA.
Neil E Kay, Department of Internal Medicine, Mayo Clinic, Rochester, MN 55905, USA.
James E Wooldridge, Department of Internal Medicine and Holden Comprehensive Cancer Center, University of Iowa, Iowa City, IA 52242. Current address: Eli Lilly and Company, Indianapolis, IN 46285, USA.
Charles W Caldwell, Department of Pathology and Anatomical Sciences, Ellis Fischel Cancer Center, University of Missouri School of Medicine, Columbia, Missouri 65212, USA.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Zent CS, Kay NE. Chronic lymphocytic leukemia: biology and current treatment. Curr Oncol Rep. 2007;9:345–352. doi: 10.1007/s11912-007-0046-8. [DOI] [PubMed] [Google Scholar]; ▪▪ Excellent review of chronic lymphocytic leukemia (CLL).
- 2.Krober A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 3.Damle RN, Wasil T, Fais F, et al. IgV gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]; ▪ Good early description of the association between candidate prognostic biomarkers.
- 4.Gentile M, Mauro FR, Calabrese E, et al. The prognostic value of CD38 expression in chronic lymphocytic leukaemia patients studied prospectively at diagnosis: a single institute experience. Br J Haematol. 2005;130:549–557. doi: 10.1111/j.1365-2141.2005.05659.x. [DOI] [PubMed] [Google Scholar]
- 5.Eisele L, Haddad T, Sellmann L, Duhrsen U, Durig J. Expression levels of CD38 on leukemic B cells but not on non-leukemic T cells are comparably stable over time and predict the course of disease in patients with chronic lymphocytic leukemia. Leuk Res. 2009;33:775–778. doi: 10.1016/j.leukres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Hamblin TJ, Orchard JA, Ibbotson RE, et al. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–1029. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]; ▪ Good discussion of the biological variability that can occur over time with CD38.
- 7.Tobin G, Rosenquist R. Prognostic usage of VH gene mutation status and its surrogate markers and the role of antigen selection in chronic lymphocytic leukemia. Med Oncol. 2005;22:217–228. doi: 10.1385/MO:22:3:217. [DOI] [PubMed] [Google Scholar]
- 8.Pittner BT, Shanafelt TD, Kay NE, Jelinek DF. CD38 expression levels in chronic lymphocytic leukemia B cells are associated with activation marker expression and differential responses to interferon stimulation. Leukemia. 2005;19:2264–2272. doi: 10.1038/sj.leu.2403975. [DOI] [PubMed] [Google Scholar]
- 9.Rahmatpanah FB, Carstens S, Guo J, et al. Differential DNA methylation patterns of small B-cell lymphoma subclasses with different clinical behavior. Leukemia. 2006;20:1855–1862. doi: 10.1038/sj.leu.2404345. [DOI] [PubMed] [Google Scholar]; ▪ Good description of the differential methylation hybridization method and early results with chronic lymphocytic leukemia.
- 10.Caldwell CW, Patterson WP. Relationship between CD45 antigen expression and putative stages of differentiation in B-cell malignancies. Am J Hematol. 1991;36:111–115. doi: 10.1002/ajh.2830360209. [DOI] [PubMed] [Google Scholar]
- 11.Estecio MR, Yan PS, Ibrahim AE, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Good discussion of the methylated CpG island amplification method used in the current study.
- 12.Walter SD. Point estimation of the odds ratio in sparse 2 × 2 contingency tables. In: MacNeill IB, Umphrey GJ, Reidel D, editors. Biostatistics. 1987. pp. 71–102. [Google Scholar]; ▪▪ Excellent description of the statistics used in the current study to assign odds ratios.
- 13.Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 14.Rassenti LZ, Jain S, Keating MJ, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ Excellent review of putative biomarkers in predicting disease aggressiveness in CLL.
- 15.Lu D, Zhao Y, Tawatao R, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]; ▪▪ An excellent bioinformatics resource for gene ontology and pathway analysis.
- 17.Wahlfors J, Hiltunen H, Heinonen K, Hamalainen E, Alhonen L, Janne J. Genomic hypomethylation in human chronic lymphocytic leukemia. Blood. 1992;80:2074–2080. [PubMed] [Google Scholar]
- 18.Lipsanen V, Leinonen P, Alhonen L, Janne J. Hypomethylation of ornithine decarboxylase gene and erb-A1 oncogene in human chronic lymphatic leukemia. Blood. 1988;72:2042–2044. [PubMed] [Google Scholar]
- 19.Yuille MR, Condie A, Stone EM, et al. TCL1 is activated by chromosomal rearrangement or by hypomethylation. Genes Chromosomes Cancer. 2001;30:336–341. doi: 10.1002/gcc.1099. [DOI] [PubMed] [Google Scholar]
- 20.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. BCL-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 21.Raval A, Tanner SM, Byrd JC, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rush LJ, Raval A, Funchain P, et al. Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res. 2004;64:2424–2433. doi: 10.1158/0008-5472.can-03-2870. [DOI] [PubMed] [Google Scholar]; ▪ Good early study using the restriction landmark genomic scanning method.
- 23.Deaglio S, Vaisitti T, Aydin S, et al. CD38 and ZAP-70 are functionally linked and mark CLL cells with high migratory potential. Blood. 2007;110:4012–4021. doi: 10.1182/blood-2007-06-094029. [DOI] [PubMed] [Google Scholar]; ▪ Describes a functional association between CD38 expression and cell migration.
- 24.Damle RN, Temburni S, Calissano C, et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood. 2007;110:3352–3359. doi: 10.1182/blood-2007-04-083832. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Describes a functional association between CD38 expression and cell proliferation.
- 25.Deaglio S, Vaisitti T, Bergui L, et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood. 2005;105:3042–3050. doi: 10.1182/blood-2004-10-3873. [DOI] [PubMed] [Google Scholar]
- 26.Malavasi F, Deaglio S, Ferrero E, et al. CD38 and CD157 as receptors of the immune system: a bridge between innate and adaptive immunity. Mol Med. 2006;12(11–12):334–341. doi: 10.2119/2006-00094.Malavasi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morabito F, Mangiola M, Stelitano C, Deaglio S, Callea V, Malavasi F. Simultaneous expression of CD38 and its ligand CD31 by chronic lymphocytic leukemia B-cells: anecdotal observation or pathogenetic hypothesis for the clinical outcome? Haematologica. 2003;88:354–355. [PubMed] [Google Scholar]
- 28.Moreno-Garcia ME, Lopez-Bojorques LN, Zentella A, Humphries LA, Rawlings DJ, Santos-Argumedo L. CD38 signaling regulates B lymphocyte activation via a phospholipase C (PLC)-γ 2-independent, protein kinase C, phosphatidylcholine-PLC, and phospholipase D-dependent signaling cascade. J Immunol. 2005;174:2687–2695. doi: 10.4049/jimmunol.174.5.2687. [DOI] [PubMed] [Google Scholar]
- 29.Batra S, Shi Y, Kuchenbecker KM, et al. Wnt inhibitory factor-1, a Wnt antagonist, is silenced by promoter hypermethylation in malignant pleural mesothelioma. Biochem Biophys Res Commun. 2006;342:1228–1232. doi: 10.1016/j.bbrc.2006.02.084. [DOI] [PubMed] [Google Scholar]
- 30.Chim CS, Pang R, Fung TK, Choi CL, Liang R. Epigenetic dysregulation of Wnt signaling pathway in multiple myeloma. Leukemia. 2007;21:2527–2536. doi: 10.1038/sj.leu.2404939. [DOI] [PubMed] [Google Scholar]
- 31.Dhir M, Montgomery EA, Glockner SC, et al. Epigenetic regulation of WNT signaling pathway genes in inflammatory bowel disease (IBD) associated neoplasia. J Gastrointest Surg. 2008;12:1745–1753. doi: 10.1007/s11605-008-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelebart P, Anand M, Armanious H, et al. Constitutive activation of the Wnt canonical pathway in mantle cell lymphoma. Blood. 2008;112:5171–5179. doi: 10.1182/blood-2008-02-139212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan NI, Bendall LJ. Role of WNT signaling in normal and malignant hematopoiesis. Histol Histopathol. 2006;21:761–774. doi: 10.14670/HH-21.761. [DOI] [PubMed] [Google Scholar]
- 34.Martin V, Agirre X, Jimenez-Velasco A, et al. Methylation status of Wnt signaling pathway genes affects the clinical outcome of Philadelphia-positive acute lymphoblastic leukemia. Cancer Sci. 2008;99(9):1865–1868. doi: 10.1111/j.1349-7006.2008.00884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- 36.Roman-Gomez J, Jimenez-Velasco A, Cordeu L, et al. WNT5A, a putative tumour suppressor of lymphoid malignancies, is inactivated by aberrant methylation in acute lymphoblastic leukaemia. Eur J Cancer. 2007;43:2736–2746. doi: 10.1016/j.ejca.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Sievers E, Endo T, Lu D, Carson D, Schmidt-Wolf IG. Targeting Wnt pathway in lymphoma and myeloma cells. Br J Haematol. 2008;144(5):796–798. doi: 10.1111/j.1365-2141.2008.07503.x. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta A, Banerjee D, Chandra S, et al. Deregulation and cross talk among Sonic hedgehog, Wnt, Hox and Notch signaling in chronic myeloid leukemia progression. Leukemia. 2007;21:949–955. doi: 10.1038/sj.leu.2404657. [DOI] [PubMed] [Google Scholar]
- 39.Wohrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol Cell Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chim CS, Fung TK, Wong KF, Lau JS, Liang R. Infrequent Wnt inhibitory factor-1 (Wif-1) methylation in chronic lymphocytic leukemia. Leuk Res. 2006;30(9):1135–1139. doi: 10.1016/j.leukres.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Chim CS, Pang R, Liang R. Epigenetic dysregulation of the Wnt signalling pathway in chronic lymphocytic leukaemia. J Clin Pathol. 2008;61:1214–1219. doi: 10.1136/jcp.2008.060152. [DOI] [PubMed] [Google Scholar]
- 42.Howe D, Bromidge T. Variation of LEF-1 mRNA expression in low-grade B-cell non-Hodgkin's lymphoma. Leuk Res. 2006;30:29–32. doi: 10.1016/j.leukres.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66:653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 44.Raval A, Lucas DM, Matkovic JJ, et al. TWIST2 demonstrates differential methylation in immunoglobulin variable heavy chain mutated and unmutated chronic lymphocytic leukemia. J Clin Oncol. 2005;23:3877–3885. doi: 10.1200/JCO.2005.02.196. [DOI] [PubMed] [Google Scholar]
- 45.Jin G, Lu D, Yao S, et al. Amide derivatives of ethacrynic acid: synthesis and evaluation as antagonists of Wnt/β-catenin signaling and CLL cell survival. Bioorg Med Chem Lett. 2009;19:606–609. doi: 10.1016/j.bmcl.2008.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geretti E, Shimizu A, Klagsbrun M. Neuropilin structure governs VEGF and semaphorin binding and regulates angiogenesis. Angiogenesis. 2008;11:31–39. doi: 10.1007/s10456-008-9097-1. [DOI] [PubMed] [Google Scholar]
- 47.Kay NE. Angiogenesis revisited in CLL. Leuk Res. 2007;31:1459–1460. doi: 10.1016/j.leukres.2007.04.001. [DOI] [PubMed] [Google Scholar]; ▪ Very good review of the association of angiogenesis with CLL and prognosis.
- 48.Molica S, Cutrona G, Vitelli G, et al. Markers of increased angiogenesis and their correlation with biological parameters identifying high-risk patients in early B-cell chronic lymphocytic leukemia. Leuk Res. 2007;31:1575–1578. doi: 10.1016/j.leukres.2007.03.009. [DOI] [PubMed] [Google Scholar]; ▪ Very good study of the association of angiogenesis with CLL prognosis.
- 49.Hammarsund M, Corcoran MM, Wilson W, et al. Characterization of a novel B-CLL candidate gene – DLEU7 – located in the 13q14 tumor suppressor locus. FEBS Lett. 2004;556(1–3):75–80. doi: 10.1016/s0014-5793(03)01371-1. [DOI] [PubMed] [Google Scholar]
- 50.Heisler LE, Torti D, Boutros PC, et al. CpG Island microarray probe sequences derived from a physical library are representative of CpG Islands annotated on the human genome. Nucleic Acids Res. 2005;33:2952–2961. doi: 10.1093/nar/gki582. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ An excellent description of the Tornonto microarray used in this study.
- 51.Laux DE, Curran EM, Welshons WV, Lubahn DB, Huang TH. Hypermethylation of the Wilms' tumor suppressor gene CpG island in human breast carcinomas. Breast Cancer Res Treat. 1999;56:35–43. doi: 10.1023/a:1006222803788. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Issa JP. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: dose, duration, and patient selection. Nat Clin Pract Oncol. 2005;2(Suppl. 1):S24–S29. doi: 10.1038/ncponc0355. [DOI] [PubMed] [Google Scholar]
- 54.Flotho C, Claus R, Batz C, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.