Summary
Dicer, which is required for the processing of both micro-RNAs (miRNAs) and small interfering RNAs (siRNAs), is essential for oocyte maturation [1, 2]. Oocytes express both miRNAs and endogenous siRNAs (endo-siRNAs) [3, 4]. To determine whether the abnormalities in Dicer knockout oocytes during meiotic maturation are secondary to the loss of endo-siRNAs and/or miRNAs, we deleted Dgcr8, which encodes an RNA-binding protein specifically required for miRNA processing. In striking contrast to Dicer, Dgcr8-deficient oocytes matured normally and, when fertilized with wild-type sperm, produced healthy-appearing offspring, even though miRNA levels were reduced to similar levels as Dicer-deficient oocytes. Furthermore, the deletion of both maternal and zygotic Dgcr8 alleles did not impair preimplantation development, including the determination of the inner cell mass and trophectoderm. Most surprisingly, the mRNA profiles of wild-type and Dgcr8 null oocytes were essentially identical, whereas Dicer null oocytes showed hundreds of misregulated transcripts. These findings show that miRNA function is globally suppressed during oocyte maturation and preimplantation development and that endo-siRNAs, rather than miRNAs, underlie the Dicer knockout phenotype in oocytes.
Results and Discussion
Dicer is an RNase III enzyme that is essential in the processing of several classes of small RNAs, including canonical micro-RNAs (miRNAs), noncanonical miRNAs, and endogenous small interfering RNAs (endo-siRNAs) [5]. These classes are differentiated based on their biogenesis upstream of Dicer (see Figure S1 available online). Canonical miRNAs are transcribed as long RNAs. The RNA-binding protein DGCR8 recognizes short hairpins within the long RNA and directs the RNase III enzyme Drosha to release the hairpin. The released hairpins are transported to the cytoplasm, where Dicer cleaves them into short double-stranded RNAs (dsRNAs). Noncanonical miRNAs bypass DGCR8/Drosha processing by using other endonucleases or direct transcription to produce the short hairpin, which again is cleaved by Dicer. Endo-siRNAs are derived from long dsRNAs in the form of either sense/antisense RNA pairs or long hairpins, which are then directly processed by Dicer consecutively along the dsRNA to produce multiple siRNAs. The differences in canonical miRNA, noncanonical miRNA, and endo-siRNA processing enable the dissection of their functions. For example, deletion of Dgcr8 specifically blocks the production of canonical miRNAs, whereas deletion of Dicer blocks production of both miRNAs and endo-siRNAs [5].
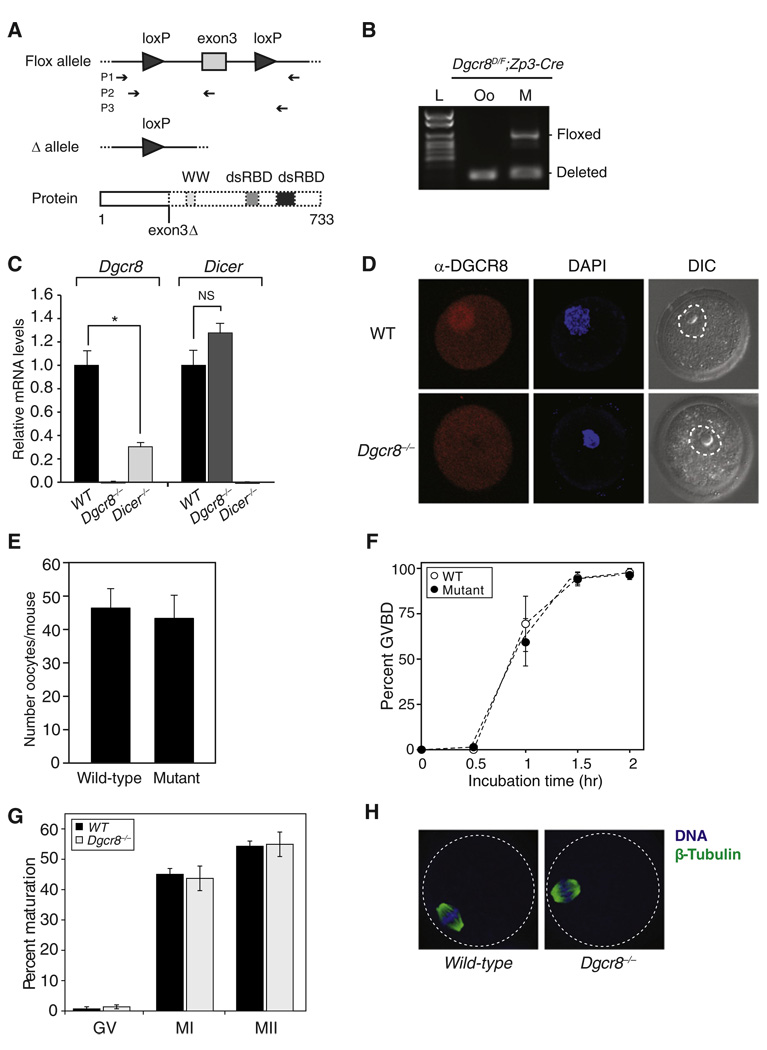
Conditional deletion of Dicer in mouse oocytes leads to infertility with severe defects in chromosomal alignment and spindle organization [1, 2]. Because numerous endo-siRNAs and miRNAs are present in oocytes [3, 4], the Dicer knockout phenotype could be the result of either of these classes of small RNAs. To test the relative contribution of canonical miRNAs versus other Dicer-dependent small RNAs to the phenotype, we crossed the same oocyte-specific Cre transgenic line (Zp3-Cre) [6] to a Dgcr8 conditional knockout model [7, 8], hence specifically removing canonical miRNAs. The conditional knockout of Dgcr8 is based on a floxed exon 3 allele. Loss of exon 3 results in a frameshift mutation with multiple downstream premature stop codons, resulting in a truncated protein missing the WW and RNA-binding domains (Figure 1A). The floxed allele in Dgcr8delta/flox; Zp3-Cre females was efficiently deleted in oocytes (Figure 1B). Furthermore, quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) showed complete loss of Dgcr8 transcripts carrying exon 3 (Figure 1C, left). It also showed a 68% reduction in Dgcr8 transcript levels in Dicer−/− oocytes (Figure 1C, left; *p = 0.0058, Student’s t test). In contrast, Zp3-Cre-induced deletion of Dgcr8 had no significant effect on Dicer transcript levels (Figure 1C, right; not significant, p = 0.1432). Immunohistochemistry showed a loss of nuclear DGCR8 protein in the knockout oocytes (Figure 1D). Together, these data show a complete loss of Dgcr8 in the conditional knockouts.
Figure 1. Dgcr8-Deficient Oocytes Mature Normally.
(A) Schematic of Dgcr8 knockout strategy. Exon 3 deletion on Dgcr8 mRNA leads to a frameshift mutation and premature stop codons, which are predicted to generate truncated DGCR8 protein. Top: box represents exon; connecting line represents intron; arrowhead represents lox sequence; arrow sets below represent primer pairs used for genotyping. Bottom: thick boxes represent the proteins that can be produced; dashed boxes represent predicted truncation; WW represents a WW protein-protein interaction domain; dsRBD represents double-stranded RNA-binding domains.
(B) Genotyping of Dgcr8delta/flox; Zp3-Cre female (M, mother) and the metaphase II (MII) oocyte (Oo) recovered from this female. Floxed alleles were detected only in mother. L denotes ladder.
(C) Dgcr8 (left) or Dicer (right) mRNA levels in wild-type, Dgcr8, and Dicer knockout oocytes. Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed. Error bars denote standard error of the mean (SEM) of three experiments. *p < 0.05; NS, not significant.
(D) Confocal microscopic images of wild-type and Dgcr8−/− oocytes stained with antibody to DGCR8. Germinal vesicle (GV) oocytes were collected, permeabilized, and incubated with DGCR8 antibody (red). DNA was visualized with DAPI (blue), and morphology was determined by differential interference contrast (DIC). Dashed line surrounds the nuclear membrane.
(E) Number of MII oocytes recovered from Dgcr8delta/flox; Zp3-Cre (mutant) or wild-type female mice, which were primed with gonadotropins. Error bar denotes SEM of seven experiments.
(F) Time course of meiotic resumption. Fully grown GV oocytes were collected from wild-type and mutant mice and scored for GV breakdown. Each point is the mean ± SEM of three experiments.
(G) The oocyte maturation status was scored by the presence of a GV (GV oocyte), a polar body (MII oocyte), or neither (MI oocyte) in wild-type and Dgcr8−/− oocytes. At least three independent pools of oocytes were scored. Error bars denote SEM of seven experiments.
(H) Spindle morphology in wild-type (left) and Dgcr8−/− (right) oocytes. Oocytes were stained with a β-tubulin antibody (green), and DNA was counterstained with DAPI (blue) (dashed line surrounds the oocyte).
To evaluate the effect of Dgcr8 loss on oocyte maturation in vivo, we collected oocytes 16 hr post-human chorionic gonadotropin (hCG) injection. Similar numbers of metaphase II (MII) oocytes were recovered from wild-type and Dgcr8delta/flox; Zp3-Cre mutant females (Figure 1E). To follow maturation of the oocytes in vitro, we isolated fully grown germinal vesicle (GV) oocytes and allowed them to mature in culture. Wild-type and Dgcr8−/− oocytes developed similarly in terms of both the kinetics of GV breakdown (GVBD) in vitro (Figure 1F) and maturation to the MI and MII stages both in vivo (Figure 1G) and in vitro (data not shown). This finding is in stark contrast to Dicer−/− oocytes, which arrest at MI, rarely extruding a polar body [1, 2]. Because Dicer−/− oocytes arrest with defects in spindle organization and chromosome condensation [1, 2], we evaluated spindle structure in Dgcr8−/− oocytes, which were collected 16 hr post-hCG. The meiotic spindles of Dgcr8−/− oocytes had a normal morphology (Figure 1H). These findings show that DGCR8, unlike Dicer, is not required for oocyte maturation.
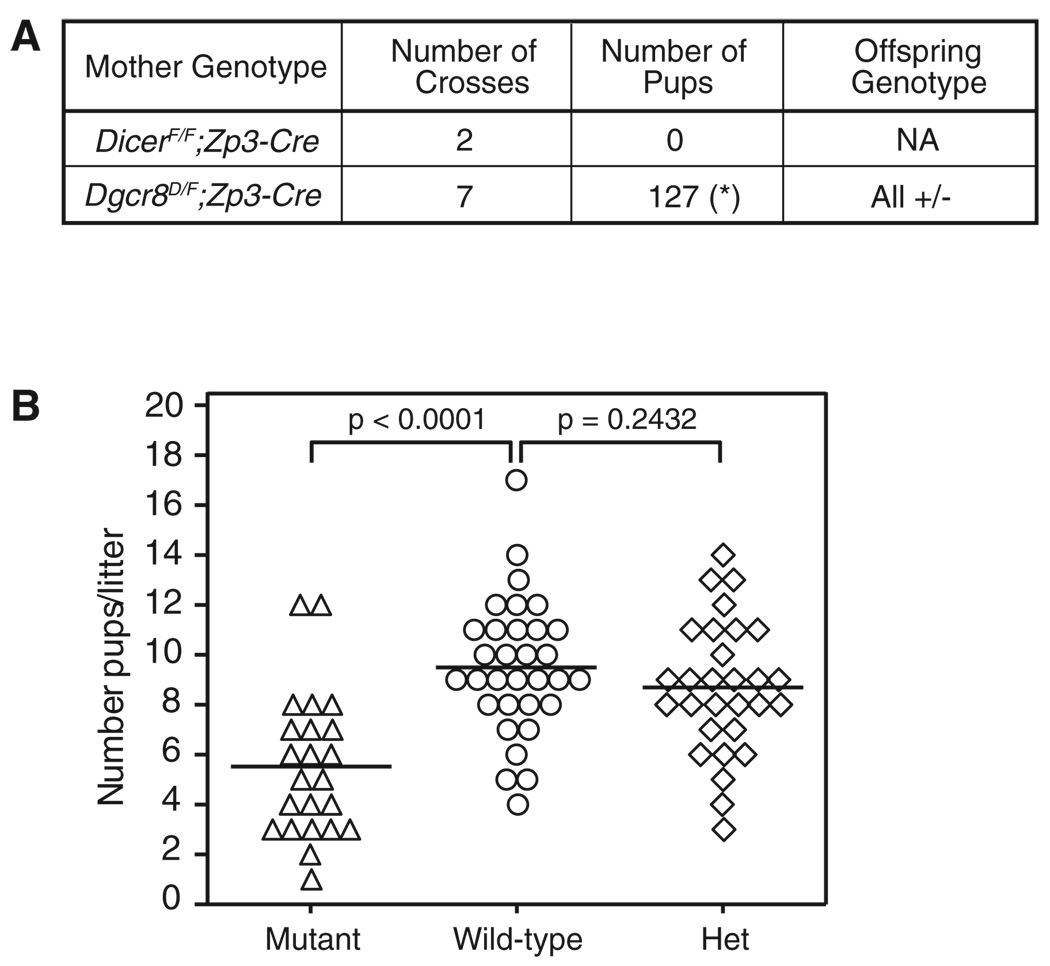
Next, we asked whether maternal loss of Dgcr8, and hence maternally contributed canonical miRNAs, would affect fertility and the development of resulting embryos. Wild-type males were crossed to the Dgcr8delta/flox; Zp3-Cre females. Initial crosses showed that unlike the Dicerflox/flox; Zp3-Cre females, Dgcr8delta/flox; Zp3-Cre females could be fertilized and produce healthy-appearing offspring (Figure 2A). However, analysis of brood size over many crosses showed that Dgcr8-deficient mothers produced fewer offspring compared to controls (5.5 ± 0.6 pups per litter, n = 23 versus 9.5 ± 0.5 pups per litter, n = 32, p < 0.0001) (Figure 2B, left). This defect was not secondary to zygotic haploinsufficiency, because control heterozygous crosses showed no significant difference (8.7 ± 0.5 pups per litter, n = 29 versus 9.5 ± 0.5 pups per litter, n = 32, p = 0.2432) (Figure 2B, right). Therefore, although maternal DGCR8 is not absolutely required for fertility, it does impact female fecundity.
Figure 2. Dgcr8-Deficient Oocytes Can Be Fertilized and Produce Healthy Pups.
(A) Table summarizes the cross between Dgcr8delta/flox; Zp3-Cre or Dicerflox/flox; Zp3-Cre with wild-type males. For Dgcr8delta/flox; Zp3-Cre, each cross produces multiple litters, denoted by asterisk (*).
(B) Brood size from wild-type, Dgcr8delta/flox; Zp3-Cre (mutant), or Dgcr8delta/+ (het) mice. Each data point represents the number of pups per litter from wild-type (○), mutant (Δ), and het (◊) mothers. Brood size is reduced approximately 36% in the mutant compared to wild-type (p < 0.0001).
The loss of zygotic Dgcr8 leads to embryonic arrest prior to E6.5 [9]. To determine whether more subtle defects could be seen at earlier stages, we analyzed preimplantation development, including cell number and distribution of cells between the inner cell mass (ICM) and trophectoderm lineages, which is the first definitive differentiation event during embryogenesis. Embryos were genotyped following morphological and immunofluorescence analysis. Crosses between Dgcr8 heterozygous mice produced normal-appearing zygotic Dgcr8 knockout blastocysts (zygotic Dgcr8−/−) (Figure 3A, left). Furthermore, knockout embryos developed to the blastocyst stage with normal Mendelian ratios (Figure S2A). Immunofluorescence showed that the total number of cells and distribution between the trophectoderm (Cdx-2-positive) and the ICM were unchanged in mutant embryos (Figure 3A, right).
Figure 3. Preimplantation Development Occurs Normally in Dgcr8 Knockout Embryos.
(A) Left: confocal microscopic images of embryos for the zygotic Dgcr8 mutant (Z Dgcr8−/−) and the control. At E3.5, embryos were collected, permeabilized, and incubated with antibodies to Oct-3/4 (green) and Cdx-2 (red). DNA was visualized with DAPI (blue), and morphology was determined by DIC. Right: average number of total cells and blastomeres that were positive for Cdx-2 in control and mutant (Z Dgcr8−/−). The data are presented as the mean ± SEM; seventeen blastocysts were analyzed.
(B) Left: confocal microscopic images of embryos for the maternal-zygotic Dgcr8 mutant (MZ Dgcr8−/−) and the control. Right: average number of total cells and blastomeres that were positive for Cdx-2 in control and mutant (MZ Dgcr8−/−). The data are presented as the mean ± SEM; twelve blastocysts were analyzed.
Maternally provided Dgcr8 and/or miRNAs may have enabled preimplantation development. Therefore, Dgcr8 conditional knockout mothers (Dgcr8delta/flox; Zp3-Cre) were crossed to heterozygous knockout males, and the resulting maternal-zygotic knockout (maternal-zygotic Dgcr8−/−) embryos were evaluated. Once again, there was no difference between the knockout and control embryos in terms of morphology (Figure 3B, left) or the number of embryos that developed to the blastocyst stage (Figure S2B). Furthermore, the number of cells per embryo and the distribution of cells between the ICM and the trophectoderm were unchanged (Figure 3B, right). Together, these findings show that the earliest differentiation event leading to epiblast and trophectoderm lineages proceed normally in the absence of canonical miRNAs.
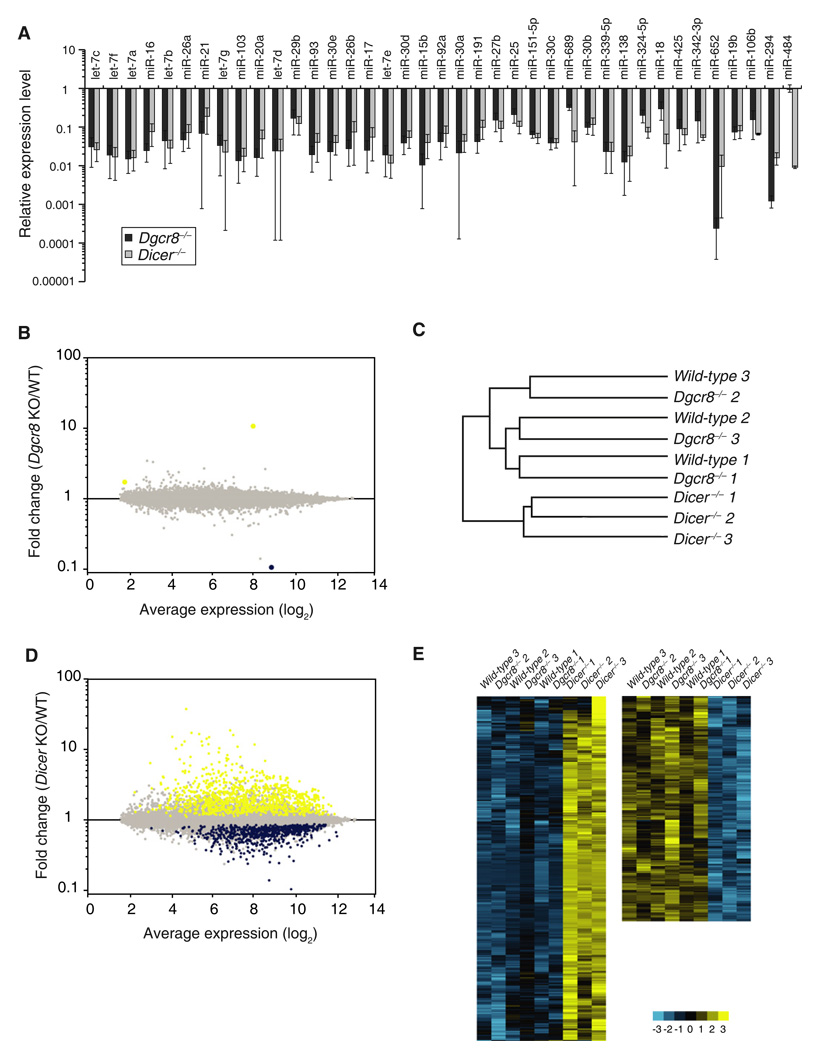
The finding that the maternal and zygotic DGCR8 were dispensable for oocyte maturation and early embryonic development was surprising considering the central role of miRNAs in other developmental decisions [10]. The dramatic difference in phenotypes between Dicer and Dgcr8 knockout oocytes could be partly explained by the loading of pre-miRNAs in the growing oocyte prior to Zp3-Cre-induced deletion, which would then be processed by Dicer at later stages without any further requirement of DGCR8. To test this possibility, we evaluated 40 highly expressed miRNAs in oocytes by performing a quantitative RT-PCR assay [11]. The levels of 39 of these 40 miRNAs were similarly depleted in the Dgcr8−/− and Dicer−/− oocytes (Figure 4A). The only miRNA not depleted, miR-484, is a noncanonical miRNA and is hence DGCR8 independent [12]. Therefore, ongoing Dgcr8 expression is indeed required for the production of canonical miRNAs in developing oocytes.
Figure 4. Molecular Effects of Dgcr8 or Dicer Loss in Oocytes.
(A) The expression levels of 40 microRNAs (miRNAs) highly expressed in oocytes were chosen for qRT-PCR TaqMan analysis. Values for wild-type level were set to 1. Error bar denotes SEM of three experiments.
(B) mRNA profiles of Dgcr8−/− versus wild-type oocytes. Yellow represents upregulated transcripts and blue represents downregulated transcripts with false discovery rate (FDR) < 0.05. Each data point represents a transcript.
(C) Dendrogram of mRNA profiles of wild-type, Dgcr8−/−, and Dicer−/− oocytes. Cluster analysis could not distinguish the wild-type from Dgcr8 knockout transcriptomes.
(D) mRNA profiles of Dicer−/− versus wild-type oocytes. Presented as in (B).
(E) Heat maps of upregulated genes (left, yellow) or downregulated genes (right, blue) in Dicer−/− oocytes compared to wild-type and Dgcr8−/− oocytes.
We next asked whether miRNAs repress mRNAs in oocytes. We reasoned that although there were no obvious phenotypic consequences, the large number of highly expressed miRNAs [1–4] in wild-type oocytes would still impact the molecular constitution of the cells. Because miRNAs function to both destabilize mRNAs as well as inhibit translation, we compared the mRNA profiles of the Dgcr8−/− and wild-type oocytes. Surprisingly, the profiles were almost identical (Figure 4B). Only three mRNAs were identified as significantly changed with a false discovery rate (FDR) cutoff of 5%, including Dgcr8 itself representing remnant exon 3 deleted transcript (0.1× relative to wild-type), MT1 (10.7×), and a predicted transcript ENSMUST00000101675 (1.7×). Furthermore, cluster analysis via Pearson’s correlation for the distance measurement could not distinguish the wild-type from Dgcr8−/− transcriptomes, whereas the Dicer−/− transcriptome was clearly diverged (Figure 4C). Similar cluster analysis on wild-type, Dgcr8−/−, and Dicer−/− embryonic stem cells showed that the transcriptomes of Dgcr8−/− and Dicer−/− were more similar to each other than to that of wild-type, as would be expected if miRNAs play a central role in the cells (Figure S3). These data show that canonical miRNAs have little effect on the transcriptome of oocytes, in stark contrast to other cell types.
Dicer−/− oocytes showed dramatic changes in their expression profile (Figure 4D). One thousand and forty nine transcripts were upregulated and 657 genes were downregulated compared to wild-type controls (FDR < 5%) (Figures 4D and 4E). These changes were likely due to DGCR8-independent, Dicer-dependent small RNAs. Indeed, qRT-PCR analysis for a number of predicted endo-siRNA mRNA targets [3, 4] showed no change in Dgcr8−/− but significant upregulation in the Dicer−/− oocytes (Figure S4A). Furthermore, qRT-PCR of the mouse transposon (MT) family of retrotransposons (also predicted targets of endo-siRNAs [3, 4]) showed upregulation in Dicer−/−, but not Dgcr8−/−, oocytes (Figure S4B). In contrast, other transposons including intracisternal A-particle transposons and short interspersed repetitive elements (SINEs) appeared unchanged in both mutants. Surprisingly, long interspersed repetitive elements (LINEs) were downregulated in Dicer−/−, but not Dgcr8−/−, oocytes. This latter finding is likely an indirect consequence of Dicer loss.
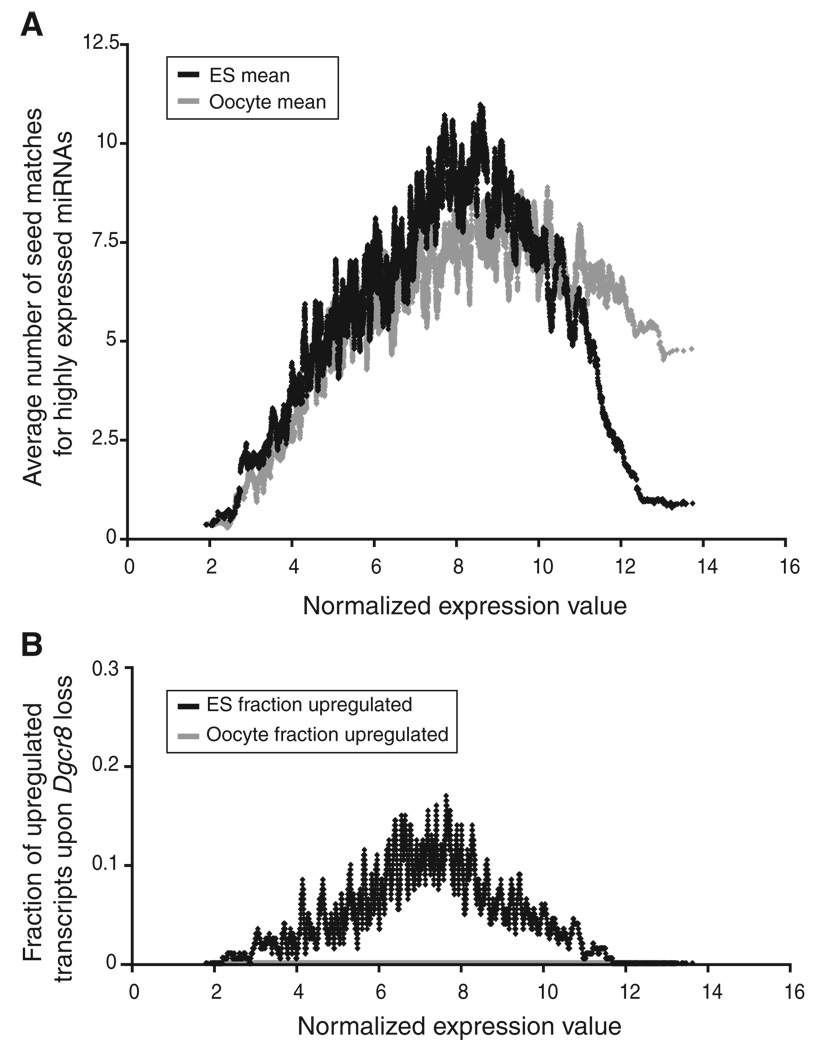
The lack of mRNA changes in the Dgcr8−/− oocytes was not simply due to the absence of mRNA targets for miRNAs, because bioinformatic analysis identified many oocyte-expressed mRNAs with multiple seed matches in their 3′ untranslated regions (3′UTRs) to highly expressed miRNAs (Figure 5A). Furthermore, a characteristic property of miRNA function in cells is the phenomenon of antitargeting, the depletion of miRNA target sites in the 3′UTRs of the most highly expressed genes [13]. In embryonic stem (ES) cells, we found evidence of antitargeting because there was a depletion of predicted ES cell miRNA target sites among the most highly expressed mRNAs and an enrichment of sites among the intermediate expressed mRNAs (Figures 5A and 5B, dark gray). In contrast, there was little depletion of predicted miRNA target sites among the most highly expressed mRNAs in oocytes (Figures 5A and 5B, light gray). This lack of antitargeting in oocytes is consistent with the finding that miRNA function—which drives the phenomenon of antitargeting—is lost in mature oocytes.
Figure 5. Highly Expressed Oocyte Transcripts Are Not Depleted for Micro-RNA Seed Matches in Their 3′UTRs.
(A) Transcripts are ranked by log2 absolute expression in embryonic stem (ES) cells and oocytes, increasing from left to right (y axis). For each transcript, a local average number of seed matches for the top 44 miRNAs expressed in ES cells (dark gray) and oocytes (light gray) is plotted.
(B) The local fraction of transcripts upregulated with FDR < 0.05 upon Dgcr8 loss in ES cells (dark gray) and oocytes (light gray) is plotted. Note that oocyte transcript levels do not change with Dgcr8 loss and, therefore, data points overlap with the x axis.
DGCR8-independent, Dicer-dependent small RNAs include the noncanonical miRNAs, mirtrons, and small hairpin RNAs (shRNAs), along with the endo-siRNAs [5]. To determine whether the noncanonical miRNAs may have a role in the Dicer−/− phenotype, we evaluated published deep-sequencing data from oocytes [3]. To identify mirtrons, we mapped sequence reads to all short introns (<500 bp). Two previously identified mirtrons [12] were expressed at low levels in oocytes: Abcf1 and Mosc2. We searched for target sites of these two mirtrons in transcripts that are present and expressed in oocytes. However, there was no enrichment for seed sequence matches in the open reading frames or 3′UTRs of transcripts that were increased in abundance in the absence of Dicer (Figure S5A), suggesting that these mirtrons were not underlying the mRNA changes. We next asked whether two previously identified shRNAs in ES cells [12], mir-320 and mir-484, could regulate mRNAs in oocytes. Once again, there was no enrichment in the complementary seed matches in the Dicer upregulated transcripts (Figure S5B). In contrast to the mirtrons and shRNA, the endo-siRNAs could be mapped to many of the upregulated transcripts [3] (Table S1). Therefore, we conclude that the dramatic gene expression changes in Dicer−/− oocytes are the result of the direct and indirect effects of endo-siRNA loss, not canonical or noncanonical miRNAs.
In summary, our findings provide two major insights into the roles of small RNAs in early embryonic development. First, miRNA function is globally suppressed in mature oocytes and early embryos. Second, endo-siRNAs, not miRNAs, are the central small RNA players at this early stage of mammalian development.
Remarkably, both mature miRNAs and siRNAs are present in the oocyte [3, 4], but only the loss of the latter impacts the transcriptome, suggesting differential regulation of these two classes downstream of Dicer maturation. This differential regulation may occur at the level of the silencing complex. For example, it may involve modifications to the Argonaute (Ago) proteins, which are key components of the silencing complex [14]. Deletion of Ago2 in the oocytes results in a Dicer knockout-like phenotype, presumably because of loss of Slicer activity and hence siRNA function [15]. The role for the remaining Argonautes in oocytes remains to be elucidated, but Argonautes appear to be highly redundant in terms of miRNA activity [16]. An alternative mechanism may involve RNA-binding proteins that suppress the capacity of miRNAs to bind mRNAs or recruit the destabilization machinery. Such a mechanism has been described for a subset of miRNAs and mRNA targets [17, 18].
Although endo-siRNAs play a central role in mammalian oocytes, it is unclear whether they play roles in other mammalian tissues. Endo-siRNAs have also been uncovered in ES cells [12]. There is no overlap between the specific endo-siRNAs expressed in oocytes and ES cells showing that they are developmentally regulated [12]. Consistent with a potential role for endo-siRNAs in ES cells, loss of Dicer has more severe phenotypes than Dgcr8 knockout cells [9, 16, 19, 20]. However, these differences may also be explained by a role for noncanonical miRNAs [12]. DGCR8 and its enzymatic partner, Drosha, have been deleted in other tissues and compared to corresponding Dicer knockout phenotypes. In particular, Dgcr8 has been knocked out in the skin and cardiomyocytes [7, 21], whereas Drosha has been knocked out in T cells [22]. In all three cases, the Dgcr8 or Drosha null phenotypes appeared to be very similar to that of the corresponding Dicer null phenotypes. These findings suggest that endo-siRNAs and/or other classes of DGCR8/Drosha-independent, Dicer-dependent small RNAs play less important roles in somatic tissues. However, because the phenotypes are all quite severe, subtle differences may have been missed. Indeed, siRNAs have been uncovered in somatic tissues of Drosophila [23]. It remains to be seen whether siRNAs are present in mammalian somatic tissues.
Consistent with our findings, Ma et al. [24] (this issue of Current Biology) found that the activities of two highly expressed miRNAs (let-7 and miR-30), as measured by reporter assays, become suppressed during oocyte development. However, it remains unclear when miRNA function is reactivated. In the absence of both maternal and zygotic Dgcr8, embryos develop normally to the blastocyst stage, although this phenotypic finding does not exclude more subtle molecular roles for miRNAs prior to this stage. Interestingly, cytoplasmic densities called P bodies containing the miRNA/mRNA inhibitory complexes disappear in the developing oocyte and only reappear around the blastocyst stage (P. Svoboda and R.M. Schultz, personal communication), consistent with suppression of miRNA activity throughout preimplantation development. Surprisingly, although maternal miRNAs do not play a phenotypic role in preimplantation development, they do appear to be important in postimplantation development as brood sizes of maternal Dgcr8 knockouts are diminished. The role of maternal miRNAs at these later stages of development remains to be elucidated.
In conclusion, our findings show an unprecedented global suppression of miRNA function at the very beginning of mammalian development, during a critical period of reprogramming of the transcriptome and epigenome [25]. An interesting possibility is that the loss of miRNA function is a key component of the dramatic reprogramming.
Supplementary Material
Acknowledgments
We would like to thank M. Conti and members of the Blelloch laboratory for critical reading of the manuscript. This work was supported by funds to R.B. from the National Institute of Child Health and Human Development and National Institutes of Health (NIH) through a pilot project funded within a cooperative agreement U54HD055764-02, part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. Further funds to R.B. came from an NIH/National Institute of Neurological Disorders and Stroke grant (K08 NS048118) and a California Institute of Regenerative Medicine New Faculty (RN-00906-01) award. R.B. is a Pew Scholar.
Footnotes
Accession Numbers
All microarray data are available at the Gene Expression Omnibus database in Minimum Information About a Microarray Experiment-compliant format with accession numbers (GSE 19894).
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at doi:10.1016/j.cub.2009.12.044.
References
- 1.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 5.Babiarz JE, Blelloch R. Small RNAs – their biogenesis, regulation and function in embryonic stem cells. StemBook. 2009 http://www.stembook.org/node/583. [PubMed] [Google Scholar]
- 6.de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- 7.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 11.Tang F, Hajkova P, Barton SC, O’Carroll D, Lee C, Lao K, Surani MA. 220-plex microRNA expression profile of a single cell. Nat. Protoc. 2006;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
- 12.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 14.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda M, Tang F, O’Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2009;2:9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 18.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS ONE. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl. Acad. Sci. USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong MM, Rasmussen JP, Rudensky AY, Rundensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr. Biol. 2010 doi: 10.1016/j.cub.2009.12.042. in press. Published online January 28, 2010. 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.